Prognostic Immune Effector Signature in Adult Acute Lymphoblastic Leukemia Patients Is Dominated by γδ T Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Flow and Mass Cytometry
2.3. Uniform Manifold Approximation and Projection Analysis
2.4. Generation of Anti-Human BTN3A mAb
2.5. Expansion of Vδ2 T Cells
2.6. Degranulation Assays
2.7. Statistics
3. Results
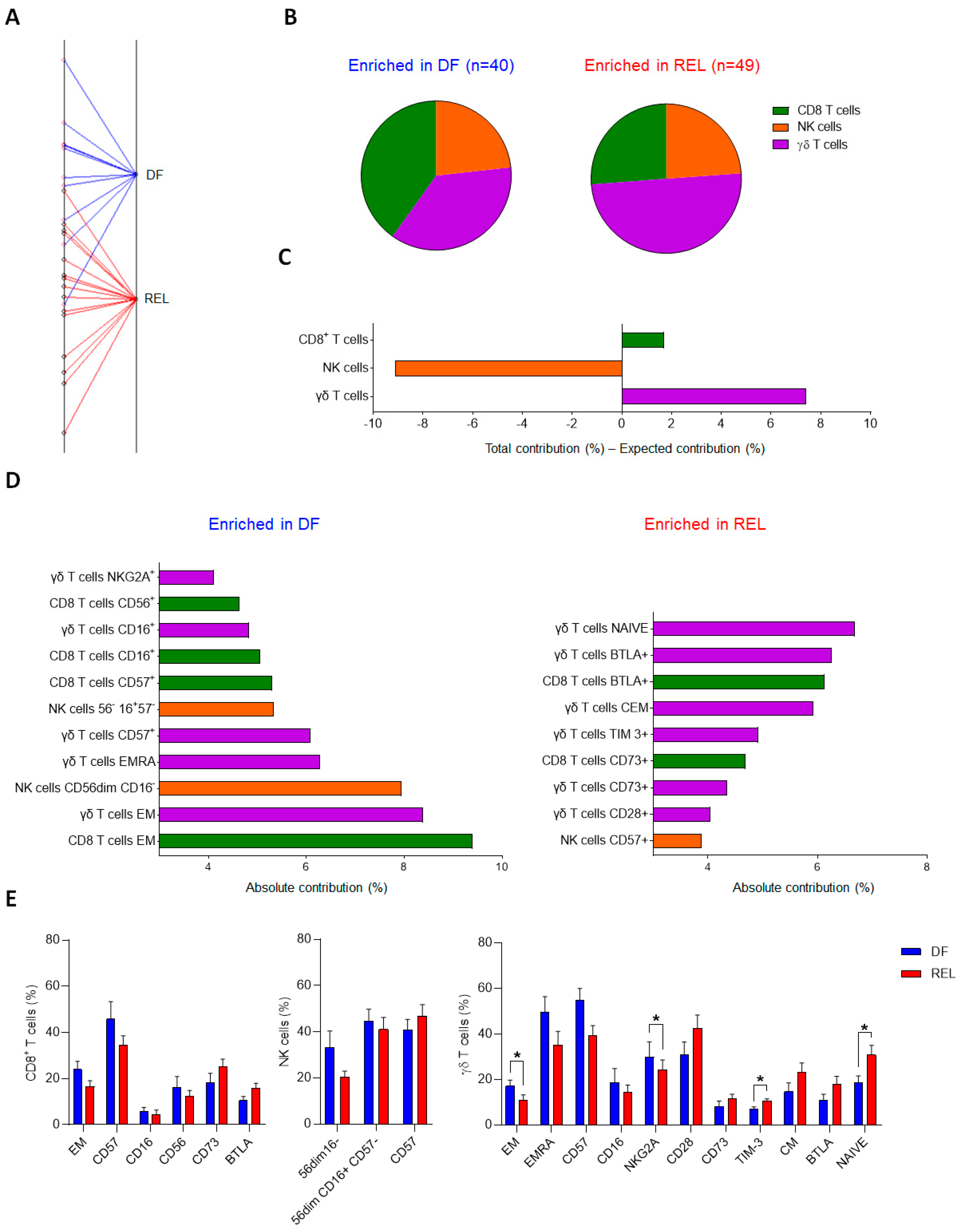
3.1. γδ T-Cell Phenotypic Variables Are Significant Contributors to the Immune Effector Signature Associated with Relapse in ALL Patients
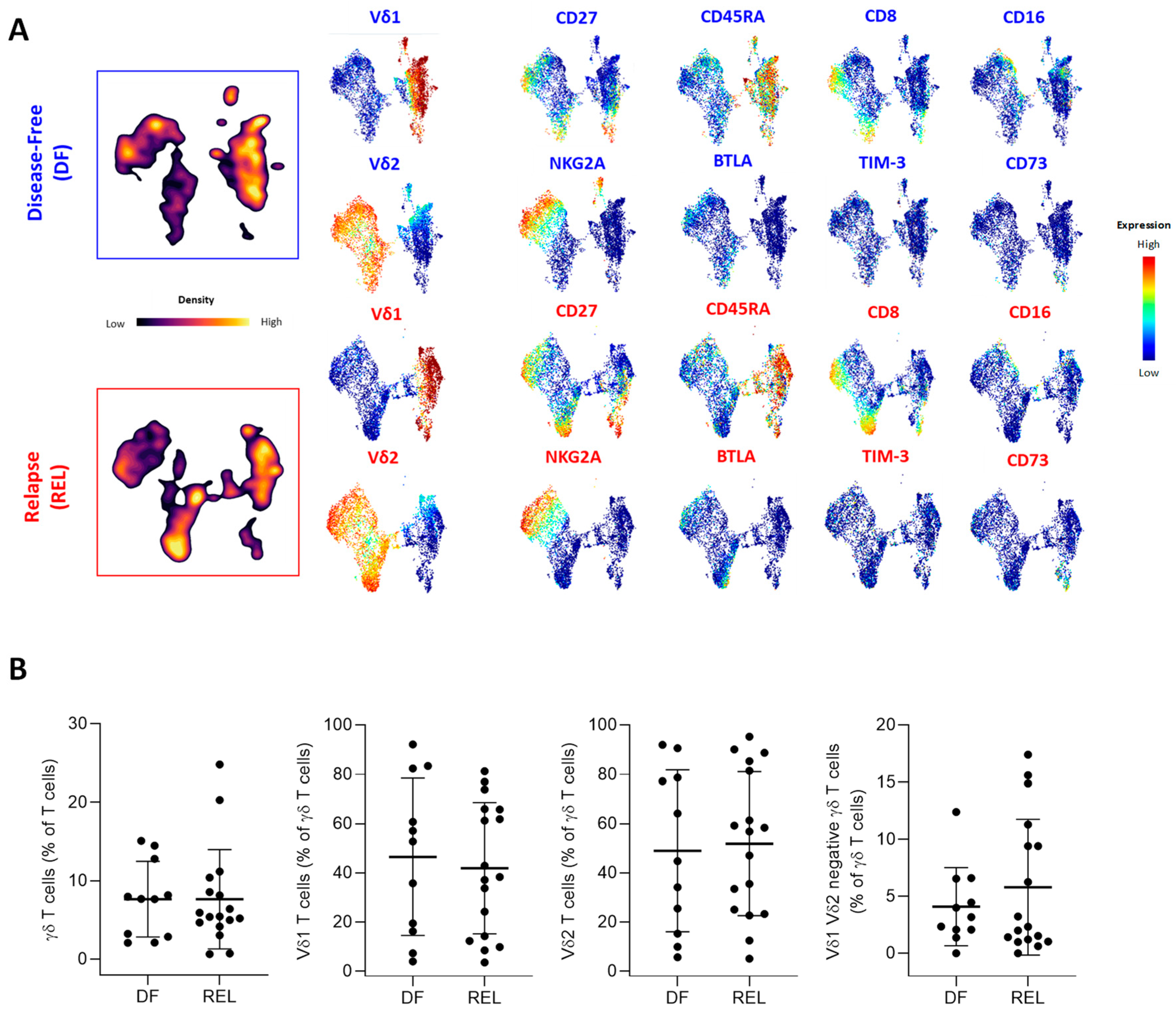
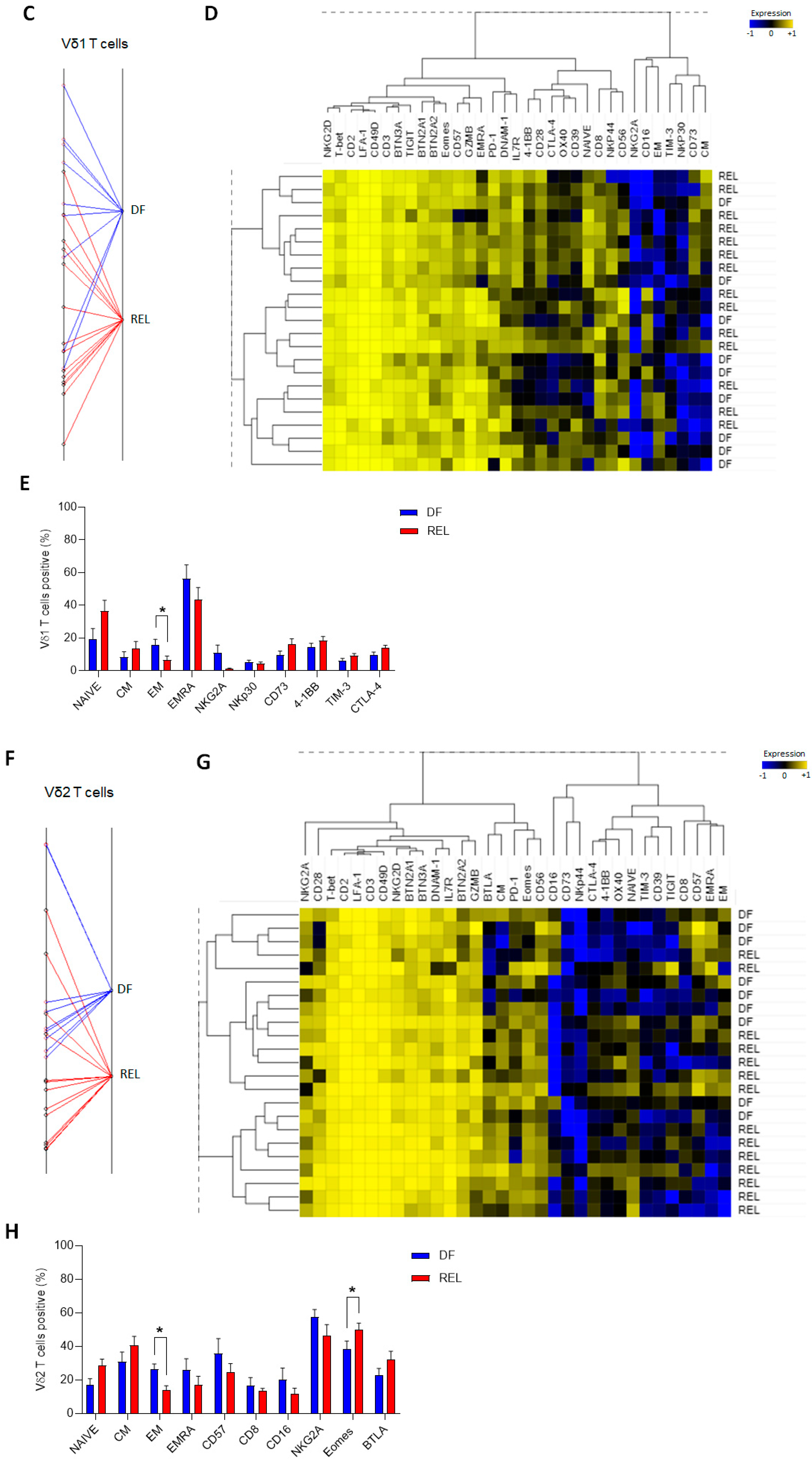
3.2. Prognostic Impact of ɣδ T-Cell Alterations in ALL Mainly Depends on Vδ2 T Cells
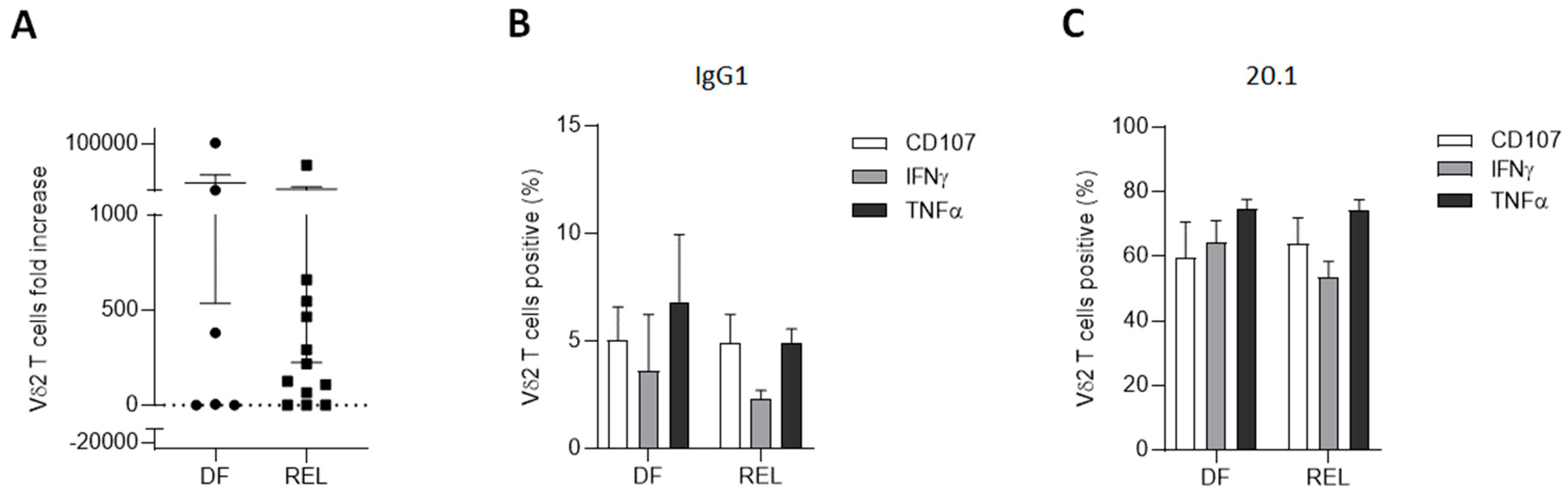
3.3. Vδ2 T Cells from Relapsed Patients Expand and Are Able to Degranulate and to Produce Th1 Cytokines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pastorczak, A.; Domka, K.; Fidyt, K.; Poprzeczko, M.; Firczuk, M. Mechanisms of Immune Evasion in Acute Lymphoblastic Leukemia. Cancers 2021, 13, 1536. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Morales, S.; Aranda-Uribe, I.S.; Pérez-Amado, C.J.; Ramírez-Bello, J.; Hidalgo-Miranda, A. Mechanisms of Immunosuppressive Tumor Evasion: Focus on Acute Lymphoblastic Leukemia. Front. Immunol. 2021, 12, 737340. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.A.; Shah, B.; Advani, A.; Aoun, P.; Boyer, M.W.; Burke, P.W.; DeAngelo, D.J.; Dinner, S.; Fathi, A.T.; Gauthier, J.; et al. Acute Lymphoblastic Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 1079–1109. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sasaki, K.; Jabbour, E.; Short, N.J.; Jain, N.; Ravandi, F.; Pui, C.-H.; Kantarjian, H. Acute Lymphoblastic Leukemia: A Population-Based Study of Outcome in the United States Based on the Surveillance, Epidemiology, and End Results (SEER) Database, 1980–2017. Am. J. Hematol. 2021, 96, 650–658. [Google Scholar] [CrossRef] [PubMed]
- Huguet, F.; Chevret, S.; Leguay, T.; Thomas, X.; Boissel, N.; Escoffre-Barbe, M.; Chevallier, P.; Hunault, M.; Vey, N.; Bonmati, C.; et al. Intensified Therapy of Acute Lymphoblastic Leukemia in Adults: Report of the Randomized GRAALL-2005 Clinical Trial. J. Clin. Oncol. 2018, 36, 2514–2523. [Google Scholar] [CrossRef] [PubMed]
- Polak, R.; de Rooij, B.; Pieters, R.; den Boer, M.L. B-Cell Precursor Acute Lymphoblastic Leukemia Cells Use Tunneling Nanotubes to Orchestrate Their Microenvironment. Blood 2015, 126, 2404–2414. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Chen, Y.-Y.; He, Y.-Y.; Wang, J.-Y.; Yang, J.-P.; Zhong, S.-L.; Jiang, N.; Zhou, P.; Jiang, H.; Zhou, J. Expansion and Activation of Granulocytic, Myeloid-Derived Suppressor Cells in Childhood Precursor B Cell Acute Lymphoblastic Leukemia. J. Leukoc. Biol. 2017, 102, 449–458. [Google Scholar] [CrossRef]
- Lyu, A.; Triplett, T.A.; Nam, S.H.; Hu, Z.; Arasappan, D.; Godfrey, W.H.; Ames, R.Y.; Sarang, A.; Selden, H.J.; Lee, C.-H.; et al. Tumor-Associated Myeloid Cells Provide Critical Support for T-ALL. Blood 2020, 136, 1837–1850. [Google Scholar] [CrossRef]
