The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth
Abstract
1. Introduction
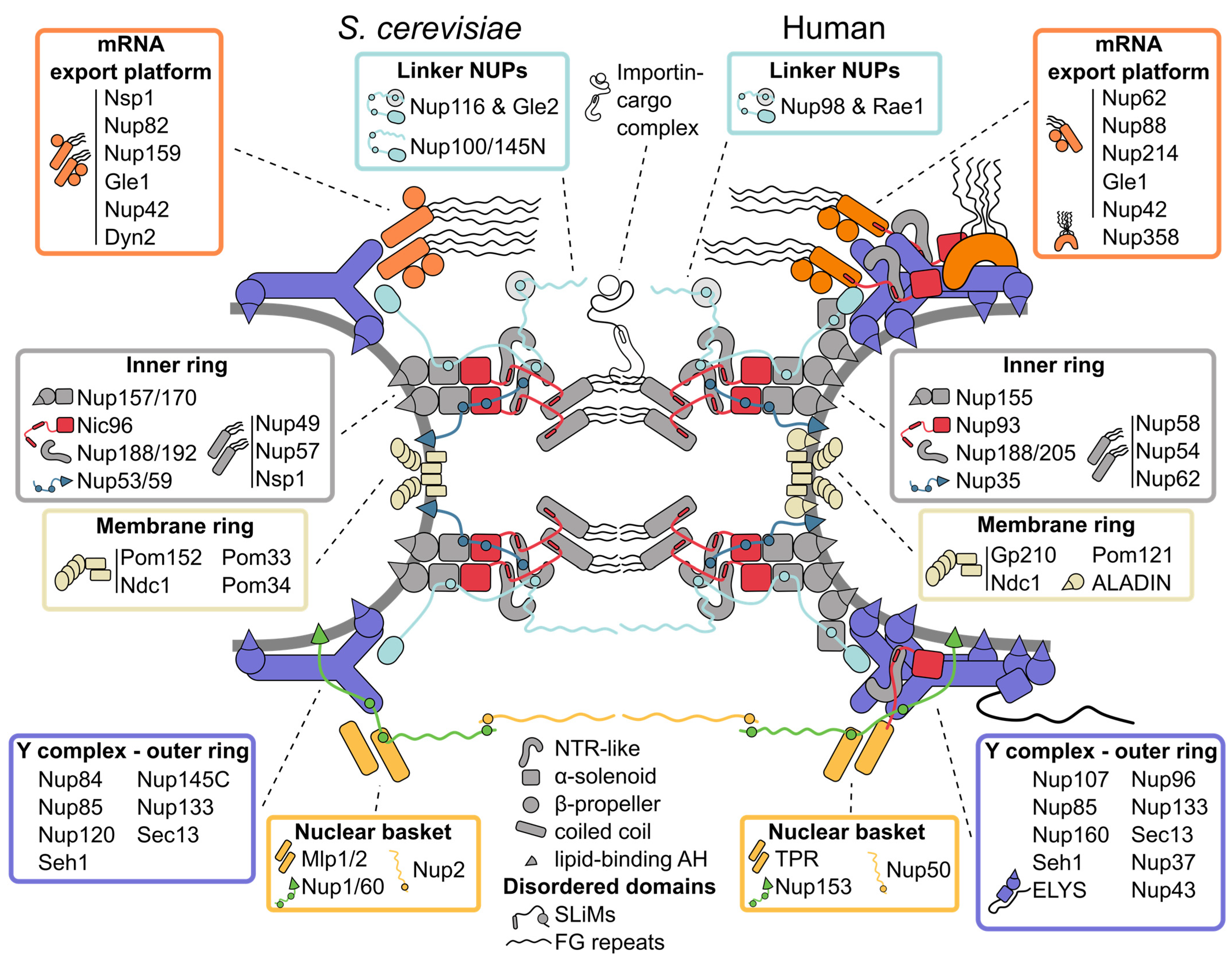
2. Tour of the Nuclear Pore Complex Architecture
2.1. Inner Ring: The Flexible Core of the Nuclear Pore Complex
2.2. Symmetric Outer Rings: The Versatile Outer Coat of the Nuclear Pore Complex
2.3. Asymmetric Appendages: Functional Extensions of the Nuclear Pore Complex
2.4. The Membrane Ring: An Enigmatic Girdle
2.5. Linker Nucleoporins: An Invisible Thread Stitching the Nuclear Pore Complex Together
3. Nuclear Pore Complex Assembly: Many Roads to the Same Destination
3.1. From Nascent Polypeptides to Nucleoporin Subcomplexes
3.2. Targeting of Nuclear Pore Complex Assembly to the Nuclear Envelope
3.3. Creating Functional Nucleocytoplasmic Conduits
3.4. Maturity: Compositional and Functional Variation of the Nuclear Pore Complex
4. Nuclear Pore Complex Remodeling and Functional Maintenance
4.1. Rejuvenation
4.2. Repair
4.3. Degradation
4.4. Disturbed Homeostasis and Disease
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAA+ | ATPases associated with diverse cellular activities |
| AH | amphipathic helix |
| ALPS | amphipathic lipid packing sensor |
| CNT | channel nucleoporin heterotrimer |
| COP | coat protein |
| ESCRT | endosomal sorting complex required for transport |
| FG | phenylalanine-glycine |
| NE | nuclear envelope |
| NPC | nuclear pore complex |
| NTM | nucleocytoplasmic transport machinery |
| NTR | nuclear transport receptor |
| NUP | nucleoporin |
| O-GlcNAc | O-linked β-N-acetylglucosamine |
| RCC1 | Regulator of chromosome condensation |
| SLiM | short linear interaction motif |
| SNARE | soluble NSF attachment protein receptor |
References
- Allegretti, M.; Zimmerli, C.E.; Rantos, V.; Wilfling, F.; Ronchi, P.; Fung, H.K.H.; Lee, C.W.; Hagen, W.; Turonova, B.; Karius, K.; et al. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 2020, 586, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Akey, C.W.; Singh, D.; Ouch, C.; Echeverria, I.; Nudelman, I.; Varberg, J.M.; Yu, Z.; Fang, F.; Shi, Y.; Wang, J.; et al. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 2022, 185, 361–378.e25. [Google Scholar] [CrossRef]
- Bui, K.H.; von Appen, A.; DiGuilio, A.L.; Ori, A.; Sparks, L.; Mackmull, M.T.; Bock, T.; Hagen, W.; Andres-Pons, A.; Glavy, J.S.; et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 2013, 155, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Mosalaganti, S.; Kosinski, J.; Albert, S.; Schaffer, M.; Strenkert, D.; Salome, P.A.; Merchant, S.S.; Plitzko, J.M.; Baumeister, W.; Engel, B.D.; et al. In situ architecture of the algal nuclear pore complex. Nat. Commun. 2018, 9, 2361. [Google Scholar] [CrossRef] [PubMed]
- Mosalaganti, S.; Obarska-Kosinska, A.; Siggel, M.; Turonova, B.; Zimmerli, C.E.; Buczak, K.; Schmidt, F.H.; Margiotta, E.; Mackmull, M.-T.; Hagen, W.; et al. Artificial intelligence reveals nuclear pore complexity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Schuller, A.P.; Wojtynek, M.; Mankus, D.; Tatli, M.; Kronenberg-Tenga, R.; Regmi, S.G.; Dip, P.V.; Lytton-Jean, A.K.R.; Brignole, E.J.; Dasso, M.; et al. The cellular environment shapes the nuclear pore complex architecture. Nature 2021, 598, 667–671. [Google Scholar] [CrossRef]
