Author Contributions
Conceptualization, P.D. and G.M.; methodology, P.D. and G.M.; software, Y.P. and D.F.; validation, Y.P., G.F. and D.F.; formal analysis, G.M.; investigation, Y.P., G.F. and D.F.; resources, P.D. and G.M.; data curation, Y.P., P.D. and G.M.; writing—original draft preparation, Y.P., G.F., D.F. and G.M.; writing—review and editing, Y.P., P.D. and G.M.; visualization, M.L.; supervision, G.M.; project administration, G.M.; funding acquisition, P.D. and G.M. All authors have read and agreed to the published version of the manuscript.
Figure 1.
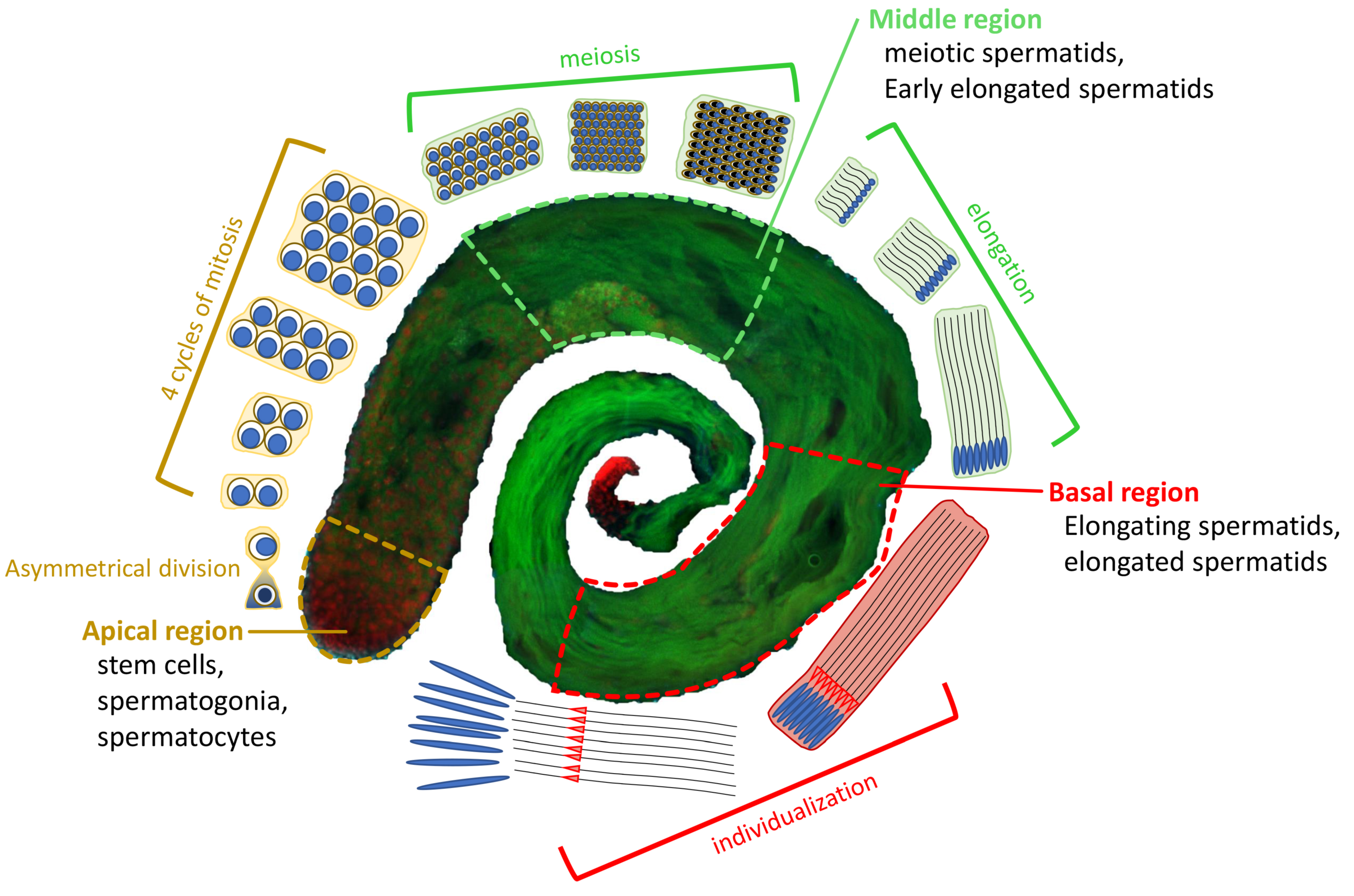
Overview of D. melanogaster spermatogenesis. At the apical region of a testis, a hub of germline stem cells (GSCs) divides asymmetrically into two cells: a daughter stem cell and a differentiating gonialblast (GB). The GB goes through 4 cycles of mitosis to form a cyst of 16 primary spermatocytes. Primary spermatocytes will proceed through meiosis, resulting in the generation of 64 roundish haploid spermatids (Middle region). The latter undergo elongation processes characterized by changes in nuclear shape and chromatin condensation to form individualized mature sperm (Basal region), which are stored in the seminal vesicle until fertilization. EGFP::α-Tubulin84D in green, and H2A.V::mRFP in red.
Figure 1.