- Witkowski, M.T.; Dolgalev, I.; Evensen, N.A.; Ma, C.; Chambers, T.; Roberts, K.G.; Sreeram, S.; Dai, Y.; Tikhonova, A.N.; Lasry, A.; et al. Extensive Remodeling of the Immune Microenvironment in B Cell Acute Lymphoblastic Leukemia. Cancer Cell 2020, 37, 867–882.e12. [Google Scholar] [CrossRef]
- D’Amico, G.; Vulcano, M.; Bugarin, C.; Bianchi, G.; Pirovano, G.; Bonamino, M.; Marin, V.; Allavena, P.; Biagi, E.; Biondi, A. CD40 Activation of BCP-ALL Cells Generates IL-10-Producing, IL-12-Defective APCs That Induce Allogeneic T-Cell Anergy. Blood 2004, 104, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-Y.; Griffith, O.L.; Griffith, M. Clinical Implications of Neoepitope Landscapes for Adult and Pediatric Cancers. Genome Med. 2017, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Rouce, R.H.; Shaim, H.; Sekine, T.; Weber, G.; Ballard, B.; Ku, S.; Barese, C.; Murali, V.; Wu, M.-F.; Liu, H.; et al. The TGF-β/SMAD Pathway Is an Important Mechanism for NK Cell Immune Evasion in Childhood B-Acute Lymphoblastic Leukemia. Leukemia 2016, 30, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chang, Y.-J.; Xu, L.-P.; Zhang, X.-H.; Wang, Y.; Liu, K.-Y.; Huang, X.-J. T Cell Exhaustion Characterized by Compromised MHC Class I and II Restricted Cytotoxic Activity Associates with Acute B Lymphoblastic Leukemia Relapse after Allogeneic Hematopoietic Stem Cell Transplantation. Clin. Immunol. 2018, 190, 32–40. [Google Scholar] [CrossRef]
- Hohtari, H.; Brück, O.; Blom, S.; Turkki, R.; Sinisalo, M.; Kovanen, P.E.; Kallioniemi, O.; Pellinen, T.; Porkka, K.; Mustjoki, S. Immune Cell Constitution in Bone Marrow Microenvironment Predicts Outcome in Adult ALL. Leukemia 2019, 33, 1570–1582. [Google Scholar] [CrossRef]
- Mansour, A.; Elkhodary, T.; Darwish, A.; Mabed, M. Increased Expression of Costimulatory Molecules CD86 and SCTLA-4 in Patients with Acute Lymphoblastic Leukemia. Leuk. Lymphoma 2014, 55, 2120–2124. [Google Scholar] [CrossRef]
- Blaeschke, F.; Willier, S.; Stenger, D.; Lepenies, M.; Horstmann, M.A.; Escherich, G.; Zimmermann, M.; Rojas Ringeling, F.; Canzar, S.; Kaeuferle, T.; et al. Leukemia-Induced Dysfunctional TIM-3+CD4+ Bone Marrow T Cells Increase Risk of Relapse in Pediatric B-Precursor ALL Patients. Leukemia 2020, 34, 2607–2620. [Google Scholar] [CrossRef]
- Niedźwiecki, M.; Budziło, O.; Adamkiewicz-Drożyńska, E.; Pawlik-Gwozdecka, D.; Zieliński, M.; Maciejka-Kembłowska, L.; Szczepański, T.; Trzonkowski, P. CD4+CD25highCD127low/-FoxP3 + Regulatory T-Cell Population in Acute Leukemias: A Review of the Literature. J. Immunol. Res. 2019, 2019, 2816498. [Google Scholar] [CrossRef]
- Duell, J.; Dittrich, M.; Bedke, T.; Mueller, T.; Eisele, F.; Rosenwald, A.; Rasche, L.; Hartmann, E.; Dandekar, T.; Einsele, H.; et al. Frequency of Regulatory T Cells Determines the Outcome of the T-Cell-Engaging Antibody Blinatumomab in Patients with B-Precursor ALL. Leukemia 2017, 31, 2181–2190. [Google Scholar] [CrossRef]