- Zimmerli, C.E.; Allegretti, M.; Rantos, V.; Goetz, S.K.; Obarska-Kosinska, A.; Zagoriy, I.; Halavatyi, A.; Hummer, G.; Mahamid, J.; Kosinski, J.; et al. Nuclear pores dilate and constrict in cellulo. Science 2021, 374, eabd9776. [Google Scholar] [CrossRef]
- Jarnik, M.; Aebi, U. Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 1991, 107, 291–308. [Google Scholar] [CrossRef]
- Goldberg, M.W.; Allen, T.D. The nuclear pore complex: Three-dimensional surface structure revealed by field emission, in-lens scanning electron microscopy, with underlying structure uncovered by proteolysis. J. Cell Sci. 1993, 106 Pt 1, 261–274. [Google Scholar] [CrossRef]
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482. [Google Scholar] [CrossRef]
- Terry, L.J.; Wente, S.R. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 2009, 8, 1814–1827. [Google Scholar] [CrossRef] [PubMed]
- Weis, K. Nucleocytoplasmic transport: Cargo trafficking across the border. Curr. Opin. Cell Biol. 2002, 14, 328–335. [Google Scholar] [CrossRef]
- DeGrasse, J.A.; DuBois, K.N.; Devos, D.; Siegel, T.N.; Sali, A.; Field, M.C.; Rout, M.P.; Chait, B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell Proteom. 2009, 8, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Stuwe, T.; Bley, C.J.; Thierbach, K.; Petrovic, S.; Schilbach, S.; Mayo, D.J.; Perriches, T.; Rundlet, E.J.; Jeon, Y.E.; Collins, L.N.; et al. Architecture of the fungal nuclear pore inner ring complex. Science 2015, 350, 56–64. [Google Scholar] [CrossRef]
- Whittle, J.R.R.; Schwartz, T.U. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J. Biol. Chem. 2009, 284, 28442–28452. [Google Scholar] [CrossRef]
- Lin, D.H.; Stuwe, T.; Schilbach, S.; Rundlet, E.J.; Perriches, T.; Mobbs, G.; Fan, Y.; Thierbach, K.; Huber, F.M.; Collins, L.N.; et al. Architecture of the symmetric core of the nuclear pore. Science 2016, 352, aaf1015. [Google Scholar] [CrossRef]
- Andersen, K.R.; Onischenko, E.; Tang, J.H.; Kumar, P.; Chen, J.Z.; Ulrich, A.; Liphardt, J.T.; Weis, K.; Schwartz, T.U. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2013, 2, e00745. [Google Scholar] [CrossRef]
- Flemming, D.; Devos, D.P.; Schwarz, J.; Amlacher, S.; Lutzmann, M.; Hurt, E. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J. Struct. Biol. 2012, 177, 99–105. [Google Scholar] [CrossRef]
- Stuwe, T.; Lin, D.H.; Collins, L.N.; Hurt, E.; Hoelz, A. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. USA 2014, 111, 2530–2535. [Google Scholar] [CrossRef]
- Li, Z.; Chen, S.; Zhao, L.; Huang, G.; Pi, X.; Sun, S.; Wang, P.; Sui, S.F. Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex. Cell Res. 2022, 1–14. [Google Scholar] [CrossRef]
- Petrovic, S.; Samanta, D.; Perriches, T.; Bley, C.J.; Thierbach, K.; Brown, B.; Nie, S.; Mobbs, G.W.; Stevens, T.A.; Liu, X.; et al. Architecture of the linker-scaffold in the nuclear pore. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bley, C.J.; Nie, S.; Mobbs, G.W.; Petrovic, S.; Gres, A.T.; Liu, X.; Mukherjee, S.; Harvey, S.; Huber, F.M.; Lin, D.H.; et al. Architecture of the cytoplasmic face of the nuclear pore. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fischer, J.; Teimer, R.; Amlacher, S.; Kunze, R.; Hurt, E. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol. 2015, 22, 774–781. [Google Scholar] [CrossRef]
- Onischenko, E.; Stanton, L.H.; Madrid, A.S.; Kieselbach, T.; Weis, K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 2009, 185, 475–491. [Google Scholar] [CrossRef]