Overview of D. melanogaster spermatogenesis. At the apical region of a testis, a hub of germline stem cells (GSCs) divides asymmetrically into two cells: a daughter stem cell and a differentiating gonialblast (GB). The GB goes through 4 cycles of mitosis to form a cyst of 16 primary spermatocytes. Primary spermatocytes will proceed through meiosis, resulting in the generation of 64 roundish haploid spermatids (Middle region). The latter undergo elongation processes characterized by changes in nuclear shape and chromatin condensation to form individualized mature sperm (Basal region), which are stored in the seminal vesicle until fertilization. EGFP::α-Tubulin84D in green, and H2A.V::mRFP in red.
Figure 2.
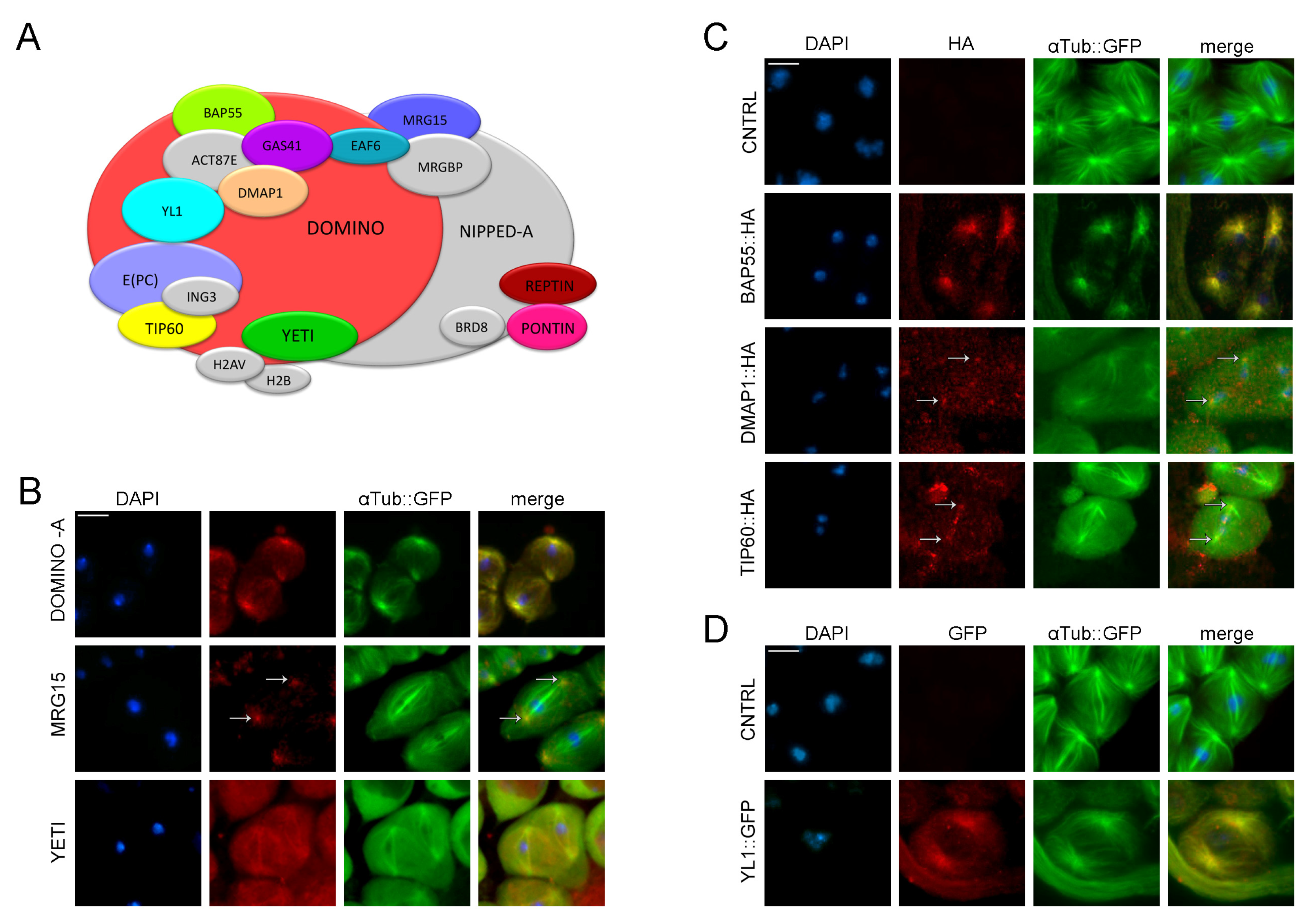
Dynamic localization of DOM/Tip60 chromatin remodeling complex subunits in meiosis. (A) Graphical representation of DOMINO/DOM/Tip60 remodeling complex. Subunits are not in scale. (B–D) Immuno-localization of DOMINO-A, MRG15, Yeti, BAP55, DMAP1, TIP60 and YL1. Testes of young adult, 1–3 days old, from EGFP::αTub; Bam>Gal4 crossed with UAS>protein-HA tag and UAS>protein-GFP tag were stained with specific-antibody (in red) while αTubulin is endogenously fluorescent (in green). DNA is stained with DAPI (in blue). DOMINO-A, MRG15, BAP55, DMAP1 and YL1 showed a specific localization to centrosomes; YETI showed a spindle localization, while Tip60 showed a signal along the meiotic spindle. Scale bar, 10 µm.
Figure 2.
