Regenerative Potential of Injured Spinal Cord in the Light of Epigenetic Regulation and Modulation
Abstract
1. Introduction
2. Epigenetic Regulation in Mammals after Spinal Cord Injury
2.1. Spinal Cord Injury: Primary and Secondary Injuries in Mammals
2.2. Mechanism of Injury Repair: Glial Scar Formation
2.3. Epigenetic Regulation during Injury Repair
2.3.1. Epigenetic Regulation of Specific Cell Types and Secondary Damage Processes
2.3.2. Histone Modification during Spinal Cord Repair
2.3.3. Histone Modification during Axon Regeneration
2.3.4. Role of DNA Methylation in Injury Repair
2.3.5. miRNA Regulation of Injury Repair
3. Epigenetic Regulation of Regeneration-Competent Animals after Spinal Cord Injury
3.1. Axolotl
3.2. Xenopus
3.3. Zebrafish
3.3.1. Spinal Cord Injury Response in Zebrafish
3.3.2. Epigenetic Regulation of Spinal Cord Regeneration in Zebrafish after Injury
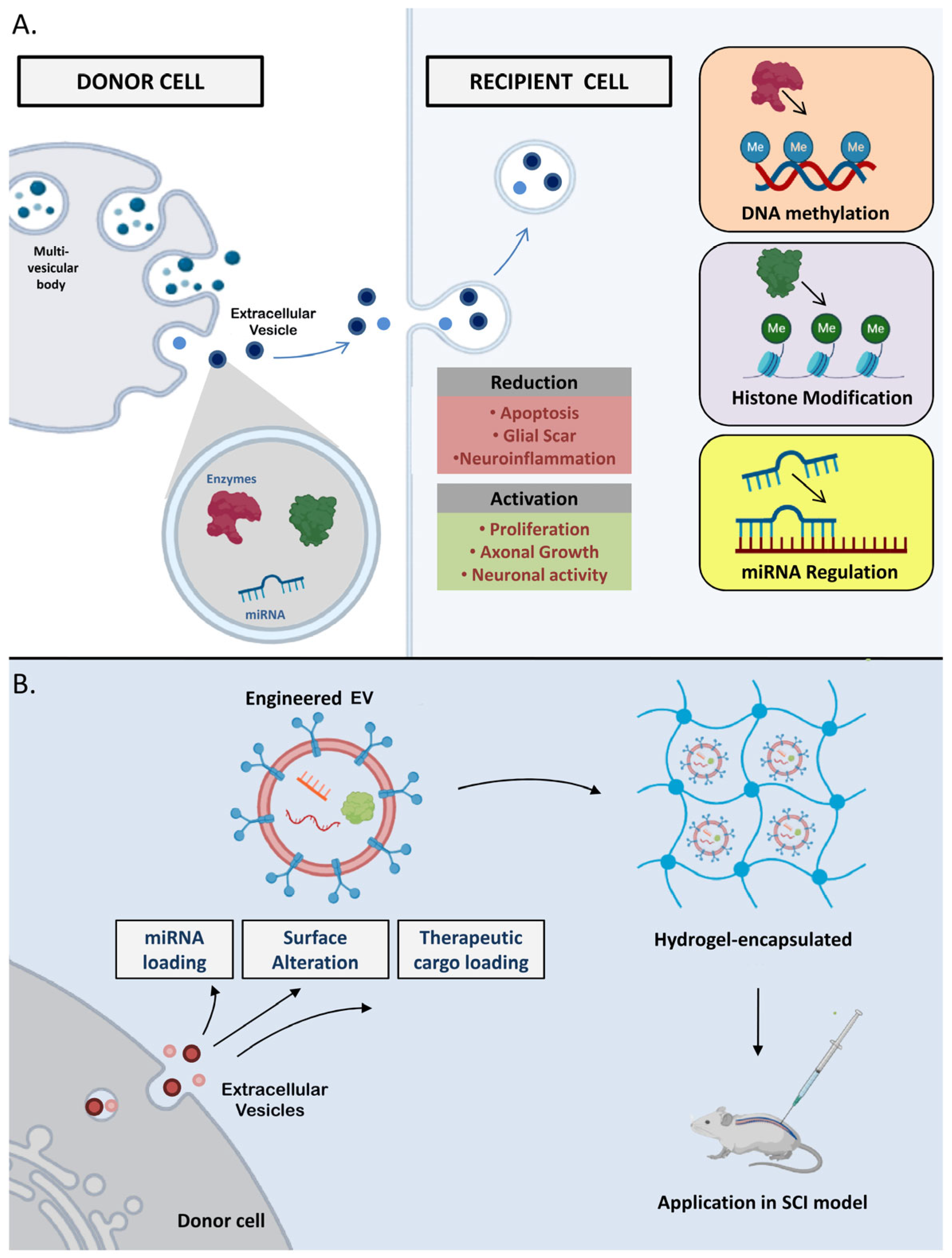
4. Extracellular Vesicles (EVs) in the Epigenetic Regulation Process
| Molecules | Functions | Examples in Extracellular Vesicles | References |
|---|---|---|---|
| microRNAs (miRNAs) | Regulate gene expression by binding to messenger RNA (mRNA) molecules, leading to mRNA degradation or inhibition of translation. They can be packaged into EVs and transferred between cells, thus influencing epigenetic processes. | miR-21: Regulates the differentiation and death of neurons in patients with SCIs | [135,136] |
| miR-30a, miR-145, miR-155, and miR-216: Higher expression in SCI patients compared to uninjured adults | [137] | ||
| miR-126: Stimulates angiogenesis and promotes regeneration of neurons while decreasing cell death in rats with SCIs | [138] | ||
| miR-29b: Heals injured spinal cords in rats | [139] | ||
| miR-133: Regenerates axons, preserves neurons | [140] | ||
| Long non-coding RNAs (lncRNAs) | Regulate gene expression by interacting with DNA, RNA, and proteins. Some lncRNAs have been detected in EVs, and their transfer can potentially affect epigenetic regulation and gene expression in recipient cells. | lncGm3749: High expression in EVs under hypoxic conditions, effective in repairing SCI by suppressing inflammatory mediators. | [141] |
| lncPTENP1: Helps in recovery from SCI by regulating the expression of miR-21 and miR-19b | [142] | ||
| lncTCTN2: Improved functional recovery after SCI | [143] | ||
| Circular RNAs (circRNAs) | Some circRNAs have been identified in EVs, and they can potentially act as carriers for miRNAs. Their transfer through EVs may influence gene expression and epigenetic processes in recipient cells. | circZFHX3: Inhibits LPS-induced BV-2 cell injury, suggesting a potential therapeutic strategy for the treatment of SCI. | [144] |
| CircRNA CDR1as (ciRS-7) has been detected in EVs and shown to act as a sponge for miR-7, thus regulating gene expression. | [145] | ||
| Histones | Though histones are primarily found within cells, recent studies have identified histones and histone-associated complexes in EVs. | Histones H3 and H4, along with the associated proteins, have been found in EVs, indicating their potential involvement in epigenetic regulation and intercellular communication. | [146] |
| DNA Methylation | While EVs have been found to contain DNA, the specific presence of methylated DNA in EVs and its role in intercellular communication and epigenetic regulation are areas of ongoing research. | DNA methyltransferases (DNMTs) and mRNA have been detected in EVs, suggesting their potential transfer and influence on gene expression in recipient cells. | [129] |
5. Discussion and Therapeutic Approaches
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinto, L.; Götz, M. Radial glial cell heterogeneity—The source of diverse progeny in the CNS. Prog. Neurobiol. 2007, 83, 2–23. [Google Scholar] [CrossRef] [PubMed]
- Hellenbrand, D.J.; Quinn, C.M.; Piper, Z.J.; Morehouse, C.N.; Fixel, J.A.; Hanna, A.S. Inflammation after spinal cord injury: A review of the critical timeline of signaling cues and cellular infiltration. J. Neuroinflamm. 2021, 18, 284. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Fouse, S.; Fan, G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 2007, 61, 58R–63R. [Google Scholar] [CrossRef] [PubMed]
- Van Den Hauwe, L.; Sundgren, P.C.; Flanders, A.E. Spinal trauma and spinal cord injury (SCI). In Diseases of the Brain, Head and Neck, Spine 2020–2023: Diagnostic Imaging; Hodler, J., Kubik-Huch, R.A., von Schulthess, G.K., Eds.; IDKD Springer Series; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Rahimi-Movaghar, V.; Sayyah, M.K.; Akbari, H.; Khorramirouz, R.; Rasouli, M.R.; Moradi-Lakeh, M.; Shokraneh, F.; Vaccaro, A.R. Epidemiology of traumatic spinal cord injury in developing countries: A systematic review. Neuroepidemiology 2013, 41, 65–85. [Google Scholar] [CrossRef]
- Hui, S.P.; Monaghan, J.R.; Voss, S.R.; Ghosh, S. Expression pattern of Nogo-A, MAG, and NgR in regenerating urodele spinal cord. Dev. Dyn. 2013, 242, 847–860. [Google Scholar] [CrossRef]