- Zila, V.; Margiotta, E.; Turonova, B.; Muller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Borner, K.; Rada, J.; Muller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e18. [Google Scholar] [CrossRef] [PubMed]
- Liashkovich, I.; Meyring, A.; Kramer, A.; Shahin, V. Exceptional structural and mechanical flexibility of the nuclear pore complex. J. Cell Physiol. 2011, 226, 675–682. [Google Scholar] [CrossRef]
- Tai, L.; Zhu, Y.; Ren, H.; Huang, X.; Zhang, C.; Sun, F. 8 Å structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI. Protein Cell 2022, 1–18. [Google Scholar] [CrossRef]
- Sampathkumar, P.; Kim, S.J.; Upla, P.; Rice, W.J.; Phillips, J.; Timney, B.L.; Pieper, U.; Bonanno, J.B.; Fernandez-Martinez, J.; Hakhverdyan, Z.; et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 2013, 21, 560–571. [Google Scholar] [CrossRef]
- Maimon, T.; Elad, N.; Dahan, I.; Medalia, O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012, 20, 998–1006. [Google Scholar] [CrossRef]
- Kelley, K.; Knockenhauer, K.E.; Kabachinski, G.; Schwartz, T.U. Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol. 2015, 22, 425–431. [Google Scholar] [CrossRef]
- Thierbach, K.; von Appen, A.; Thoms, M.; Beck, M.; Flemming, D.; Hurt, E. Protein interfaces of the conserved Nup84 complex from Chaetomium thermophilum shown by crosslinking mass spectrometry and electron microscopy. Structure 2013, 21, 1672–1682. [Google Scholar] [CrossRef]
- Liu, H.L.; De Souza, C.P.; Osmani, A.H.; Osmani, S.A. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol. Biol. Cell 2009, 20, 616–630. [Google Scholar] [CrossRef]
- Kampmann, M.; Blobel, G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 2009, 16, 782–788. [Google Scholar] [CrossRef]
- Lutzmann, M.; Kunze, R.; Buerer, A.; Aebi, U.; Hurt, E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002, 21, 387–397. [Google Scholar] [CrossRef]
- Nordeen, S.A.; Turman, D.L.; Schwartz, T.U. Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif. Nat. Commun. 2020, 11, 6060. [Google Scholar] [CrossRef]
- Siniossoglou, S.; Lutzmann, M.; Santos-Rosa, H.; Leonard, K.; Mueller, S.; Aebi, U.; Hurt, E. Structure and assembly of the Nup84p complex. J. Cell Biol. 2000, 149, 41–54. [Google Scholar] [CrossRef]
- von Appen, A.; Kosinski, J.; Sparks, L.; Ori, A.; DiGuilio, A.L.; Vollmer, B.; Mackmull, M.T.; Banterle, N.; Parca, L.; Kastritis, P.; et al. In situ structural analysis of the human nuclear pore complex. Nature 2015, 526, 140–143. [Google Scholar] [CrossRef]
- Eibauer, M.; Pellanda, M.; Turgay, Y.; Dubrovsky, A.; Wild, A.; Medalia, O. Structure and gating of the nuclear pore complex. Nat. Commun. 2015, 6, 7532. [Google Scholar] [CrossRef]
- Huang, G.; Zhang, Y.; Zhu, X.; Zeng, C.; Wang, Q.; Zhou, Q.; Tao, Q.; Liu, M.; Lei, J.; Yan, C.; et al. Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis. Cell Res. 2020, 30, 520–531. [Google Scholar] [CrossRef]
- Berke, I.C.; Boehmer, T.; Blobel, G.; Schwartz, T.U. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol. 2004, 167, 591–597. [Google Scholar] [CrossRef]
- Drin, G.; Casella, J.F.; Gautier, R.; Boehmer, T.; Schwartz, T.U.; Antonny, B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 2007, 14, 138–146. [Google Scholar] [CrossRef]
- Leksa, N.C.; Brohawn, S.G.; Schwartz, T.U. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 2009, 17, 1082–1091. [Google Scholar] [CrossRef][Green Version]