Dynamic localization of DOM/Tip60 chromatin remodeling complex subunits in meiosis. (A) Graphical representation of DOMINO/DOM/Tip60 remodeling complex. Subunits are not in scale. (B–D) Immuno-localization of DOMINO-A, MRG15, Yeti, BAP55, DMAP1, TIP60 and YL1. Testes of young adult, 1–3 days old, from EGFP::αTub; Bam>Gal4 crossed with UAS>protein-HA tag and UAS>protein-GFP tag were stained with specific-antibody (in red) while αTubulin is endogenously fluorescent (in green). DNA is stained with DAPI (in blue). DOMINO-A, MRG15, BAP55, DMAP1 and YL1 showed a specific localization to centrosomes; YETI showed a spindle localization, while Tip60 showed a signal along the meiotic spindle. Scale bar, 10 µm.
Figure 3.
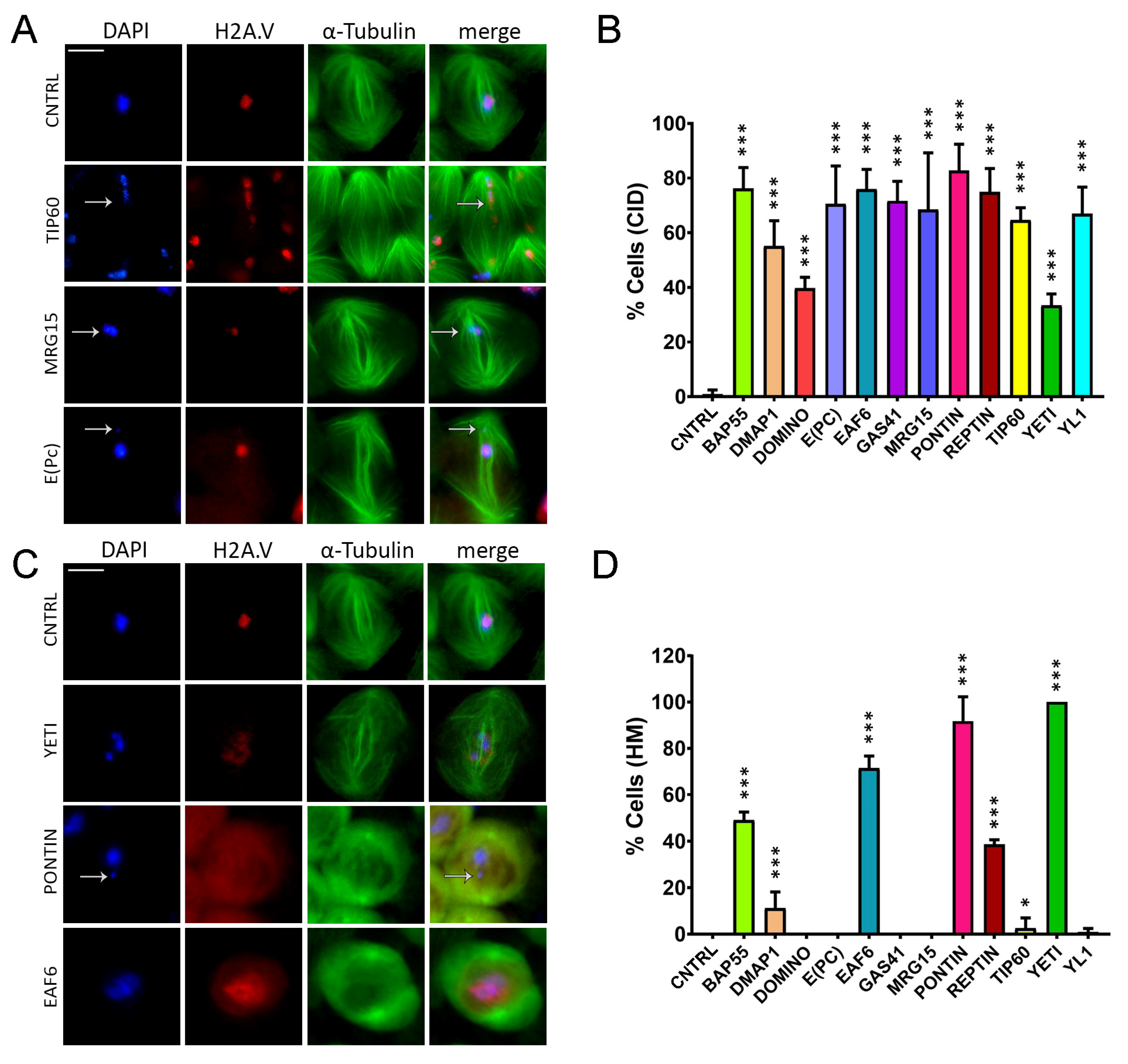
Chromatin integrity defects (CID) and H2A.V mislocalization (HM) defects induced by RNAi in meiosis. Cytological analysis of testis squashes prepared from EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver crossed with specific remodelers of the RNAi construct. DNA is stained with DAPI (in blue), EGFP::αTub (in green) and H2A.V::mRFP (in red). Scale bar, 10 µm. (A) For Tip60, MRG15 and E(pc), the white arrow indicates chromatin fragments probably lost during segregation. (B) Quantification analysis of CID after RNAi knockdown effects activated by the EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 72), BAP55 (n = 128), DMAP1 (n = 260), DOMINO (n = 98), E(PC) (n = 113), EAF6 (n = 289), GAS41 (n = 196), MRG15 (n = 82), PONTIN (n = 281), REPTIN (n = 339), TIP60 (n = 116), YETI (n = 79) and YL1 (n = 102). (C) H2A.V mislocalization is reported for YETI, PONTIN and EAF6 as a widespread nuclear signal compared to Control sample. (D) Quantification analysis of HM after RNAi knockdown effects activated by the EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 148), BAP55 (n = 138), DMAP1 (n = 132), DOMINO (n = 64), E(PC) (n = 124), EAF6 (n = 184), GAS41 (n = 195), MRG15 (n = 70), PONTIN (n = 119), REPTIN (n = 85), TIP60 (n = 43), YETI (n = 154) and YL1 (n = 82). The statistical analysis is performed by using two-tailed Fisher’s exact test (* = p value ≤ 0.05, ** = p value ≤ 0.005, *** = p value ≤ 0.0005).