- An, F.; Wang, H.; Liu, Z.; Wu, F.; Zhang, J.; Tao, Q.; Li, Y.; Shen, Y.; Ruan, Y.; Zhang, Q.; et al. Influence of Patient Characteristics on Chimeric Antigen Receptor T Cell Therapy in B-Cell Acute Lymphoblastic Leukemia. Nat. Commun. 2020, 11, 5928. [Google Scholar] [CrossRef]
- Silva-Santos, B.; Mensurado, S.; Coffelt, S.B. Γδ T Cells: Pleiotropic Immune Effectors with Therapeutic Potential in Cancer. Nat. Rev. Cancer 2019, 19, 392–404. [Google Scholar] [CrossRef]
- Motulsky, A.G. The 1985 Nobel Prize in Physiology or Medicine. Science 1986, 231, 126–129. [Google Scholar] [CrossRef]
- Iguchi-Manaka, A.; Kai, H.; Yamashita, Y.; Shibata, K.; Tahara-Hanaoka, S.; Honda, S.; Yasui, T.; Kikutani, H.; Shibuya, K.; Shibuya, A. Accelerated Tumor Growth in Mice Deficient in DNAM-1 Receptor. J. Exp. Med. 2008, 205, 2959–2964. [Google Scholar] [CrossRef]
- Guerra, N.; Tan, Y.X.; Joncker, N.T.; Choy, A.; Gallardo, F.; Xiong, N.; Knoblaugh, S.; Cado, D.; Greenberg, N.R.; Raulet, D.H. NKG2D-Deficient Mice Are Defective in Tumor Surveillance in Models of Spontaneous Malignancy. Immunity 2008, 28, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Yazdanifar, M.; Cocco, C.; Bertaina, A.; Airoldi, I. Engineering the Bridge between Innate and Adaptive Immunity for Cancer Immunotherapy: Focus on Γδ T and NK Cells. Cells 2020, 9, 1757. [Google Scholar] [CrossRef] [PubMed]
- Holtmeier, W.; Kabelitz, D. Γδ T Cells Link Innate and Adaptive Immune Responses. Mech. Epithel. Def. 2005, 86, 151–183. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, W.; Pan, M.; Scully, E.; Girardi, M.; Augenlicht, L.H.; Craft, J.; Yin, Z. Γδ T Cells Provide an Early Source of Interferon γ in Tumor Immunity. J. Exp. Med. 2003, 198, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.F.; Duarte, J.D.G.; Ostrouska, S.; Behren, A. Γδ T Cells in the Tumor Microenvironment—Interactions with Other Immune Cells. Front. Immunol. 2022, 13, 894315. [Google Scholar] [CrossRef]
- Yazdanifar, M.; Barbarito, G.; Bertaina, A.; Airoldi, I. Γδ T Cells: The Ideal Tool for Cancer Immunotherapy. Cells 2020, 9, 1305. [Google Scholar] [CrossRef]
- Kabelitz, D.; Serrano, R.; Kouakanou, L.; Peters, C.; Kalyan, S. Cancer Immunotherapy with Γδ T Cells: Many Paths Ahead of Us. Cell Mol. Immunol. 2020, 17, 925–939. [Google Scholar] [CrossRef]
- De Libero, G. Sentinel Function of Broadly Reactive Human Γδ T Cells. Immunol. Today 1997, 18, 22–26. [Google Scholar] [CrossRef] [PubMed]
- LeFranc, M.P.; Forster, A.; Baer, R.; Stinson, M.A.; Rabbitts, T.H. Diversity and Rearrangement of the Human T Cell Rearranging Gamma Genes: Nine Germ-Line Variable Genes Belonging to Two Subgroups. Cell 1986, 45, 237–246. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. Regulatory and Effector Functions of Gamma-Delta (Γδ) T Cells and Their Therapeutic Potential in Adoptive Cellular Therapy for Cancer. Int. J. Cancer 2016, 139, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Niu, C.; Cui, J. Gamma-Delta (Γδ) T Cells: Friend or Foe in Cancer Development? J. Transl. Med. 2018, 16, 3. [Google Scholar] [CrossRef]