- Brohawn, S.G.; Leksa, N.C.; Spear, E.D.; Rajashankar, K.R.; Schwartz, T.U. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 2008, 322, 1369–1373. [Google Scholar] [CrossRef]
- Debler, E.W.; Ma, Y.; Seo, H.S.; Hsia, K.C.; Noriega, T.R.; Blobel, G.; Hoelz, A. A fence-like coat for the nuclear pore membrane. Mol. Cell 2008, 32, 815–826. [Google Scholar] [CrossRef]
- Hsia, K.C.; Stavropoulos, P.; Blobel, G.; Hoelz, A. Architecture of a coat for the nuclear pore membrane. Cell 2007, 131, 1313–1326. [Google Scholar] [CrossRef]
- Field, M.C.; Rout, M.P. Pore timing: The evolutionary origins of the nucleus and nuclear pore complex. F1000Res 2019, 8. [Google Scholar] [CrossRef]
- Hinshaw, J.E.; Milligan, R.A. Nuclear pore complexes exceeding eightfold rotational symmetry. J. Struct. Biol. 2003, 141, 259–268. [Google Scholar] [CrossRef]
- Loschberger, A.; Franke, C.; Krohne, G.; van de Linde, S.; Sauer, M. Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution. J. Cell Sci. 2014, 127, 4351–4355. [Google Scholar] [CrossRef]
- Onischenko, E.; Tang, J.H.; Andersen, K.R.; Knockenhauer, K.E.; Vallotton, P.; Derrer, C.P.; Kralt, A.; Mugler, C.F.; Chan, L.Y.; Schwartz, T.U.; et al. Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell 2017, 171, 904–917.e19. [Google Scholar] [CrossRef]
- Franz, C.; Walczak, R.; Yavuz, S.; Santarella, R.; Gentzel, M.; Askjaer, P.; Galy, V.; Hetzer, M.; Mattaj, I.W.; Antonin, W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007, 8, 165–172. [Google Scholar] [CrossRef]
- Rasala, B.A.; Orjalo, A.V.; Shen, Z.; Briggs, S.; Forbes, D.J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA 2006, 103, 17801–17806. [Google Scholar] [CrossRef]
- Rasala, B.A.; Ramos, C.; Harel, A.; Forbes, D.J. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell 2008, 19, 3982–3996. [Google Scholar] [CrossRef]
- Watson, M.L. Further observations on the nuclear envelope of the animal cell. J. Biophys Biochem. Cytol. 1959, 6, 147–156. [Google Scholar] [CrossRef]
- Fernandez-Martinez, J.; Kim, S.J.; Shi, Y.; Upla, P.; Pellarin, R.; Gagnon, M.; Chemmama, I.E.; Wang, J.; Nudelman, I.; Zhang, W.; et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 2016, 167, 1215–1228.e25. [Google Scholar] [CrossRef]
- Walther, T.C.; Pickersgill, H.S.; Cordes, V.C.; Goldberg, M.W.; Allen, T.D.; Mattaj, I.W.; Fornerod, M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 2002, 158, 63–77. [Google Scholar] [CrossRef]
- Vallotton, P.; Rajoo, S.; Wojtynek, M.; Onischenko, E.; Kralt, A.; Derrer, C.P.; Weis, K. Mapping the native organization of the yeast nuclear pore complex using nuclear radial intensity measurements. Proc. Natl. Acad. Sci. USA 2019, 116, 14606–14613. [Google Scholar] [CrossRef]
- Lin, D.H.; Hoelz, A. The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem. 2019, 88, 725–783. [Google Scholar] [CrossRef]
- Obado, S.O.; Brillantes, M.; Uryu, K.; Zhang, W.; Ketaren, N.E.; Chait, B.T.; Field, M.C.; Rout, M.P. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol. 2016, 14, e1002365. [Google Scholar] [CrossRef]
- Goldberg, M.W.; Allen, T.D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 1992, 119, 1429–1440. [Google Scholar] [CrossRef]
- Kiseleva, E.; Allen, T.D.; Rutherford, S.; Bucci, M.; Wente, S.R.; Goldberg, M.W. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 2004, 145, 272–288. [Google Scholar] [CrossRef]