Figure 3.
Chromatin integrity defects (CID) and H2A.V mislocalization (HM) defects induced by RNAi in meiosis. Cytological analysis of testis squashes prepared from EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver crossed with specific remodelers of the RNAi construct. DNA is stained with DAPI (in blue), EGFP::αTub (in green) and H2A.V::mRFP (in red). Scale bar, 10 µm. (A) For Tip60, MRG15 and E(pc), the white arrow indicates chromatin fragments probably lost during segregation. (B) Quantification analysis of CID after RNAi knockdown effects activated by the EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 72), BAP55 (n = 128), DMAP1 (n = 260), DOMINO (n = 98), E(PC) (n = 113), EAF6 (n = 289), GAS41 (n = 196), MRG15 (n = 82), PONTIN (n = 281), REPTIN (n = 339), TIP60 (n = 116), YETI (n = 79) and YL1 (n = 102). (C) H2A.V mislocalization is reported for YETI, PONTIN and EAF6 as a widespread nuclear signal compared to Control sample. (D) Quantification analysis of HM after RNAi knockdown effects activated by the EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 148), BAP55 (n = 138), DMAP1 (n = 132), DOMINO (n = 64), E(PC) (n = 124), EAF6 (n = 184), GAS41 (n = 195), MRG15 (n = 70), PONTIN (n = 119), REPTIN (n = 85), TIP60 (n = 43), YETI (n = 154) and YL1 (n = 82). The statistical analysis is performed by using two-tailed Fisher’s exact test (* = p value ≤ 0.05, ** = p value ≤ 0.005, *** = p value ≤ 0.0005).
![Cells 12 01348 g003 Cells 12 01348 g003]()
Figure 4.
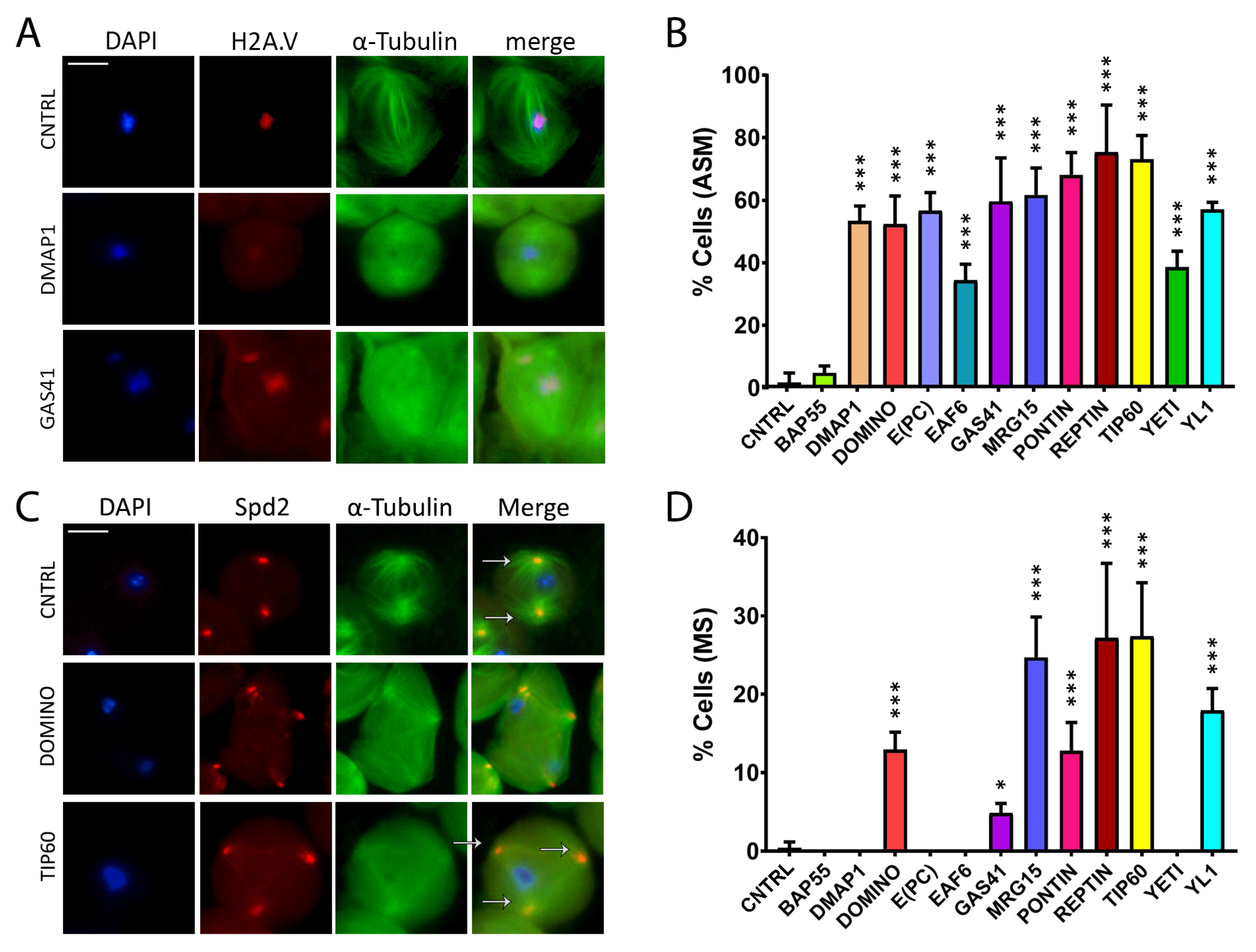
Abnormal spindle morphology (ASM) and multipolar spindle (MS) defects induced by RNAi in meiosis. Testis squashes preparation from Bam>Gal4 driver strain crossed with specific subunit RNAi construct. DNA is stained with DAPI (in blue), EGFP::αTub (in green) and H2A.V::mRFP (in red). Scale bar, 10 µm. (A) Early chromatin decondensation effects are shown for DMAP1 knockdown, with a widespread signal for DNA and H2A.V as well, and for GAS41 knockdown, with detached chromatin fragments from the central plate as well. (B) Quantification analysis of ASM after RNAi knockdown effects activated by the EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 85), BAP55 (n = 74), DMAP1 (n = 58), DOMINO (n = 113), E(PC) (n = 28), EAF6 (n = 32), GAS41 (n = 31), MRG15 (n = 68), PONTIN (n = 53), REPTIN (n = 69), TIP60 (n = 65), YETI (n = 87) and YL1 (n = 63). (C) Alteration of spindle structure, shown here for DOMINO and TIP60 subunits from squashed testes of young adult flies, 1–3 days old, from EGFP::αTub, Spd2::mRFP; Bam>Gal4 driver crossed with specific subunit RNAi construct. DNA