- Diaz Quiroz, J.F.; Echeverri, K. Spinal cord regeneration: Where fish, frogs and salamanders lead the way, can we follow? Biochem. J. 2013, 451, 353–364. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef]
- York, E.M.; Petit, A.; Roskams, A.J. Epigenetics of neural repair following spinal cord injury. Neurotherapeutics 2013, 10, 757–770. [Google Scholar] [CrossRef]
- Robertson, K.D.; Wolffe, A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000, 1, 11–19. [Google Scholar] [CrossRef]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef]
- Fan, G.; Hutnick, L. Methyl-CpG binding proteins in the nervous system. Cell Res. 2005, 15, 255–261. [Google Scholar] [CrossRef]
- Kameda, T.; Imamura, T.; Nakashima, K. Epigenetic regulation of neural stem cell differentiation towards spinal cord regeneration. Cell Tissue Res. 2018, 371, 189–199. [Google Scholar] [CrossRef]
- Zhu, X.; Xiao, C.; Xiong, J.-W. Epigenetic regulation of organ regeneration in zebrafish. J. Cardiovasc. Dev. Dis. 2018, 5, 57. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Cavalieri, V.; Spinelli, G. Environmental epigenetics in zebrafish. Epigenetics Chromatin 2017, 10, 46. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Best, C.; Ikert, H.; Kostyniuk, D.J.; Craig, P.M.; Navarro-Martin, L.; Marandel, L.; Mennigen, J.A. Epigenetics in teleost fish: From molecular mechanisms to physiological phenotypes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 224, 210–244. [Google Scholar] [CrossRef]
- Kawasaki, H.; Schiltz, L.; Chiu, R.; Itakura, K.; Taira, K.; Nakatani, Y.; Yokoyama, K.K. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 2000, 405, 195–200. [Google Scholar] [CrossRef]
- Ng, S.S.; Yue, W.W.; Oppermann, U.; Klose, R.J. Dynamic protein methylation in chromatin biology. Cell. Mol. Life Sci. 2009, 66, 407–422. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.P.; Jaenisch, R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lee, H.; Henle, S.J.; Cheever, T.R.; Ekker, S.C.; Henley, J.R. Primary neuron culture for nerve growth and axon guidance studies in zebrafish (Danio rerio). PLoS ONE 2013, 8, e57539. [Google Scholar] [CrossRef]
- Pal, D.; Ghatak, S.; Sen, C.K. Epigenetic modification of micrornas. In Microrna in Regenerative Medicine; Elsevier: Amsterdam, The Netherlands, 2015; pp. 77–109. [Google Scholar]
- Norenberg, M.D.; Smith, J.; Marcillo, A. The pathology of human spinal cord injury: Defining the problems. J. Neurotrauma 2004, 21, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Al Mamun, A.; Yuan, Y.; Lu, Q.; Xiong, J.; Yang, S.; Wu, C.; Wu, Y.; Wang, J. Acute spinal cord injury: Pathophysiology and pharmacological intervention (Review). Mol. Med. Rep. 2021, 23, 417. [Google Scholar] [CrossRef] [PubMed]
- Bunge, R.P.; Puckett, W.R.; Becerra, J.L.; Marcillo, A.; Quencer, R.M. Observations on the pathology of human spinal cord injury. A review and classification of 22 new cases with details from a case of chronic cord compression with extensive focal demyelination. Adv. Neurol. 1993, 59, 75–89. [Google Scholar]
- Dumont, R.J.; Okonkwo, D.O.; Verma, S.; Hurlbert, R.J.; Boulos, P.T.; Ellegala, D.B.; Dumont, A.S. Acute spinal cord injury, part I: Pathophysiologic mechanisms. Clin. Neuropharmacol. 2001, 24, 254–264. [Google Scholar] [CrossRef]
- Anjum, A.; Yazid, M.D.; Fauzi Daud, M.; Idris, J.; Ng, A.M.H.; Selvi Naicker, A.; Ismail, O.H.R.; Athi Kumar, R.K.; Lokanathan, Y. Spinal cord injury: Pathophysiology, multimolecular interactions, and underlying recovery mechanisms. Int. J. Mol. Sci. 2020, 21, 7533. [Google Scholar] [CrossRef]
- Sharma, H.S.; Patnaik, R.; Sharma, A.; Sjöquist, P.-O.; Lafuente, J.V. Silicon dioxide nanoparticles (SiO2, 40–50 nm) exacerbate pathophysiology of traumatic spinal cord injury and deteriorate functional outcome in the rat. An experimental study using pharmacological and morphological approaches. J. Nanosci. Nanotechnol. 2009, 9, 4970–4980. [Google Scholar] [CrossRef]
- Xiong, Y.; Hall, E.D. Pharmacological evidence for a role of peroxynitrite in the pathophysiology of spinal cord injury. Exp. Neurol. 2009, 216, 105–114. [Google Scholar] [CrossRef]
- Agrawal, S.K.; Nashmi, R.; Fehlings, M.G. Role of L- and N-type calcium channels in the pathophysiology of traumatic spinal cord white matter injury. Neuroscience 2000, 99, 179–188. [Google Scholar] [CrossRef]
- Clifford, T.; Finkel, Z.; Rodriguez, B.; Joseph, A.; Cai, L. Current Advancements in Spinal Cord Injury Research-Glial Scar Formation and Neural Regeneration. Cells 2023, 12, 853. [Google Scholar] [CrossRef]
- Hayta, E.; Elden, H. Acute spinal cord injury: A review of pathophysiology and potential of non-steroidal anti-inflammatory drugs for pharmacological intervention. J. Chem. Neuroanat. 2018, 87, 25–31. [Google Scholar] [CrossRef]
- Hall, E.D. Pathophysiology of spinal cord injury. Current and future therapies. Minerva Anestesiol. 1989, 55, 63–66. [Google Scholar]
- Thuret, S.; Moon, L.D.F.; Gage, F.H. Therapeutic interventions after spinal cord injury. Nat. Rev. Neurosci. 2006, 7, 628–643. [Google Scholar] [CrossRef]