- Krull, S.; Thyberg, J.; Bjorkroth, B.; Rackwitz, H.R.; Cordes, V.C. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 2004, 15, 4261–4277. [Google Scholar] [CrossRef]
- Niepel, M.; Molloy, K.R.; Williams, R.; Farr, J.C.; Meinema, A.C.; Vecchietti, N.; Cristea, I.M.; Chait, B.T.; Rout, M.P.; Strambio-De-Castillia, C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 2013, 24, 3920–3938. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Aksenova, V.; Tingey, M.; Yu, J.; Ma, P.; Arnaoutov, A.; Chen, S.; Dasso, M.; Yang, W. Distinct roles of nuclear basket proteins in directing the passage of mRNA through the nuclear pore. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Goryaynov, A.; Lindquist, A.; Rexach, M.; Yang, W. Quantifying nucleoporin stoichiometry inside single nuclear pore complexes in vivo. Sci. Rep. 2015, 5, 9372. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Banterle, N.; Iskar, M.; Andres-Pons, A.; Escher, C.; Khanh Bui, H.; Sparks, L.; Solis-Mezarino, V.; Rinner, O.; Bork, P.; et al. Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013, 9, 648. [Google Scholar] [CrossRef]
- Rajoo, S.; Vallotton, P.; Onischenko, E.; Weis, K. Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc. Natl. Acad. Sci. USA 2018, 115, E3969–E3977. [Google Scholar] [CrossRef]
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.M.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731. [Google Scholar] [CrossRef]
- Cibulka, J.; Bisaccia, F.; Radisavljevic, K.; Gudino Carrillo, R.M.; Kohler, A. Assembly principle of a membrane-anchored nuclear pore basket scaffold. Sci. Adv. 2022, 8, eabl6863. [Google Scholar] [CrossRef]
- Mészáros, N.; Cibulka, J.; Mendiburo, M.J.; Romanauska, A.; Schneider, M.; Köhler, A. Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev. Cell 2015, 33, 285–298. [Google Scholar] [CrossRef]
- Vollmer, B.; Lorenz, M.; Moreno-Andres, D.; Bodenhofer, M.; De Magistris, P.; Astrinidis, S.A.; Schooley, A.; Flotenmeyer, M.; Leptihn, S.; Antonin, W. Nup153 Recruits the Nup107-160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly. Dev. Cell 2015, 33, 717–728. [Google Scholar] [CrossRef]
- Hase, M.E.; Cordes, V.C. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 2003, 14, 1923–1940. [Google Scholar] [CrossRef] [PubMed]
- Makise, M.; Mackay, D.R.; Elgort, S.; Shankaran, S.S.; Adam, S.A.; Ullman, K.S. The Nup153-Nup50 protein interface and its role in nuclear import. J. Biol. Chem. 2012, 287, 38515–38522. [Google Scholar] [CrossRef] [PubMed]
- Bensidoun, P.; Zenklusen, D.; Oeffinger, M. Choosing the right exit: How functional plasticity of the nuclear pore drives selective and efficient mRNA export. Wiley Interdiscip. Rev. RNA 2021, 12, e1660. [Google Scholar] [CrossRef] [PubMed]
- Neumann, N.; Lundin, D.; Poole, A.M. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 2010, 5, e13241. [Google Scholar] [CrossRef]
- Field, M.C.; Koreny, L.; Rout, M.P. Enriching the pore: Splendid complexity from humble origins. Traffic 2014, 15, 141–156. [Google Scholar] [CrossRef]
- Chadrin, A.; Hess, B.; San Roman, M.; Gatti, X.; Lombard, B.; Loew, D.; Barral, Y.; Palancade, B.; Doye, V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 2010, 189, 795–811. [Google Scholar] [CrossRef]