is stained with DAPI (in blue), EGFP::αTub (in green) and Spd2::mRFP (in red). White arrows indicate two centrosomes in the control sample, while in the interfered samples for TIP60 and DOMINO, multiple centrosomes are noticeable. (D) Quantification analysis of MS after RNAi knockdown effects activated by the EGFP::αTub, Spd2::mRFP; Bam>Gal4 driver. N = number of analyzed cells: CNTRL (n = 199), BAP55 (n = 131), DMAP1 (n = 246), DOMINO (n = 328), E(PC) (n = 189), EAF6 (n = 147), GAS41 (n = 181), MRG15 (n = 96), PONTIN (n = 119), REPTIN (n = 152), TIP60 (n = 162), YETI (n = 130) and YL1 (n = 192). The statistical analysis is performed by using two-tailed Fisher’s exact test (* = p value ≤ 0.05, ** = p value ≤ 0.005, *** = p value ≤ 0.0005).
Figure 4.
Abnormal spindle morphology (ASM) and multipolar spindle (MS) defects induced by RNAi in meiosis. Testis squashes preparation from Bam>Gal4 driver strain crossed with specific subunit RNAi construct. DNA is stained with DAPI (in blue), EGFP::αTub (in green) and H2A.V::mRFP (in red). Scale bar, 10 µm. (A) Early chromatin decondensation effects are shown for DMAP1 knockdown, with a widespread signal for DNA and H2A.V as well, and for GAS41 knockdown, with detached chromatin fragments from the central plate as well. (B) Quantification analysis of ASM after RNAi knockdown effects activated by the EGFP::αTub, H2A.V::mRFP; Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 85), BAP55 (n = 74), DMAP1 (n = 58), DOMINO (n = 113), E(PC) (n = 28), EAF6 (n = 32), GAS41 (n = 31), MRG15 (n = 68), PONTIN (n = 53), REPTIN (n = 69), TIP60 (n = 65), YETI (n = 87) and YL1 (n = 63). (C) Alteration of spindle structure, shown here for DOMINO and TIP60 subunits from squashed testes of young adult flies, 1–3 days old, from EGFP::αTub, Spd2::mRFP; Bam>Gal4 driver crossed with specific subunit RNAi construct. DNA is stained with DAPI (in blue), EGFP::αTub (in green) and Spd2::mRFP (in red). White arrows indicate two centrosomes in the control sample, while in the interfered samples for TIP60 and DOMINO, multiple centrosomes are noticeable. (D) Quantification analysis of MS after RNAi knockdown effects activated by the EGFP::αTub, Spd2::mRFP; Bam>Gal4 driver. N = number of analyzed cells: CNTRL (n = 199), BAP55 (n = 131), DMAP1 (n = 246), DOMINO (n = 328), E(PC) (n = 189), EAF6 (n = 147), GAS41 (n = 181), MRG15 (n = 96), PONTIN (n = 119), REPTIN (n = 152), TIP60 (n = 162), YETI (n = 130) and YL1 (n = 192). The statistical analysis is performed by using two-tailed Fisher’s exact test (* = p value ≤ 0.05, ** = p value ≤ 0.005, *** = p value ≤ 0.0005).