- Rigau, M.; Ostrouska, S.; Fulford, T.S.; Johnson, D.N.; Woods, K.; Ruan, Z.; McWilliam, H.E.G.; Hudson, C.; Tutuka, C.; Wheatley, A.K.; et al. Butyrophilin 2A1 Is Essential for Phosphoantigen Reactivity by Γδ T Cells. Science 2020, 367, eaay5516. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, M.M.; Willcox, C.R.; Salim, M.; Paletta, D.; Fichtner, A.S.; Noll, A.; Starick, L.; Nöhren, A.; Begley, C.R.; Berwick, K.A.; et al. Butyrophilin-2A1 Directly Binds Germline-Encoded Regions of the Vγ9Vδ2 TCR and Is Essential for Phosphoantigen Sensing. Immunity 2020, 52, 487–498.e6. [Google Scholar] [CrossRef]
- Cano, C.E.; Pasero, C.; De Gassart, A.; Kerneur, C.; Gabriac, M.; Fullana, M.; Granarolo, E.; Hoet, R.; Scotet, E.; Rafia, C.; et al. BTN2A1, an Immune Checkpoint Targeting Vγ9Vδ2 T Cell Cytotoxicity against Malignant Cells. Cell Rep. 2021, 36, 109359. [Google Scholar] [CrossRef]
- Yuan, L.; Ma, X.; Yang, Y.; Li, X.; Ma, W.; Yang, H.; Huang, J.-W.; Xue, J.; Yi, S.; Zhang, M.; et al. Phosphoantigens Are Molecular Glues That Promote Butyrophilin 3A1/2A1 Association Leading to Vγ9Vδ2 T Cell Activation. BioRxiv 2022, 2022.01.02.474068. [Google Scholar]
- Duval, M.; Yotnda, P.; Bensussan, A.; Oudhiri, N.; Guidal, C.; Rohrlich, P.; Boumsell, L.; Grandchamp, B.; Vilmer, E. Potential Antileukemic Effect of Gamma Delta T Cells in Acute Lymphoblastic Leukemia. Leukemia 1995, 9, 863–868. [Google Scholar]
- Lança, T.; Correia, D.V.; Moita, C.F.; Raquel, H.; Neves-Costa, A.; Ferreira, C.; Ramalho, J.S.; Barata, J.T.; Moita, L.F.; Gomes, A.Q.; et al. The MHC Class Ib Protein ULBP1 Is a Nonredundant Determinant of Leukemia/Lymphoma Susceptibility to Γδ T-Cell Cytotoxicity. Blood 2010, 115, 2407–2411. [Google Scholar] [CrossRef]
- Gomes, A.Q.; Correia, D.V.; Grosso, A.R.; Lança, T.; Ferreira, C.; Lacerda, J.F.; Barata, J.T.; da Silva, M.G.; Silva-Santos, B. Identification of a Panel of Ten Cell Surface Protein Antigens Associated with Immunotargeting of Leukemias and Lymphomas by Peripheral Blood Gammadelta T Cells. Haematologica 2010, 95, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Lamb, L., Jr.; Musk, P.; Ye, Z.; van Rhee, F.; Geier, S.S.; Tong, J.-J.; King, K.M.; Henslee-Downey, P.J. Human Γδ+ T Lymphocytes Have in Vitro Graft vs Leukemia Activity in the Absence of an Allogeneic Response. Bone Marrow Transplant. 2001, 27, 601–606. [Google Scholar] [CrossRef] [PubMed]
- D’Asaro, M.; Mendola, C.L.; Liberto, D.D.; Orlando, V.; Todaro, M.; Spina, M.; Guggino, G.; Meraviglia, S.; Caccamo, N.; Messina, A.; et al. Vγ9Vδ2 T Lymphocytes Efficiently Recognize and Kill Zoledronate-Sensitized, Imatinib-Sensitive, and Imatinib-Resistant Chronic Myelogenous Leukemia Cells. J. Immunol. 2010, 184, 3260–3268. [Google Scholar] [CrossRef] [PubMed]
- Siegers, G.M.; Felizardo, T.C.; Mathieson, A.M.; Kosaka, Y.; Wang, X.-H.; Medin, J.A.; Keating, A. Anti-Leukemia Activity of in Vitro-Expanded Human Gamma Delta T Cells in a Xenogeneic Ph+ Leukemia Model. PLoS ONE 2011, 6, e16700. [Google Scholar] [CrossRef] [PubMed]