- Guest, J.D.; Hiester, E.D.; Bunge, R.P. Demyelination and Schwann cell responses adjacent to injury epicenter cavities following chronic human spinal cord injury. Exp. Neurol. 2005, 192, 384–393. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Wang, Y.; Qian, T. Inflammatory response to spinal cord injury and its treatment. World Neurosurg. 2021, 155, 19–31. [Google Scholar] [CrossRef]
- Tran, A.P.; Warren, P.M.; Silver, J. New insights into glial scar formation after spinal cord injury. Cell Tissue Res. 2022, 387, 319–336. [Google Scholar] [CrossRef]
- Silver, J.; Miller, J.H. Regeneration beyond the glial scar. Nat. Rev. Neurosci. 2004, 5, 146–156. [Google Scholar] [CrossRef]
- Fawcett, J.W. Overcoming inhibition in the damaged spinal cord. J. Neurotrauma 2006, 23, 371–383. [Google Scholar] [CrossRef]
- Henke, A.M.; Billington, Z.J.; Gater, D.R. Autonomic Dysfunction and Management after Spinal Cord Injury: A Narrative Review. J. Pers. Med. 2022, 12, 1110. [Google Scholar] [CrossRef]
- Sandvig, A.; Berry, M.; Barrett, L.B.; Butt, A.; Logan, A. Myelin-, reactive glia-, and scar-derived CNS axon growth inhibitors: Expression, receptor signaling, and correlation with axon regeneration. Glia 2004, 46, 225–251. [Google Scholar] [CrossRef]
- Busch, S.A.; Horn, K.P.; Cuascut, F.X.; Hawthorne, A.L.; Bai, L.; Miller, R.H.; Silver, J. Adult NG2+ cells are permissive to neurite outgrowth and stabilize sensory axons during macrophage-induced axonal dieback after spinal cord injury. J. Neurosci. 2010, 30, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Lubińska, L. Patterns of Wallerian degeneration of myelinated fibres in short and long peripheral stumps and in isolated segments of rat phrenic nerve. Interpretation of the role of axoplasmic flow of the trophic factor. Brain Res. 1982, 233, 227–240. [Google Scholar] [CrossRef]
- Kerschensteiner, M.; Schwab, M.E.; Lichtman, J.W.; Misgeld, T. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat. Med. 2005, 11, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Barres, B.A. Axon degeneration: Where the Wlds things are. Curr. Biol. 2012, 22, R221–R223. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, M.G.; Wang, Z.; Lerch, J.K.; Motti, D.; Zhang, Y.P.; Shields, C.B.; Lee, J.K.; Goldberg, J.L.; Lemmon, V.P.; Bixby, J.L. Krüppel-like Factor 7 engineered for transcriptional activation promotes axon regeneration in the adult corticospinal tract. Proc. Natl. Acad. Sci. USA 2012, 109, 7517–7522. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Deng, K.; Hou, J.; Bryson, J.B.; Barco, A.; Nikulina, E.; Spencer, T.; Mellado, W.; Kandel, E.R.; Filbin, M.T. Activated CREB is sufficient to overcome inhibitors in myelin and promote spinal axon regeneration in vivo. Neuron 2004, 44, 609–621. [Google Scholar] [CrossRef]
- Park, K.K.; Liu, K.; Hu, Y.; Smith, P.D.; Wang, C.; Cai, B.; Xu, B.; Connolly, L.; Kramvis, I.; Sahin, M.; et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 2008, 322, 963–966. [Google Scholar] [CrossRef]
- Smith, P.D.; Sun, F.; Park, K.K.; Cai, B.; Wang, C.; Kuwako, K.; Martinez-Carrasco, I.; Connolly, L.; He, Z. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron 2009, 64, 617–623. [Google Scholar] [CrossRef]
- James, G.; Butt, A.M. P2Y and P2X purinoceptor mediated Ca2+ signalling in glial cell pathology in the central nervous system. Eur. J. Pharmacol. 2002, 447, 247–260. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef]
- Wanner, I.B.; Anderson, M.A.; Song, B.; Levine, J.; Fernandez, A.; Gray-Thompson, Z.; Ao, Y.; Sofroniew, M.V. Glial scar borders are formed by newly proliferated, elongated astrocytes that interact to corral inflammatory and fibrotic cells via STAT3-dependent mechanisms after spinal cord injury. J. Neurosci. 2013, 33, 12870–12886. [Google Scholar] [CrossRef]
- Khakh, B.S.; Sofroniew, M.V. Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci. 2015, 18, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Dai, Y.; Chen, G.; Cui, S. Dissecting the Dual Role of the Glial Scar and Scar-Forming Astrocytes in Spinal Cord Injury. Front. Cell. Neurosci. 2020, 14, 78. [Google Scholar] [CrossRef]
- Azari, M.F.; Profyris, C.; Zang, D.W.; Petratos, S.; Cheema, S.S. Induction of endogenous neural precursors in mouse models of spinal cord injury and disease. Eur. J. Neurol. 2005, 12, 638–648. [Google Scholar] [CrossRef]
- Horky, L.L.; Galimi, F.; Gage, F.H.; Horner, P.J. Fate of endogenous stem/progenitor cells following spinal cord injury. J. Comp. Neurol. 2006, 498, 525–538. [Google Scholar] [CrossRef]
- Yang, H.; Lu, P.; McKay, H.M.; Bernot, T.; Keirstead, H.; Steward, O.; Gage, F.H.; Edgerton, V.R.; Tuszynski, M.H. Endogenous neurogenesis replaces oligodendrocytes and astrocytes after primate spinal cord injury. J. Neurosci. 2006, 26, 2157–2166. [Google Scholar] [CrossRef]
- Takizawa, T.; Nakashima, K.; Namihira, M.; Ochiai, W.; Uemura, A.; Yanagisawa, M.; Fujita, N.; Nakao, M.; Taga, T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 2001, 1, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Alzayady, K.; Stewart, R.; Ye, P.; Yang, S.; Li, W.; Shi, Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 2010, 30, 1997–2005. [Google Scholar] [CrossRef]