![Cells 12 01348 g004 Cells 12 01348 g004]()
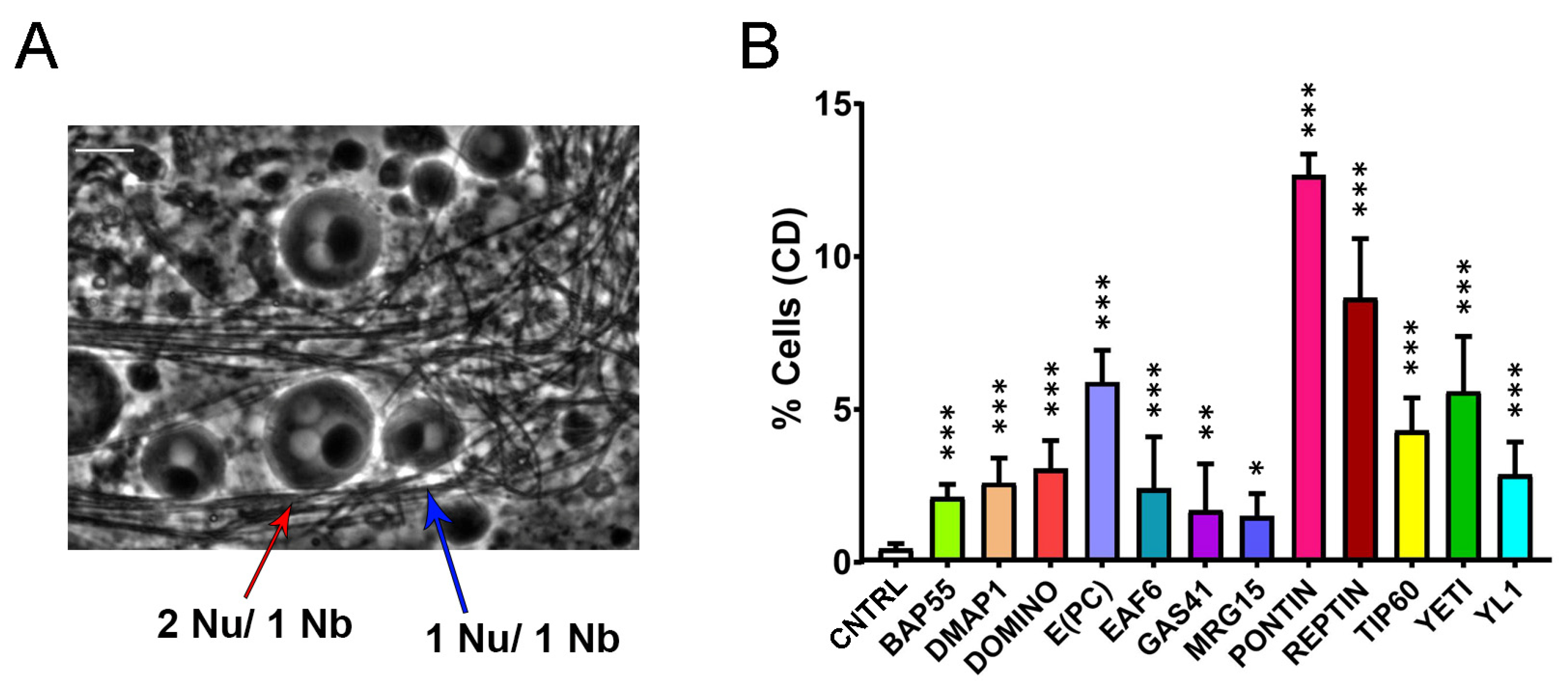
Figure 5.
Cytokinesis defects (CD) induced by RNAi-knockdown of DOM/TIP60 complex subunits in meiosis. (A) Normal onion-stage cells, in which the ratio between Nucleus (lighter grey circles) and Nebenkern (black circle) is equal to 1:1 with the same volume (blue arrow), while in the case of cytokinesis defects, the ratio become 2:1 or more and the nucleus volume become smaller than the Nebenkern (red arrow). Phase contrast microscopy. Scale bar, 10 µm. (B) Quantification analysis of CD after RNAi knockdown effects activated by the Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 1231), BAP55 (n = 772), DMAP1 (n = 1327), DOMINO (n = 1061), E(PC) (n = 595), EAF6 (n = 1118), GAS41 (n = 1242), MRG15 (n = 993), PONTIN (n = 922), REPTIN (n = 968), TIP60 (n = 1013), YETI (n = 940) and YL1 (n = 1228). The statistical analysis is performed using two-tailed Fisher’s exact test (* = p value ≤ 0.05, ** = p value ≤ 0.005, *** = p value ≤ 0.0005).
Figure 5.
Cytokinesis defects (CD) induced by RNAi-knockdown of DOM/TIP60 complex subunits in meiosis. (A) Normal onion-stage cells, in which the ratio between Nucleus (lighter grey circles) and Nebenkern (black circle) is equal to 1:1 with the same volume (blue arrow), while in the case of cytokinesis defects, the ratio become 2:1 or more and the nucleus volume become smaller than the Nebenkern (red arrow). Phase contrast microscopy. Scale bar, 10 µm. (B) Quantification analysis of CD after RNAi knockdown effects activated by the Bam>Gal4 driver. n = number of analyzed cells: CNTRL (n = 1231), BAP55 (n = 772), DMAP1 (n = 1327), DOMINO (n = 1061), E(PC) (n = 595), EAF6 (n = 1118), GAS41 (n = 1242), MRG15 (n = 993), PONTIN (n = 922), REPTIN (n = 968), TIP60 (n = 1013), YETI (n = 940) and YL1 (n = 1228). The statistical analysis is performed using two-tailed Fisher’s exact test (* = p value ≤ 0.05, ** = p value ≤ 0.005, *** = p value ≤ 0.0005).