- Lamb, L.S.; Henslee-Downey, P.J.; Parrish, R.S.; Godder, K.; Thompson, J.; Lee, C.; Gee, A.P. Increased Frequency of TCR Gamma Delta + T Cells in Disease-Free Survivors Following T Cell-Depleted, Partially Mismatched, Related Donor Bone Marrow Transplantation for Leukemia. J. Hematother. 1996, 5, 503–509. [Google Scholar] [CrossRef]
- Godder, K.T.; Henslee-Downey, P.J.; Mehta, J.; Park, B.S.; Chiang, K.-Y.; Abhyankar, S.; Lamb, L.S. Long Term Disease-Free Survival in Acute Leukemia Patients Recovering with Increased Γδ T Cells after Partially Mismatched Related Donor Bone Marrow Transplantation. Bone Marrow Transpl. 2007, 39, 751–757. [Google Scholar] [CrossRef]
- Marks, D.I.; Clifton-Hadley, L.; Copland, M.; Hussain, J.; Menne, T.F.; McMillan, A.; Moorman, A.V.; Morley, N.; Okasha, D.; Patel, B.; et al. In-Vivo T-Cell Depleted Reduced-Intensity Conditioned Allogeneic Haematopoietic Stem-Cell Transplantation for Patients with Acute Lymphoblastic Leukaemia in First Remission: Results from the Prospective, Single-Arm Evaluation of the UKALL14 Trial. Lancet Haematol. 2022, 9, e276–e288. [Google Scholar] [CrossRef]
- Merli, P.; Algeri, M.; Galaverna, F.; Milano, G.M.; Bertaina, V.; Biagini, S.; Girolami, E.; Palumbo, G.; Sinibaldi, M.; Becilli, M.; et al. Immune Modulation Properties of Zoledronic Acid on TcRγδ T-Lymphocytes After TcRαβ/CD19-Depleted Haploidentical Stem Cell Transplantation: An Analysis on 46 Pediatric Patients Affected by Acute Leukemia. Front. Immunol. 2020, 11, 699. [Google Scholar] [CrossRef]
- Saura-Esteller, J.; de Jong, M.; King, L.A.; Ensing, E.; Winograd, B.; de Gruijl, T.D.; Parren, P.W.H.I.; van der Vliet, H.J. Gamma Delta T-Cell Based Cancer Immunotherapy: Past-Present-Future. Front. Immunol. 2022, 13, 915837. [Google Scholar] [CrossRef]
- Tosolini, M.; Pont, F.; Poupot, M.; Vergez, F.; Nicolau-Travers, M.-L.; Vermijlen, D.; Sarry, J.-E.; Dieli, F.; Fournié, J.-J. Assessment of Tumor-Infiltrating TCRVγ9Vδ2 Γδ Lymphocyte Abundance by Deconvolution of Human Cancers Microarrays. Oncoimmunology 2017, 6, e1284723. [Google Scholar] [CrossRef]
- Kang, S.H.; Hwang, H.J.; Yoo, J.W.; Kim, H.; Choi, E.S.; Hwang, S.-H.; Cho, Y.-U.; Jang, S.; Park, C.-J.; Im, H.J.; et al. Expression of Immune Checkpoint Receptors on T-Cells and Their Ligands on Leukemia Blasts in Childhood Acute Leukemia. Anticancer Res. 2019, 39, 5531–5539. [Google Scholar] [CrossRef] [PubMed]
- Pawlik-Gwozdecka, D.; Zieliński, M.; Sakowska, J.; Adamkiewicz-Drożyńska, E.; Trzonkowski, P.; Niedźwiecki, M. CD8+ Γδ T Cells Correlate with Favorable Prognostic Factors in Childhood Acute Lymphoblastic Leukemia. Arch. Med. Sci. 2021, 17, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Ben Amara, A.; Rouviere, M.-S.; Fattori, S.; Wlosik, J.; Gregori, E.; Boucherit, N.; Bernard, P.-L.; Nunès, J.A.; Vey, N.; Luche, H.; et al. High-Throughput Mass Cytometry Staining for Deep Phenotyping of Human Natural Killer Cells. STAR Protoc. 2022, 3, 101768. [Google Scholar] [CrossRef] [PubMed]