- Adefuin, A.M.D.; Kimura, A.; Noguchi, H.; Nakashima, K.; Namihira, M. Epigenetic mechanisms regulating differentiation of neural stem/precursor cells. Epigenomics 2014, 6, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, S.T. Gene expression changes after focal stroke, traumatic brain and spinal cord injuries. Curr. Opin. Neurol. 2003, 16, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Kigerl, K.A.; Gensel, J.C.; Ankeny, D.P.; Alexander, J.K.; Donnelly, D.J.; Popovich, P.G. Identification of two distinct macrophage subsets with divergent effects causing either neurotoxicity or regeneration in the injured mouse spinal cord. J. Neurosci. 2009, 29, 13435–13444. [Google Scholar] [CrossRef] [PubMed]
- Halili, M.A.; Andrews, M.R.; Sweet, M.J.; Fairlie, D.P. Histone deacetylase inhibitors in inflammatory disease. Curr. Top. Med. Chem. 2009, 9, 309–319. [Google Scholar] [CrossRef]
- Mullican, S.E.; Gaddis, C.A.; Alenghat, T.; Nair, M.G.; Giacomin, P.R.; Everett, L.J.; Feng, D.; Steger, D.J.; Schug, J.; Artis, D.; et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011, 25, 2480–2488. [Google Scholar] [CrossRef] [PubMed]
- Takeuch, O.; Akira, S. Epigenetic control of macrophage polarization. Eur. J. Immunol. 2011, 41, 2490–2493. [Google Scholar] [CrossRef] [PubMed]
- Schomberg, D.; Olson, J.K. Immune responses of microglia in the spinal cord: Contribution to pain states. Exp. Neurol. 2012, 234, 262–270. [Google Scholar] [CrossRef]
- Chaudhry, N.; Filbin, M.T. Myelin-associated inhibitory signaling and strategies to overcome inhibition. J. Cereb. Blood Flow Metab. 2007, 27, 1096–1107. [Google Scholar] [CrossRef]
- Liu, J.; Casaccia, P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010, 33, 193–201. [Google Scholar] [CrossRef]
- Stam, F.J.; MacGillavry, H.D.; Armstrong, N.J.; de Gunst, M.C.M.; Zhang, Y.; van Kesteren, R.E.; Smit, A.B.; Verhaagen, J. Identification of candidate transcriptional modulators involved in successful regeneration after nerve injury. Eur. J. Neurosci. 2007, 25, 3629–3637. [Google Scholar] [CrossRef]
- Zhang, B.-Y.; Chang, P.-Y.; Zhu, Q.-S.; Zhu, Y.-H.; Saijilafu. Decoding epigenetic codes: New frontiers in exploring recovery from spinal cord injury. Neural Regen. Res. 2020, 15, 1613–1622. [Google Scholar]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef]
- Broide, R.S.; Redwine, J.M.; Aftahi, N.; Young, W.; Bloom, F.E.; Winrow, C.J. Distribution of histone deacetylases 1-11 in the rat brain. J. Mol. Neurosci. 2007, 31, 47–58. [Google Scholar] [CrossRef]
- Wong, J.K.; Zou, H. Reshaping the chromatin landscape after spinal cord injury. Front. Biol. 2014, 9, 356–366. [Google Scholar] [CrossRef]
- Liu, K.; Tedeschi, A.; Park, K.K.; He, Z. Neuronal intrinsic mechanisms of axon regeneration. Annu. Rev. Neurosci. 2011, 34, 131–152. [Google Scholar] [CrossRef]
- Wang, Z.; Reynolds, A.; Kirry, A.; Nienhaus, C.; Blackmore, M.G. Overexpression of Sox11 promotes corticospinal tract regeneration after spinal injury while interfering with functional recovery. J. Neurosci. 2015, 35, 3139–3145. [Google Scholar] [CrossRef]
- Venkatesh, I.; Mehra, V.; Wang, Z.; Califf, B.; Blackmore, M.G. Developmental Chromatin Restriction of Pro-Growth Gene Networks Acts as an Epigenetic Barrier to Axon Regeneration in Cortical Neurons. Dev. Neurobiol. 2018, 78, 960–977. [Google Scholar] [CrossRef]
- Li, S.; Xue, C.; Yuan, Y.; Zhang, R.; Wang, Y.; Wang, Y.; Yu, B.; Liu, J.; Ding, F.; Yang, Y.; et al. The transcriptional landscape of dorsal root ganglia after sciatic nerve transection. Sci. Rep. 2015, 5, 16888. [Google Scholar] [CrossRef]
- Cho, Y.; Sloutsky, R.; Naegle, K.M.; Cavalli, V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell 2013, 155, 894–908. [Google Scholar] [CrossRef]
- Neumann, S.; Woolf, C.J. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron 1999, 23, 83–91. [Google Scholar] [CrossRef]
- Finelli, M.J.; Wong, J.K.; Zou, H. Epigenetic regulation of sensory axon regeneration after spinal cord injury. J. Neurosci. 2013, 33, 19664–19676. [Google Scholar] [CrossRef]
- Puttagunta, R.; Tedeschi, A.; Sória, M.G.; Hervera, A.; Lindner, R.; Rathore, K.I.; Gaub, P.; Joshi, Y.; Nguyen, T.; Schmandke, A.; et al. PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat. Commun. 2014, 5, 3527. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.-H.E.; Koemeter-Cox, A.; Finelli, M.J.; Shen, L.; Friedel, R.H.; Zou, H. Comprehensive mapping of 5-hydroxymethylcytosine epigenetic dynamics in axon regeneration. Epigenetics 2017, 12, 77–92. [Google Scholar] [CrossRef] [PubMed]
- VandenBosch, L.S.; Reh, T.A. Epigenetics in neuronal regeneration. Semin. Cell Dev. Biol. 2020, 97, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Raafat, A.; Pak, E.; Clemens, S.; Murashov, A.K. Dicer-microRNA pathway is critical for peripheral nerve regeneration and functional recovery in vivo and regenerative axonogenesis in vitro. Exp. Neurol. 2012, 233, 555–565. [Google Scholar] [CrossRef]
- Li, P.; Teng, Z.-Q.; Liu, C.-M. Extrinsic and Intrinsic Regulation of Axon Regeneration by MicroRNAs after Spinal Cord Injury. Neural Plast. 2016, 2016, 1279051. [Google Scholar] [CrossRef]