- Stavru, F.; Hulsmann, B.B.; Spang, A.; Hartmann, E.; Cordes, V.C.; Gorlich, D. NDC1: A crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 2006, 173, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Tcheperegine, S.E.; Marelli, M.; Wozniak, R.W. Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 1999, 274, 5252–5258. [Google Scholar] [CrossRef]
- Upla, P.; Kim, S.J.; Sampathkumar, P.; Dutta, K.; Cahill, S.M.; Chemmama, I.E.; Williams, R.; Bonanno, J.B.; Rice, W.J.; Stokes, D.L.; et al. Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure 2017, 25, 434–445. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, B.; Yuan, K.; Shi, H.; Blobel, G. Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string. Cell Discov. 2018, 4, 56. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Zeng, C.; Huang, G.; Zhu, X.; Wang, Q.; Wang, K.; Zhou, Q.; Yan, C.; Zhang, W.; et al. Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res. 2020, 30, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Stavru, F.; Nautrup-Pedersen, G.; Cordes, V.C.; Gorlich, D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J. Cell Biol. 2006, 173, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, R.W.; Blobel, G.; Rout, M.P. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 1994, 125, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.; Scheele, S.; Ekblom, P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 2004, 292, 359–370. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, M.A.; Raices, M.; Panowski, S.H.; Hetzer, M.W. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009, 136, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Rabut, G.; Doye, V.; Ellenberg, J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004, 6, 1114–1121. [Google Scholar] [CrossRef]
- Savas, J.N.; Toyama, B.H.; Xu, T.; Yates, J.R., 3rd; Hetzer, M.W. Extremely long-lived nuclear pore proteins in the rat brain. Science 2012, 335, 942. [Google Scholar] [CrossRef]
- Toyama, B.H.; Arrojo, E.D.R.; Lev-Ram, V.; Ramachandra, R.; Deerinck, T.J.; Lechene, C.; Ellisman, M.H.; Hetzer, M.W. Visualization of long-lived proteins reveals age mosaicism within nuclei of postmitotic cells. J. Cell Biol. 2019, 218, 433–444. [Google Scholar] [CrossRef]
- Toyama, B.H.; Savas, J.N.; Park, S.K.; Harris, M.S.; Ingolia, N.T.; Yates, J.R., 3rd; Hetzer, M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013, 154, 971–982. [Google Scholar] [CrossRef]
- Knockenhauer, K.E.; Schwartz, T.U. The nuclear pore complex as a flexible and dynamic gate. Cell 2016, 164, 1162–1171. [Google Scholar] [CrossRef]
- Dultz, E.; Zanin, E.; Wurzenberger, C.; Braun, M.; Rabut, G.; Sironi, L.; Ellenberg, J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008, 180, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Amlacher, S.; Sarges, P.; Flemming, D.; van Noort, V.; Kunze, R.; Devos, D.P.; Arumugam, M.; Bork, P.; Hurt, E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 2011, 146, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Teimer, R.; Kosinski, J.; von Appen, A.; Beck, M.; Hurt, E. A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat. Commun. 2017, 8, 1107. [Google Scholar] [CrossRef] [PubMed]
- Hamed, M.; Antonin, W. Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly. Cells 2021, 10, 3601. [Google Scholar] [CrossRef] [PubMed]