- Compte, E.; Pontarotti, P.; Collette, Y.; Lopez, M.; Olive, D. Frontline: Characterization of BT3 Molecules Belonging to the B7 Family Expressed on Immune Cells. Eur. J. Immunol. 2004, 34, 2089–2099. [Google Scholar] [CrossRef]
- García, V.E.; Jullien, D.; Song, M.; Uyemura, K.; Shuai, K.; Morita, C.T.; Modlin, R.L. IL-15 Enhances the Response of Human Gamma Delta T Cells to Nonpeptide [Correction of Nonpetide] Microbial Antigens. J. Immunol. 1998, 160, 4322–4329. [Google Scholar] [CrossRef]
- Aehnlich, P.; Carnaz Simões, A.M.; Skadborg, S.K.; Holmen Olofsson, G.; thor Straten, P. Expansion With IL-15 Increases Cytotoxicity of Vγ9Vδ2 T Cells and Is Associated with Higher Levels of Cytotoxic Molecules and T-Bet. Front. Immunol. 2020, 11, 1868. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, H.H.; Anguille, S.; Willemen, Y.; Van den Bergh, J.M.; Berneman, Z.N.; Lion, E.; Smits, E.L.; Van Tendeloo, V.F. Interleukin-15 Enhances the Proliferation, Stimulatory Phenotype, and Antitumor Effector Functions of Human Gamma Delta T Cells. J. Hematol. Oncol. 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Benyamine, A.; Le Roy, A.; Mamessier, E.; Gertner-Dardenne, J.; Castanier, C.; Orlanducci, F.; Pouyet, L.; Goubard, A.; Collette, Y.; Vey, N.; et al. BTN3A Molecules Considerably Improve Vγ9Vδ2T Cells-Based Immunotherapy in Acute Myeloid Leukemia. Oncoimmunology 2016, 5, e1146843. [Google Scholar] [CrossRef]
- Kong, Y.; Jia, B.; Zhao, C.; Claxton, D.F.; Sharma, A.; Annageldiyev, C.; Fotos, J.S.; Zeng, H.; Paulson, R.F.; Prabhu, K.S.; et al. Downregulation of CD73 Associates with T Cell Exhaustion in AML Patients. J. Hematol. Oncol. 2019, 12, 40. [Google Scholar] [CrossRef]
- Zhou, Y.; Tse, E.W.-C.; Leung, R.; Cheung, E.; Li, H.; Sun, H. Multiplex Single-Cell Analysis of Cancer Cells Enables Unbiased Uncovering Subsets Associated with Cancer Relapse: Heterogeneity of Multidrug Resistance in Precursor B-ALL. ChemMedChem 2021, 17, e202100638. [Google Scholar] [CrossRef]
- Ning, Z.; Liu, K.; Xiong, H. Roles of BTLA in Immunity and Immune Disorders. Front. Immunol. 2021, 12, 654960. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.; Lan, X.; Meng, Y.; Guo, X.; Guo, Y.; Zhao, L.; Chen, X.; Liu, A. BTLA Marks a Less Cytotoxic T-Cell Subset in Diffuse Large B-Cell Lymphoma with High Expression of Checkpoints. Exp. Hematol. 2018, 60, 47–56.e1. [Google Scholar] [CrossRef] [PubMed]
- Bekiaris, V.; Šedý, J.R.; Macauley, M.G.; Rhode-Kurnow, A.; Ware, C.F. The Inhibitory Receptor BTLA Controls Γδ T Cell Homeostasis and Inflammatory Responses. Immunity 2013, 39, 1082–1094. [Google Scholar] [CrossRef] [PubMed]