- Hachisuka, S.; Kamei, N.; Ujigo, S.; Miyaki, S.; Yasunaga, Y.; Ochi, M. Circulating microRNAs as biomarkers for evaluating the severity of acute spinal cord injury. Spinal Cord 2014, 52, 596–600. [Google Scholar] [CrossRef]
- Yunta, M.; Nieto-Díaz, M.; Esteban, F.J.; Caballero-López, M.; Navarro-Ruíz, R.; Reigada, D.; Pita-Thomas, D.W.; del Águila, A.; Muñoz-Galdeano, T.; Maza, R.M. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS ONE 2012, 7, e34534. [Google Scholar] [CrossRef]
- Hu, J.-R.; Lv, G.-H.; Yin, B.-L. Altered microRNA expression in the ischemic-reperfusion spinal cord with atorvastatin therapy. J. Pharmacol. Sci. 2013, 121, 343–346. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Jiang, J.-J.; Yan-Gao; Wang, R.-Y.; Tu, G.-J. MicroRNA-210 promotes sensory axon regeneration of adult mice in vivo and in vitro. Neurosci. Lett. 2016, 622, 61–66. [Google Scholar] [CrossRef]
- Ujigo, S.; Kamei, N.; Hadoush, H.; Fujioka, Y.; Miyaki, S.; Nakasa, T.; Tanaka, N.; Nakanishi, K.; Eguchi, A.; Sunagawa, T.; et al. Administration of microRNA-210 promotes spinal cord regeneration in mice. Spine 2014, 39, 1099–1107. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, S.; Ding, Y.; Nong, L.; Li, H.; Gao, G.; Zhou, D.; Xu, N. MicroRNA-21 promotes neurite outgrowth by regulating PDCD4 in a rat model of spinal cord injury. Mol. Med. Rep. 2017, 16, 2522–2528. [Google Scholar] [CrossRef]
- Lin, C.A.; Duan, K.Y.; Wang, X.W.; Zhang, Z.S. MicroRNA-409 promotes recovery of spinal cord injury by regulating ZNF366. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3649–3655. [Google Scholar]
- Liu, C.-M.; Wang, R.-Y.; Saijilafu; Jiao, Z.-X.; Zhang, B.-Y.; Zhou, F.-Q. MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 2013, 27, 1473–1483. [Google Scholar] [CrossRef]
- Slack, J.M.W.; Lin, G.; Chen, Y. The Xenopus tadpole: A new model for regeneration research. Cell. Mol. Life Sci. 2008, 65, 54–63. [Google Scholar] [CrossRef]
- Tanaka, E.M.; Ferretti, P. Considering the evolution of regeneration in the central nervous system. Nat. Rev. Neurosci. 2009, 10, 713–723. [Google Scholar] [CrossRef]
- Egar, M.; Singer, M. The role of ependyma in spinal cord regeneration in the urodele, Triturus. Exp. Neurol. 1972, 37, 422–430. [Google Scholar] [CrossRef]
- Géraudie, J.; Nordlander, R.; Singer, M.; Singer, J. Early stages of spinal ganglion formation during tail regeneration in the newt, Notophthalmus viridescens. Am. J. Anat. 1988, 183, 359–370. [Google Scholar] [CrossRef]
- Clarke, J.D.; Alexander, R.; Holder, N. Regeneration of descending axons in the spinal cord of the axolotl. Neurosci. Lett. 1988, 89, 1–6. [Google Scholar] [CrossRef]
- Diaz Quiroz, J.F.; Tsai, E.; Coyle, M.; Sehm, T.; Echeverri, K. Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: A cross-species comparison between salamander and rat. Dis. Model. Mech. 2014, 7, 601–611. [Google Scholar]
- Voss, S.R.; Ponomareva, L.V.; Dwaraka, V.B.; Pardue, K.E.; Baddar, N.W.A.H.; Rodgers, A.K.; Woodcock, M.R.; Qiu, Q.; Crowner, A.; Blichmann, D.; et al. HDAC regulates transcription at the outset of axolotl tail regeneration. Sci. Rep. 2019, 9, 6751. [Google Scholar] [CrossRef] [PubMed]
- Sehm, T.; Sachse, C.; Frenzel, C.; Echeverri, K. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev. Biol. 2009, 334, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Sabin, K.Z.; Jiang, P.; Gearhart, M.D.; Stewart, R.; Echeverri, K. AP-1cFos/JunB/miR-200a regulate the pro-regenerative glial cell response during axolotl spinal cord regeneration. Commun. Biol. 2019, 2, 91. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Takagi, C.; Miura, S.; Sakane, Y.; Suzuki, M.; Sakuma, T.; Sakamoto, N.; Endo, T.; Kamei, Y.; Sato, Y.; et al. In vivo tracking of histone H3 lysine 9 acetylation in Xenopus laevis during tail regeneration. Genes Cells 2016, 21, 358–369. [Google Scholar] [CrossRef]
- Tseng, A.-S.; Carneiro, K.; Lemire, J.M.; Levin, M. HDAC activity is required during Xenopus tail regeneration. PLoS ONE 2011, 6, e26382. [Google Scholar] [CrossRef]
- Pentagna, N.; Soares dos Santos, F.; Martins de Almeida, F.; Garcia Abreu, J.; Levin, M.; Carneiro, K. Epigenetic immune-modulation by Histone Deacetylase Activity (HDAC) of tissue and organ regeneration in Xenopsu laevis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yu, Y.-M.; Gibbs, K.M.; Davila, J.; Campbell, N.; Sung, S.; Todorova, T.I.; Otsuka, S.; Sabaawy, H.E.; Hart, R.P.; Schachner, M. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur. J. Neurosci. 2011, 33, 1587–1597. [Google Scholar] [CrossRef]
- Huang, R.; Chen, M.; Yang, L.; Wagle, M.; Guo, S.; Hu, B. MicroRNA-133b Negatively Regulates Zebrafish Single Mauthner-Cell Axon Regeneration through Targeting tppp3 In Vivo. Front. Mol. Neurosci. 2017, 10, 375. [Google Scholar] [CrossRef]
- McCann, T. The Role of Histone Deacetylase 1 Inneuroregeneration in the Zebrafishspinal Cord. Ph.D. Dissertation, The University of Edinburgh, Edinburgh, UK, 2019. [Google Scholar]