- Maul, G.G. Nuclear pore complexes. Elimination and reconstruction during mitosis. J. Cell Biol. 1977, 74, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.Y.; Upadhyayula, S.; Houser, J.; He, K.; Skillern, W.; Scanavachi, G.; Dang, S.; Sanyal, A.; Ohashi, K.G.; Di Caprio, G.; et al. Inherited nuclear pore substructures template post-mitotic pore assembly. Dev. Cell 2021, 56, 1786–1803.e9. [Google Scholar] [CrossRef]
- Maeshima, K.; Iino, H.; Hihara, S.; Funakoshi, T.; Watanabe, A.; Nishimura, M.; Nakatomi, R.; Yahata, K.; Imamoto, F.; Hashikawa, T. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat. Struct. Mol. Biol. 2010, 17, 1065–1071. [Google Scholar] [CrossRef]
- Zeligs, J.D.; Wollman, S.H. Mitosis in rat thyroid epithelial cells in vivo. I. Ultrastructural changes in cytoplasmic organelles during the mitotic cycle. J. Ultrastruct Res. 1979, 66, 53–77. [Google Scholar] [CrossRef]
- Maul, G.G.; Price, J.W.; Lieberman, M.W. Formation and distribution of nuclear pore complexes in interphase. J. Cell Biol. 1971, 51, 405–418. [Google Scholar] [CrossRef]
- D’Angelo, M.A.; Anderson, D.J.; Richard, E.; Hetzer, M.W. Nuclear pores form de novo from both sides of the nuclear envelope. Science 2006, 312, 440–443. [Google Scholar] [CrossRef]
- Dultz, E.; Ellenberg, J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 2010, 191, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Onischenko, E.; Noor, E.; Fischer, J.S.; Gillet, L.; Wojtynek, M.; Vallotton, P.; Weis, K. Maturation Kinetics of a Multiprotein Complex Revealed by Metabolic Labeling. Cell 2020, 183, 1785–1800.e26. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, S.; Steyer, A.M.; Schorb, M.; Heriche, J.K.; Hossain, M.J.; Sethi, S.; Kueblbeck, M.; Schwab, Y.; Beck, M.; Ellenberg, J. Postmitotic nuclear pore assembly proceeds by radial dilation of small membrane openings. Nat. Struct. Mol. Biol. 2018, 25, 21–28. [Google Scholar] [CrossRef]
- Otsuka, S.; Tempkin, J.O.B.; Politi, A.Z.; Rybina, A.; Hossain, M.J.; Kueblbeck, M.; Callegari, A.; Koch, B.; Sali, A.; Ellenberg, J. A quantitative map of nuclear pore assembly reveals two distinct mechanisms. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hampoelz, B.; Mackmull, M.T.; Machado, P.; Ronchi, P.; Bui, K.H.; Schieber, N.; Santarella-Mellwig, R.; Necakov, A.; Andres-Pons, A.; Philippe, J.M.; et al. Pre-assembled Nuclear Pores Insert into the Nuclear Envelope during Early Development. Cell 2016, 166, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Hampoelz, B.; Schwarz, A.; Ronchi, P.; Bragulat-Teixidor, H.; Tischer, C.; Gaspar, I.; Ephrussi, A.; Schwab, Y.; Beck, M. Nuclear Pores Assemble from Nucleoporin Condensates During Oogenesis. Cell 2019, 179, 671–686.e17. [Google Scholar] [CrossRef]
- Kutay, U.; Juhlen, R.; Antonin, W. Mitotic disassembly and reassembly of nuclear pore complexes. Trends Cell Biol. 2021, 31, 1019–1033. [Google Scholar] [CrossRef]
- Walther, T.C.; Alves, A.; Pickersgill, H.; Loiodice, I.; Hetzer, M.; Galy, V.; Hulsmann, B.B.; Kocher, T.; Wilm, M.; Allen, T.; et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 2003, 113, 195–206. [Google Scholar] [CrossRef]
- Otsuka, S.; Bui, K.H.; Schorb, M.; Hossain, M.J.; Politi, A.Z.; Koch, B.; Eltsov, M.; Beck, M.; Ellenberg, J. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife 2016, 5, e19071. [Google Scholar] [CrossRef]
- Thaller, D.J.; Patrick Lusk, C. Fantastic nuclear envelope herniations and where to find them. Biochem. Soc. Trans. 2018, 46, 877–889. [Google Scholar] [CrossRef]