- Gertner-Dardenne, J.; Fauriat, C.; Orlanducci, F.; Thibult, M.-L.; Pastor, S.; Fitzgibbon, J.; Bouabdallah, R.; Xerri, L.; Olive, D. The Co-Receptor BTLA Negatively Regulates Human Vγ9Vδ2 T-Cell Proliferation: A Potential Way of Immune Escape for Lymphoma Cells. Blood 2013, 122, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Radwan, S.M.; Elleboudy, N.S.; Nabih, N.A.; El-kholy, A.; Kamal, A.M. The Prospective Prognostic Value of the Immune Checkpoint BTLA Expression in Adult Acute Myeloid Leukemia Patients. Egypt. J. Med. Hum. Genet. 2021, 22, 78. [Google Scholar] [CrossRef]
- Geng, H.; Chen, Z.; Anderson, S.; Fraser, E.; Lu, M.; Lingjing, C.; Collins, C.; Markus, M.; Rubenstein, J.L. Expression of B and T Lymphocyte Attenuator (BTLA) Correlates with CNS Metastasis and Adverse Prognosis in Activated B-Cell Lymphoma and Acute Lymphoblastic Leukemia. Blood 2015, 126, 3900. [Google Scholar] [CrossRef]
- Anand, P.; Guillaumet-Adkins, A.; Dimitrova, V.; Yun, H.; Drier, Y.; Sotudeh, N.; Rogers, A.; Ouseph, M.M.; Nair, M.; Potdar, S.; et al. Single-Cell RNA-Seq Reveals Developmental Plasticity with Coexisting Oncogenic States and Immune Evasion Programs in ETP-ALL. Blood 2021, 137, 2463–2480. [Google Scholar] [CrossRef]
- Li, X.; Lu, H.; Gu, Y.; Zhang, X.; Zhang, G.; Shi, T.; Chen, W. Tim-3 Suppresses the Killing Effect of Vγ9Vδ2 T Cells on Colon Cancer Cells by Reducing Perforin and Granzyme B Expression. Exp. Cell Res. 2020, 386, 111719. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, L.; Tian, X.; Xiang, X.; Yu, Y.; Zeng, Z.; Cao, Y.; Chen, S.; Sun, A. Identification of Immune Subtypes of Ph-Neg B-ALL with Ferroptosis Related Genes and the Potential Implementation of Sorafenib. BMC Cancer 2021, 21, 1331. [Google Scholar] [CrossRef]
- Yadav, B.D.; Samuels, A.L.; Wells, J.E.; Sutton, R.; Venn, N.C.; Bendak, K.; Anderson, D.; Marshall, G.M.; Cole, C.H.; Beesley, A.H.; et al. Heterogeneity in Mechanisms of Emergent Resistance in Pediatric T-Cell Acute Lymphoblastic Leukemia. Oncotarget 2016, 7, 58728–58742. [Google Scholar] [CrossRef]
- Dufva, O.; Pölönen, P.; Brück, O.; Keränen, M.A.I.; Klievink, J.; Mehtonen, J.; Huuhtanen, J.; Kumar, A.; Malani, D.; Siitonen, S.; et al. Immunogenomic Landscape of Hematological Malignancies. Cancer Cell 2020, 38, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Pearce, E.L.; Mullen, A.C.; Martins, G.A.; Krawczyk, C.M.; Hutchins, A.S.; Zediak, V.P.; Banica, M.; DiCioccio, C.B.; Gross, D.A.; Mao, C.; et al. Control of Effector CD8+ T Cell Function by the Transcription Factor Eomesodermin. Science 2003, 302, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ji, Z.; Ngiow, S.F.; Manne, S.; Cai, Z.; Huang, A.C.; Johnson, J.; Staupe, R.P.; Bengsch, B.; Xu, C.; et al. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 2019, 51, 840–855.e5. [Google Scholar] [CrossRef] [PubMed]
- Lino, C.N.R.; Barros-Martins, J.; Oberdörfer, L.; Walzer, T.; Prinz, I. Eomes Expression Reports the Progressive Differentiation of IFN-γ-Producing Th1-like Γδ T Cells. Eur. J. Immunol. 2017, 47, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Cheung, L.C.; Tickner, J.; Hughes, A.M.; Skut, P.; Howlett, M.; Foley, B.; Oommen, J.; Wells, J.E.; He, B.; Singh, S.; et al. New Therapeutic Opportunities from Dissecting the Pre-B Leukemia Bone Marrow Microenvironment. Leukemia 2018, 32, 2326–2338. [Google Scholar] [CrossRef] [PubMed]