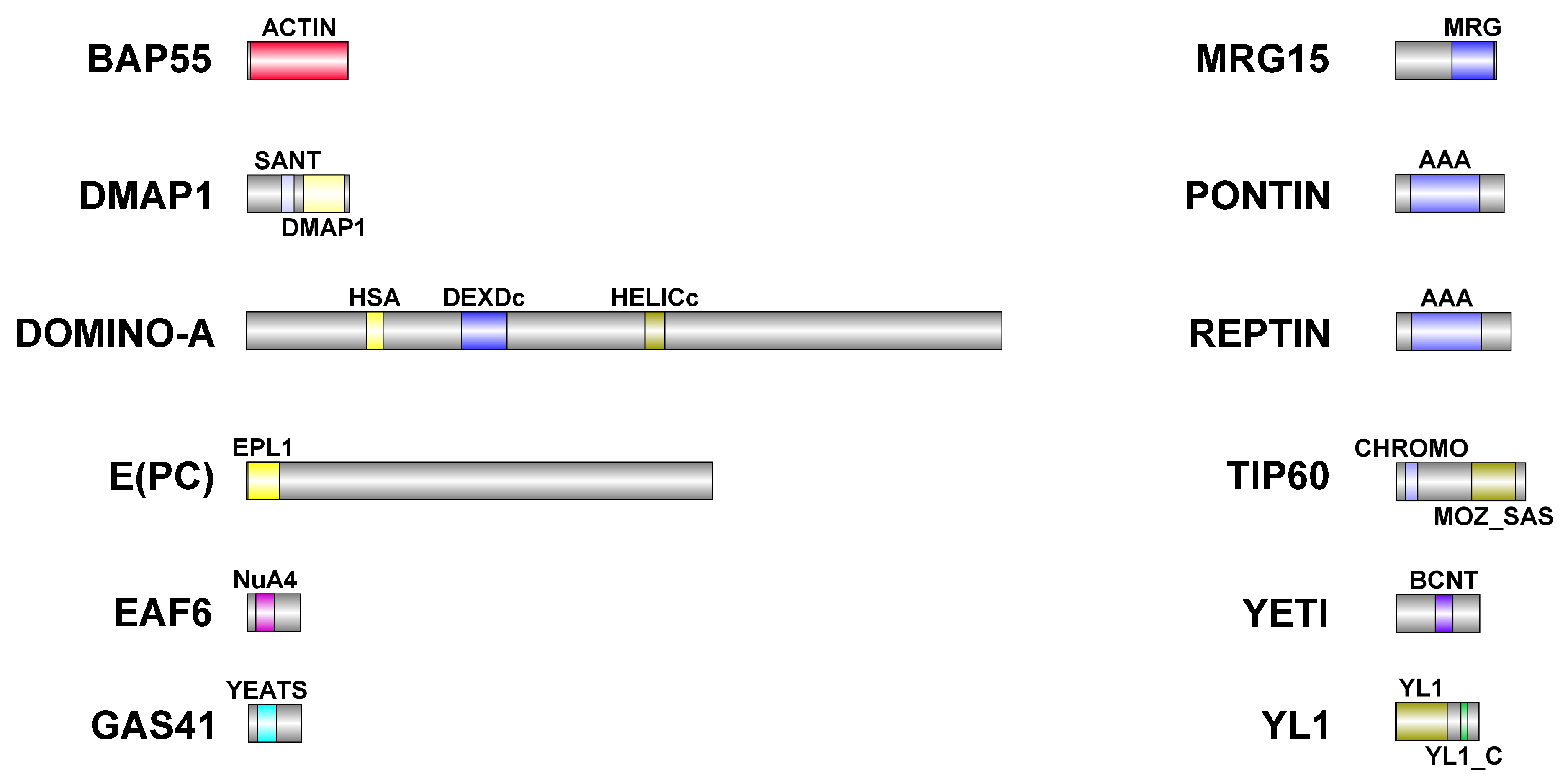
Figure 6.
Schematic representation of DOM/TIP60 chromatin remodeling subunits. Functional domains are color-coded and details about their conservation in human orthologues are described in
Table 6. Dimensions of proteins and domains are in scale.
Figure 6.
Schematic representation of DOM/TIP60 chromatin remodeling subunits. Functional domains are color-coded and details about their conservation in human orthologues are described in
Table 6. Dimensions of proteins and domains are in scale.
Table 1.
Quantification of chromatin integrity defects (CID).
Table 1.
Quantification of chromatin integrity defects (CID).
| CNTRL | BAP55 | DMAP1 | DOMINO | E(PC) | EAF6 | GAS41 |
|---|
| 0.90 ± 1.56 | 76.20 ± 7.64 | 55.14 ± 9.23 | 39.69 ± 4.00 | 70.47 ± 13.93 | 75.96 ± 7.20 | 71.59 ± 7.21 |
| MRG15 | PONTIN | REPTIN | TIP60 | YETI | YL1 |
| 68.49 ± 20.73 | 82.80 ± 9.58 | 74.88 ± 8.63 | 64.63 ± 4.50 | 33.37 ± 4.23 | 66.93 ± 9.64 |
Table 2.
H2A.V mislocalization defects (HM).
Table 2.