- Taylor, A.J.; Beck, C.W. Histone deacetylases are required for amphibian tail and limb regeneration but not development. Mech. Dev. 2012, 129, 208–218. [Google Scholar] [CrossRef]
- Hui, S.P.; Sengupta, D.; Lee, S.G.P.; Sen, T.; Kundu, S.; Mathavan, S.; Ghosh, S. Genome wide expression profiling during spinal cord regeneration identifies comprehensive cellular responses in zebrafish. PLoS ONE 2014, 9, e84212. [Google Scholar] [CrossRef]
- Gupta, S.; Adhikary, S.; Hui, S.P. Decoding the proregenerative competence of regulatory T cells through complex tissue regeneration in zebrafish. Clin. Exp. Immunol. 2021, 206, 346–353. [Google Scholar] [CrossRef]
- Hui, S.P.; Dutta, A.; Ghosh, S. Cellular response after crush injury in adult zebrafish spinal cord. Dev. Dyn. 2010, 239, 2962–2979. [Google Scholar] [CrossRef]
- Becker, T.; Lieberoth, B.C.; Becker, C.G.; Schachner, M. Differences in the regenerative response of neuronal cell populations and indications for plasticity in intraspinal neurons after spinal cord transection in adult zebrafish. Mol. Cell. Neurosci. 2005, 30, 265–278. [Google Scholar] [CrossRef]
- Becker, T.; Becker, C.G. Regenerating descending axons preferentially reroute to the gray matter in the presence of a general macrophage/microglial reaction caudal to a spinal transection in adult zebrafish. J. Comp. Neurol. 2001, 433, 131–147. [Google Scholar] [CrossRef]
- Horn, K.P.; Busch, S.A.; Hawthorne, A.L.; van Rooijen, N.; Silver, J. Another barrier to regeneration in the CNS: Activated macrophages induce extensive retraction of dystrophic axons through direct physical interactions. J. Neurosci. 2008, 28, 9330–9341. [Google Scholar] [CrossRef]
- Ghosh, S.; Hui, S.P. Regeneration of zebrafish CNS: Adult neurogenesis. Neural Plast. 2016, 2016, 5815439. [Google Scholar] [CrossRef]
- Gemberling, M.; Bailey, T.J.; Hyde, D.R.; Poss, K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013, 29, 611–620. [Google Scholar] [CrossRef]
- Theis, T.; Yoo, M.; Park, C.S.; Chen, J.; Kügler, S.; Gibbs, K.M.; Schachner, M. Lentiviral Delivery of miR-133b Improves Functional Recovery After Spinal Cord Injury in Mice. Mol. Neurobiol. 2017, 54, 4659–4671. [Google Scholar] [CrossRef]
- Dutta, S.; Bitan, G.; Van Keuren-Jensen, K.; Nimse, S.B. Exosomes: Message in a vesicle. Front. Pharmacol. 2022, 13, 1018928. [Google Scholar] [CrossRef]
- Mukherjee, A.; Bisht, B.; Dutta, S.; Paul, M.K. Current advances in the use of exosomes, liposomes, and bioengineered hybrid nanovesicles in cancer detection and therapy. Acta Pharmacol. Sin. 2022, 43, 2759–2776. [Google Scholar] [CrossRef]
- Yan, Z.; Dutta, S.; Liu, Z.; Yu, X.; Mesgarzadeh, N.; Ji, F.; Bitan, G.; Xie, Y.-H. A Label-Free Platform for Identification of Exosomes from Different Sources. ACS Sens. 2019, 4, 488–497. [Google Scholar] [CrossRef]
- Hornung, S.; Dutta, S.; Bitan, G. CNS-Derived Blood Exosomes as a Promising Source of Biomarkers: Opportunities and Challenges. Front. Mol. Neurosci. 2020, 13, 38. [Google Scholar] [CrossRef]
- Dutta, S.; Reamtong, O.; Panvongsa, W.; Kitdumrongthum, S.; Janpipatkul, K.; Sangvanich, P.; Piyachaturawat, P.; Chairoungdua, A. Proteomics profiling of cholangiocarcinoma exosomes: A potential role of oncogenic protein transferring in cancer progression. Biochim. Biophys. Acta 2015, 1852, 1989–1999. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Shen, Q.; Yang, X.; Qiu, Y.; Zhang, W. The role of extracellular vesicles: An epigenetic view of the cancer microenvironment. Biomed Res. Int. 2015, 2015, 649161. [Google Scholar] [CrossRef] [PubMed]
- Munir, J.; Yoon, J.K.; Ryu, S. Therapeutic miRNA-Enriched Extracellular Vesicles: Current Approaches and Future Prospects. Cells 2020, 9, 2271. [Google Scholar] [CrossRef]
- Dakhlallah, D.A.; Wisler, J.; Gencheva, M.; Brown, C.M.; Leatherman, E.R.; Singh, K.; Brundage, K.; Karsies, T.; Dakhlallah, A.; Witwer, K.W.; et al. Circulating extracellular vesicle content reveals de novo DNA methyltransferase expression as a molecular method to predict septic shock. J. Extracell. Vesicles 2019, 8, 1669881. [Google Scholar] [CrossRef]
- Schiano, C.; Balbi, C.; Burrello, J.; Ruocco, A.; Infante, T.; Fiorito, C.; Panella, S.; Barile, L.; Mauro, C.; Vassalli, G.; et al. De novo DNA methylation induced by circulating extracellular vesicles from acute coronary syndrome patients. Atherosclerosis 2022, 354, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Khan, N.; Wu, J.; Jay, S.M. Extracellular vesicles as an emerging frontier in spinal cord injury pathobiology and therapy. Trends Neurosci. 2021, 44, 492–506. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, Y.; Zhu, Z.; Gu, C.; Waqas, A.; Chen, L. Emerging exosomes and exosomal mirnas in spinal cord injury. Front. Cell Dev. Biol. 2021, 9, 703989. [Google Scholar] [CrossRef]
- Malvandi, A.M.; Rastegar-Moghaddam, S.H.; Ebrahimzadeh-Bideskan, S.; Lombardi, G.; Ebrahimzadeh-Bideskan, A.; Mohammadipour, A. Targeting miR-21 in spinal cord injuries: A game-changer? Mol. Med. 2022, 28, 118. [Google Scholar] [CrossRef]