- Bertaina, A.; Zorzoli, A.; Petretto, A.; Barbarito, G.; Inglese, E.; Merli, P.; Lavarello, C.; Brescia, L.P.; De Angelis, B.; Tripodi, G.; et al. Zoledronic Acid Boosts Γδ T-Cell Activity in Children Receiving Aβ+ T and CD19+ Cell-Depleted Grafts from an HLA-Haplo-Identical Donor. Oncoimmunology 2016, 6, e1216291. [Google Scholar] [CrossRef]
- Chew, V.; Lai, L.; Pan, L.; Lim, C.J.; Li, J.; Ong, R.; Chua, C.; Leong, J.Y.; Lim, K.H.; Toh, H.C.; et al. Delineation of an Immunosuppressive Gradient in Hepatocellular Carcinoma Using High-Dimensional Proteomic and Transcriptomic Analyses. Proc. Natl. Acad. Sci. USA 2017, 114, E5900–E5909. [Google Scholar] [CrossRef]
- Xu, B.; Yuan, L.; Gao, Q.; Yuan, P.; Zhao, P.; Yuan, H.; Fan, H.; Li, T.; Qin, P.; Han, L.; et al. Circulating and Tumor-Infiltrating Tim-3 in Patients with Colorectal Cancer. Oncotarget 2015, 6, 20592–20603. [Google Scholar] [CrossRef]
- Brauneck, F.; Weimer, P.; Schulze Zur Wiesch, J.; Weisel, K.; Leypoldt, L.; Vohwinkel, G.; Fritzsche, B.; Bokemeyer, C.; Wellbrock, J.; Fiedler, W. Bone Marrow-Resident Vδ1 T Cells Co-Express TIGIT With PD-1, TIM-3 or CD39 in AML and Myeloma. Front. Med. (Lausanne) 2021, 8, 763773. [Google Scholar] [CrossRef]
- De Vries, N.L.; van de Haar, J.; Veninga, V.; Chalabi, M.; Ijsselsteijn, M.E.; van der Ploeg, M.; van den Bulk, J.; Ruano, D.; van den Berg, J.G.; Haanen, J.B.; et al. Γδ T Cells Are Effectors of Immunotherapy in Cancers with HLA Class I Defects. Nature 2023, 613, 743–750. [Google Scholar] [CrossRef]
- Koay, H.-F.; Lynch, L. Γδ T Cells Unveil Invisible Tumors. Trends Immunol. 2023, 44, 159–161. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Floch, A.-C.; Rouvière, M.-S.; Salem, N.; Ben Amara, A.; Orlanducci, F.; Vey, N.; Gorvel, L.; Chretien, A.-S.; Olive, D. Prognostic Immune Effector Signature in Adult Acute Lymphoblastic Leukemia Patients Is Dominated by γδ T Cells. Cells 2023, 12, 1693. https://doi.org/10.3390/cells12131693
Le Floch A-C, Rouvière M-S, Salem N, Ben Amara A, Orlanducci F, Vey N, Gorvel L, Chretien A-S, Olive D. Prognostic Immune Effector Signature in Adult Acute Lymphoblastic Leukemia Patients Is Dominated by γδ T Cells. Cells. 2023; 12(13):1693. https://doi.org/10.3390/cells12131693
Chicago/Turabian StyleLe Floch, Anne-Charlotte, Marie-Sarah Rouvière, Nassim Salem, Amira Ben Amara, Florence Orlanducci, Norbert Vey, Laurent Gorvel, Anne-Sophie Chretien, and Daniel Olive. 2023. "Prognostic Immune Effector Signature in Adult Acute Lymphoblastic Leukemia Patients Is Dominated by γδ T Cells" Cells 12, no. 13: 1693. https://doi.org/10.3390/cells12131693
APA StyleLe Floch, A.-C., Rouvière, M.-S., Salem, N., Ben Amara, A., Orlanducci, F., Vey, N., Gorvel, L., Chretien, A.-S., & Olive, D. (2023). Prognostic Immune Effector Signature in Adult Acute Lymphoblastic Leukemia Patients Is Dominated by γδ T Cells. Cells, 12(13), 1693. https://doi.org/10.3390/cells12131693