H2A.V mislocalization defects (HM).
| CNTRL | BAP55 | DMAP1 | DOMINO | E(PC) | EAF6 | GAS41 |
|---|
| 0 ± 0 | 0 ± 0 | 7.69 ± 4.44 | 0 ± 0 | 0 ± 0 | 71.41 ± 5.27 | 0 ± 0 |
| MRG15 | PONTIN | REPTIN | TIP60 | YETI | YL1 |
| 0 ± 0 | 91.79 ± 10.47 | 38.66 ± 2.01 | 2.56 ± 4.44 | 100 ± 0 | 0.90 ± 1.56 |
Table 3.
Aberrant Spindle Morphology (ASM).
Table 3.
Aberrant Spindle Morphology (ASM).
| CNTRL | BAP55 | DMAP1 | DOMINO | E(PC) | EAF6 | GAS41 |
|---|
| 1.75 ± 3.04 | 4.74 ± 2.26 | 53.42 ± 4.74 | 52.37 ± 8.98 | 56.67 ± 5.77 | 34.44 ± 5.09 | 59.60 ± 13.94 |
| MRG15 | PONTIN | REPTIN | TIP60 | YETI | YL1 |
| 61.69 ± 8.63 | 68.14 ± 7.09 | 75.32 ± 15.10 | 73.04 ± 7.65 | 38.53 ± 5.18 | 56.98 ± 2.36 |
Table 4.
Multipolar Spindle (MS).
Table 4.
Multipolar Spindle (MS).
| CNTRL | BAP55 | DMAP1 | DOMINO | E(PC) | EAF6 | GAS41 |
|---|
| 0.43 ± 0.74 | 0 ± 0 | 0 ± 0 | 12.93 ± 2.25 | 0 ± 0 | 0 ± 0 | 4.84 ± 1.25 |
| MRG15 | PONTIN | REPTIN | TIP60 | YETI | YL1 |
| 24.72 ± 5.14 | 12.81 ± 3.60 | 27.18 ± 9.54 | 27.37 ± 6.88 | 0 ± 0 | 17.93 ± 2.81 |
Table 5.
Cytokinesis defects (CD).
Table 5.
Cytokinesis defects (CD).
| Control | BAP55 | DMAP1 | DOMINO | E(PC) | EAF6 | GAS41 |
|---|
| 0.44 ± 0.16 | 2.13 ± 0.41 | 2.60 ± 0.81 | 3.05 ± 0.93 | 5.89 ± 1.04 | 2.41 ± 0.63 | 1.70 ± 1.51 |
| MRG15 | PONTIN | REPTIN | TIP60 | YETI | YL1 |
| 1.52 ± 0.72 | 12.68 ± 0.68 | 8.64 ± 1.94 | 4.31 ± 1.07 | 5.59 ± 1.80 | 2.87 ± 1.06 |
Table 6.
Fertility Test (FT).
Table 6.
Fertility Test (FT).
| CNTRL | BAP55 | DMAP1 | DOMINO | E(PC) | EAF6 | GAS41 |
|---|
| 100 ± 10.60 | 77.08 ± 9.59 | 65.64 ± 9.76 * | 71.26 ± 15.13 | 75.64 ± 7.18 | 94.78 ± 12.86 | 63.19 ± 9.19 ** |
| MRG15 | PONTIN | REPTIN | TIP60 | YETI | YL1 |
| 79.17 ± 14.31 | 0 ± 0 *** | 0 ± 0 *** | 55.42 ± 15.42 ** | 54.44 ± 2.78 ** | 56.56 ± 6.23 ** |
Table 7.
Conserved domains in analyzed DOM/TIP60 remodeling complex subunits.
Table 7.
Conserved domains in analyzed DOM/TIP60 remodeling complex subunits.
| D. melanogaster | H. sapiens | Domain | Identity (%) | Similarity (%) |
|---|
| BAP55 (425aa) | ACTL6A (429aa) | ACTIN | 54 | 71.2 |
| DMAP1 (433aa) | DMAP1 (467aa) | SANT | 55.6 | 79.6 |
| DMAP1 | 47.4 | 68.6 |
| DOMINO-A (3198aa) | SRCAP (3230aa) | HSA | 47.2 | 72.2 |
| DEXDc | 86.1 | 93.3 |
| HELICc | 91.7 | 95.2 |
| E(PC) (1974aa) | EPC1 (834aa) | EPL1 | 1.4 | 2.8 |
| EPC2 (807aa) | EPL1 | 5.7 | 17.8 |
| EAF6 (225aa) | MEAF6 (191aa) | NuA4 | 77.5 | 90 |
| GAS41 (227aa) | YEATS (227aa) | YEATS | 77.8 | 86.4 |
| MRG15 (424aa) | MORF4L1 (362aa) | MRG | 51.4 | 71.8 |
| Tudor-knot | 46.3 | 59.3 |
| PONTIN (456aa) | RUVBL1 (456aa) | AAA | 85.5 | 92.1 |
| REPTIN (481aa) | RUVBL2 (467aa) | AAA | 82.3 | 93.2 |
| TIP60 (543aa) | KAT5 (513aa) | CHROMO | 59.6 | 82.7 |
| MOZ_SAS | 81.1 | 86.5 |
| YETI (241aa) | CFDP1 (299aa) | BCNT | 49.3 | 76 |
| YL1 (351aa) | VPS72 (364aa) | YL1 | 46.6 | 60.2 |
| YL1_C | 50 | 70 |