- Kar, A.N.; Lee, S.-J.; Sahoo, P.K.; Thames, E.; Yoo, S.; Houle, J.D.; Twiss, J.L. MicroRNAs 21 and 199a-3p Regulate Axon Growth Potential through Modulation of Pten and mTor mRNAs. eNeuro 2021, 8, ENEURO.0155-21.2021. [Google Scholar] [CrossRef]
- Kang, J.; Li, Z.; Zhi, Z.; Wang, S.; Xu, G. MiR-21 derived from the exosomes of MSCs regulates the death and differentiation of neurons in patients with spinal cord injury. Gene Ther. 2019, 26, 491–503. [Google Scholar] [CrossRef]
- Xu, G.; Ao, R.; Zhi, Z.; Jia, J.; Yu, B. miR-21 and miR-19b delivered by hMSC-derived EVs regulate the apoptosis and differentiation of neurons in patients with spinal cord injury. J. Cell. Physiol. 2019, 234, 10205–10217. [Google Scholar] [CrossRef]
- Park, A.J.; Fandl, H.K.; Garcia, V.P.; Coombs, G.B.; DeSouza, N.M.; Greiner, J.J.; Barak, O.F.; Mijacika, T.; Dujic, Z.; Ainslie, P.N.; et al. Differential Expression of Vascular-Related MicroRNA in Circulating Endothelial Microvesicles in Adults With Spinal Cord Injury: A Pilot Study. Top. Spinal Cord Inj. Rehabil. 2023, 29, 34–42. [Google Scholar] [CrossRef]
- Huang, J.-H.; Xu, Y.; Yin, X.-M.; Lin, F.-Y. Exosomes Derived from miR-126-modified MSCs Promote Angiogenesis and Neurogenesis and Attenuate Apoptosis after Spinal Cord Injury in Rats. Neuroscience 2020, 424, 133–145. [Google Scholar] [CrossRef]
- Yu, T.; Zhao, C.; Hou, S.; Zhou, W.; Wang, B.; Chen, Y. Exosomes secreted from miRNA-29b-modified mesenchymal stem cells repaired spinal cord injury in rats. Braz. J. Med. Biol. Res. 2019, 52, e8735. [Google Scholar] [CrossRef]
- Li, D.; Zhang, P.; Yao, X.; Li, H.; Shen, H.; Li, X.; Wu, J.; Lu, X. Exosomes Derived From miR-133b-Modified Mesenchymal Stem Cells Promote Recovery After Spinal Cord Injury. Front. Neurosci. 2018, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Jin, M.; Xu, S.; Zheng, C.; Zhu, W.; Ma, X.; Lv, F. Exosomes from Long Noncoding RNA-Gm37494-ADSCs Repair Spinal Cord Injury via Shifting Microglial M1/M2 Polarization. Inflammation 2020, 43, 1536–1547. [Google Scholar] [CrossRef]
- Wang, Z.; Song, Y.; Han, X.; Qu, P.; Wang, W. Long noncoding RNA PTENP1 affects the recovery of spinal cord injury by regulating the expression of miR-19b and miR-21. J. Cell. Physiol. 2020, 235, 3634–3645. [Google Scholar] [CrossRef]
- Liu, J.; Lin, M.; Qiao, F.; Zhang, C. Exosomes Derived from lncRNA TCTN2-Modified Mesenchymal Stem Cells Improve Spinal Cord Injury by miR-329-3p/IGF1R Axis. J. Mol. Neurosci. 2022, 72, 482–495. [Google Scholar] [CrossRef]
- Tian, F.; Yang, J.; Xia, R. Exosomes Secreted from circZFHX3-modified Mesenchymal Stem Cells Repaired Spinal Cord Injury through mir-16-5p/IGF-1 in Mice. Neurochem. Res. 2022, 47, 2076–2089. [Google Scholar] [CrossRef] [PubMed]
- Fanale, D.; Taverna, S.; Russo, A.; Bazan, V. Circular RNA in exosomes. Adv. Exp. Med. Biol. 2018, 1087, 109–117. [Google Scholar] [PubMed]
- Ortiz, A. Not all extracellular vesicles were created equal: Clinical implications. Ann. Transl. Med. 2017, 5, 111. [Google Scholar] [CrossRef]
- Collino, F.; Pomatto, M.; Bruno, S.; Lindoso, R.S.; Tapparo, M.; Sicheng, W.; Quesenberry, P.; Camussi, G. Exosome and Microvesicle-Enriched Fractions Isolated from Mesenchymal Stem Cells by Gradient Separation Showed Different Molecular Signatures and Functions on Renal Tubular Epithelial Cells. Stem Cell Rev. Rep. 2017, 13, 226–243. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Augustine, G.J.; Ammar, M.-R.; Tashiro, A.; Cohen, S.M. A neuroprotective role for microRNA miR-1000 mediated by limiting glutamate excitotoxicity. Nat. Neurosci. 2015, 18, 379–385. [Google Scholar] [CrossRef]
- Xin, H.; Li, Y.; Liu, Z.; Wang, X.; Shang, X.; Cui, Y.; Zhang, Z.G.; Chopp, M. MiR-133b promotes neural plasticity and functional recovery after treatment of stroke with multipotent mesenchymal stromal cells in rats via transfer of exosome-enriched extracellular particles. Stem Cells 2013, 31, 2737–2746. [Google Scholar] [CrossRef]
- Pusic, A.D.; Kraig, R.P. Youth and environmental enrichment generate serum exosomes containing miR-219 that promote CNS myelination. Glia 2014, 62, 284–299. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Y.; Gong, F.; Rong, Y.; Luo, Y.; Tang, P.; Zhou, Z.; Zhou, Z.; Xu, T.; Jiang, T.; et al. Exosomes Derived from Bone Mesenchymal Stem Cells Repair Traumatic Spinal Cord Injury by Suppressing the Activation of A1 Neurotoxic Reactive Astrocytes. J. Neurotrauma 2019, 36, 469–484. [Google Scholar] [CrossRef]
- Guo, S.; Redenski, I.; Levenberg, S. Spinal cord repair: From cells and tissue engineering to extracellular vesicles. Cells 2021, 10, 1872. [Google Scholar] [CrossRef]
- Ohgo, S.; Sakamoto, T.; Nakajima, W.; Matsunaga, S.; Wada, N. Visualization of extracellular vesicles in the regenerating caudal fin blastema of zebrafish using in vivo electroporation. Biochem. Biophys. Res. Commun. 2020, 533, 1371–1377. [Google Scholar] [CrossRef]
- McDonald, J.W.; Liu, X.Z.; Qu, Y.; Liu, S.; Mickey, S.K.; Turetsky, D.; Gottlieb, D.I.; Choi, D.W. Transplanted embryonic stem cells survive, differentiate and promote recovery in injured rat spinal cord. Nat. Med. 1999, 5, 1410–1412. [Google Scholar] [CrossRef] [PubMed]
- Setoguchi, T.; Nakashima, K.; Takizawa, T.; Yanagisawa, M.; Ochiai, W.; Okabe, M.; Yone, K.; Komiya, S.; Taga, T. Treatment of spinal cord injury by transplantation of fetal neural precursor cells engineered to express BMP inhibitor. Exp. Neurol. 2004, 189, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Abematsu, M.; Tsujimura, K.; Yamano, M.; Saito, M.; Kohno, K.; Kohyama, J.; Namihira, M.; Komiya, S.; Nakashima, K. Neurons derived from transplanted neural stem cells restore disrupted neuronal circuitry in a mouse model of spinal cord injury. J. Clin. Investig. 2010, 120, 3255–3266. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Abematsu, M.; Falk, A.; Tsujimura, K.; Sanosaka, T.; Juliandi, B.; Semi, K.; Namihira, M.; Komiya, S.; Smith, A.; et al. Treatment of a mouse model of spinal cord injury by transplantation of human induced pluripotent stem cell-derived long-term self-renewing neuroepithelial-like stem cells. Stem Cells 2012, 30, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Okano, H.; Yamanaka, S. iPS cell technologies: Significance and applications to CNS regeneration and disease. Mol. Brain 2014, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, N.; Okano, H.; Nakamura, M. Regenerative therapy for spinal cord injury using iPSC technology. Inflamm. Regen. 2020, 40, 40. [Google Scholar] [CrossRef]
- Nagoshi, N.; Tsuji, O.; Nakamura, M.; Okano, H. Cell therapy for spinal cord injury using induced pluripotent stem cells. Regen. Ther. 2019, 11, 75–80. [Google Scholar] [CrossRef]
- Li, Y.; Shen, P.-P.; Wang, B. Induced pluripotent stem cell technology for spinal cord injury: A promising alternative therapy. Neural Regen. Res. 2021, 16, 1500–1509. [Google Scholar]
- De Gioia, R.; Biella, F.; Citterio, G.; Rizzo, F.; Abati, E.; Nizzardo, M.; Bresolin, N.; Comi, G.P.; Corti, S. Neural stem cell transplantation for neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 3103. [Google Scholar] [CrossRef]
- Fischer, I.; Dulin, J.N.; Lane, M.A. Transplanting neural progenitor cells to restore connectivity after spinal cord injury. Nat. Rev. Neurosci. 2020, 21, 366–383. [Google Scholar] [CrossRef]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release 2013, 172, 962–974. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef]
- Hu, M.; Cao, Z.; Jiang, D. The Effect of miRNA-Modified Exosomes in Animal Models of Spinal Cord Injury: A meta-Analysis. Front. Bioeng. Biotechnol. 2021, 9, 819651. [Google Scholar] [CrossRef]
- Galieva, L.R.; James, V.; Mukhamedshina, Y.O.; Rizvanov, A.A. Therapeutic potential of extracellular vesicles for the treatment of nerve disorders. Front. Neurosci. 2019, 13, 163. [Google Scholar] [CrossRef]
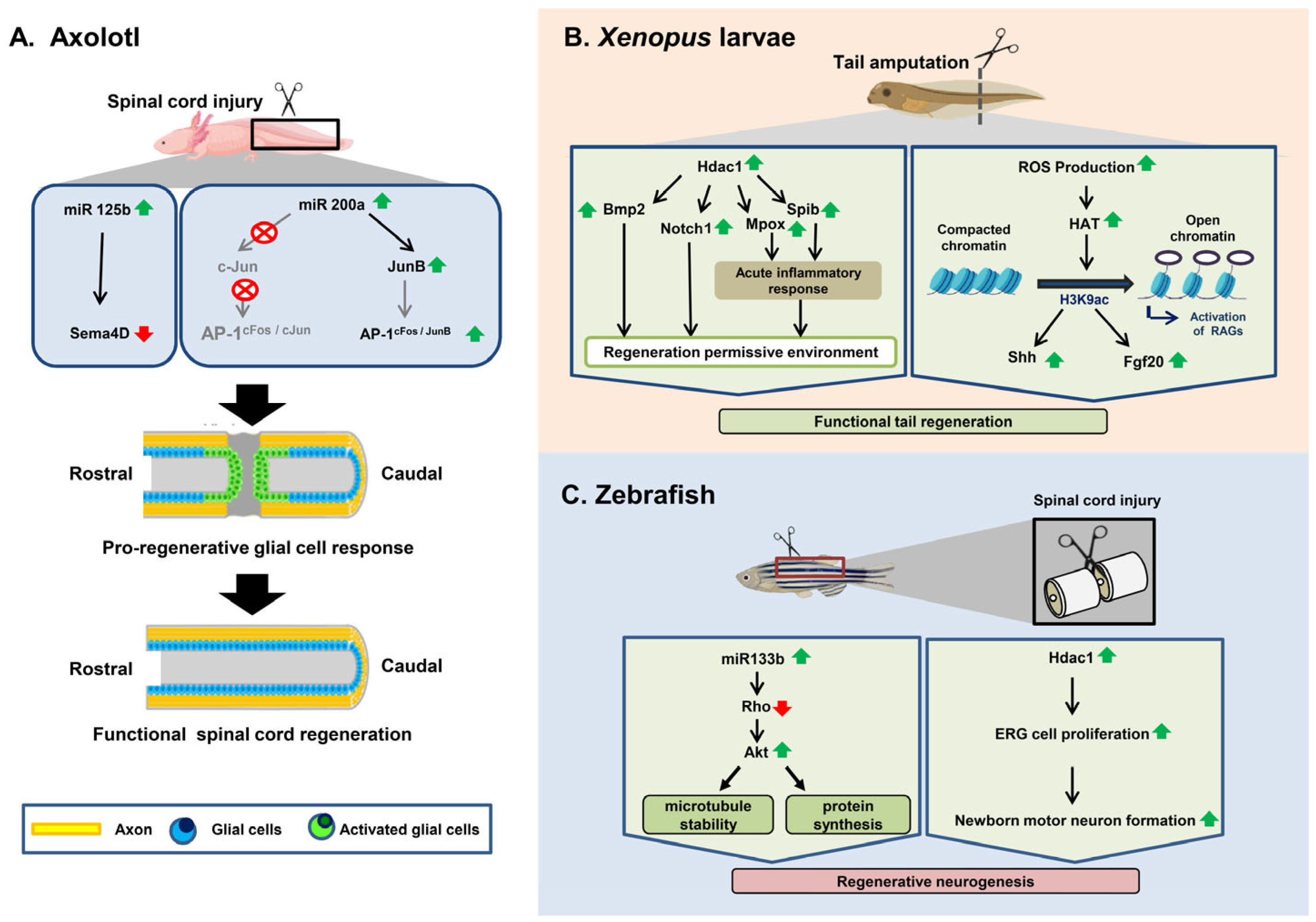

| Different Regenerating Organisms | Types of Epigenetic Modification | Involved Genes/Mediators | Role | Reference |
|---|---|---|---|---|
| Axolotl | Histone deacetylation | hdac1 | To control the initial transcriptional response to injury and regeneration in axolotls at the time of tail amputation | [103] |
| miRNA regulation | miR-125b | Guides proper axon regeneration involving a direct downstream target, Sema4D, a member of the Semaphorin gene class | [102] | |
| miR-196 | Acts upstream of the main patterning events within the spinal cord based on BMP4 and Pax7 | [104] | ||
| miR–200b | Upregulates JunB after SCI, preventing glial cells around the injury site from upregulating the expression of AP-1cFos/c-Jun and inducing glial cells to proliferate | [105] | ||
| Xenopus | Histone Acetylation | h3k9ac | Induces shh and fgf20 | [106] |
| Histone Deacetylation | hdac1 | Induces the expression of two genes, notch1 and bmp2 | [107] | |
| hdac | Induces the expression of mpox and spib, which create an inflammatory response favoring tail regeneration | [108] | ||
| Zebrafish | miRNA regulation | miR-133b | Exogenous overexpression of miR-133b increases motor function recovery after SCI by regulating the RhoA signaling pathway | [109] |
| Inhibition of miR-133b promotes axon outgrowth via the modulation of tppp3 | [110] | |||
| Histone Deacetylation | hdac1 | Induces ependymo-radial glial cell proliferation and increases formation of newborn motor neurons | [111] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, S.; Dutta, S.; Hui, S.P. Regenerative Potential of Injured Spinal Cord in the Light of Epigenetic Regulation and Modulation. Cells 2023, 12, 1694. https://doi.org/10.3390/cells12131694
Gupta S, Dutta S, Hui SP. Regenerative Potential of Injured Spinal Cord in the Light of Epigenetic Regulation and Modulation. Cells. 2023; 12(13):1694. https://doi.org/10.3390/cells12131694
Chicago/Turabian StyleGupta, Samudra, Suman Dutta, and Subhra Prakash Hui. 2023. "Regenerative Potential of Injured Spinal Cord in the Light of Epigenetic Regulation and Modulation" Cells 12, no. 13: 1694. https://doi.org/10.3390/cells12131694
APA StyleGupta, S., Dutta, S., & Hui, S. P. (2023). Regenerative Potential of Injured Spinal Cord in the Light of Epigenetic Regulation and Modulation. Cells, 12(13), 1694. https://doi.org/10.3390/cells12131694