- Doucet, C.M.; Talamas, J.A.; Hetzer, M.W. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 2010, 141, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Golchoubian, B.; Brunner, A.; Bragulat-Teixidor, H.; Neuner, A.; Akarlar, B.A.; Ozlu, N.; Schlaitz, A.L. Reticulon-like REEP4 at the inner nuclear membrane promotes nuclear pore complex formation. J. Cell Biol. 2022, 221, e202101049. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, T.; Clever, M.; Watanabe, A.; Imamoto, N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol. Biol. Cell 2011, 22, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, B.; Schooley, A.; Sachdev, R.; Eisenhardt, N.; Schneider, A.M.; Sieverding, C.; Madlung, J.; Gerken, U.; Macek, B.; Antonin, W. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 2012, 31, 4072–4084. [Google Scholar] [CrossRef]
- Gillespie, P.J.; Khoudoli, G.A.; Stewart, G.; Swedlow, J.R.; Blow, J.J. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 2007, 17, 1657–1662. [Google Scholar] [CrossRef]
- Hausser, J.; Mayo, A.; Keren, L.; Alon, U. Central dogma rates and the trade-off between precision and economy in gene expression. Nat. Commun. 2019, 10, 68. [Google Scholar] [CrossRef]
- Fontoura, B.M.; Blobel, G.; Matunis, M.J. A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 1999, 144, 1097–1112. [Google Scholar] [CrossRef]
- Ratner, G.A.; Hodel, A.E.; Powers, M.A. Molecular determinants of binding between Gly-Leu-Phe-Gly nucleoporins and the nuclear pore complex. J. Biol. Chem. 2007, 282, 33968–33976. [Google Scholar] [CrossRef]
- Lautier, O.; Penzo, A.; Rouviere, J.O.; Chevreux, G.; Collet, L.; Loiodice, I.; Taddei, A.; Devaux, F.; Collart, M.A.; Palancade, B. Co-translational assembly and localized translation of nucleoporins in nuclear pore complex biogenesis. Mol. Cell 2021, 81, 2417–2427.e5. [Google Scholar] [CrossRef]
- Seidel, M.; Becker, A.; Pereira, F.; Landry, J.J.M.; de Azevedo, N.T.D.; Fusco, C.M.; Kaindl, E.; Romanov, N.; Baumbach, J.; Langer, J.D.; et al. Co-translational assembly orchestrates competing biogenesis pathways. Nat. Commun. 2022, 13, 1224. [Google Scholar] [CrossRef]
- Kamenova, I.; Mukherjee, P.; Conic, S.; Mueller, F.; El-Saafin, F.; Bardot, P.; Garnier, J.M.; Dembele, D.; Capponi, S.; Timmers, H.T.M.; et al. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 2019, 10, 1740. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.O.; Somasekharan, S.P.; Villanyi, Z.; Zagatti, M.; Bezrukov, F.; Rashpa, R.; Cornut, J.; Iqbal, J.; Longis, M.; Carl, S.H.; et al. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol. 2019, 26, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Shiber, A.; Doring, K.; Friedrich, U.; Klann, K.; Merker, D.; Zedan, M.; Tippmann, F.; Kramer, G.; Bukau, B. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 2018, 561, 268–272. [Google Scholar] [CrossRef]
- Schwarz, A.; Beck, M. The Benefits of Cotranslational Assembly: A Structural Perspective. Trends Cell Biol. 2019, 29, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.; Dokudovskaya, S.; Alber, F.; Williams, R.; Chait, B.T.; Sali, A.; Rout, M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004, 2, e380. [Google Scholar] [CrossRef]
- Mans, B.J.; Anantharaman, V.; Aravind, L.; Koonin, E.V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 2004, 3, 1612–1637. [Google Scholar] [CrossRef] [PubMed]
- Rampello, A.J.; Laudermilch, E.; Vishnoi, N.; Prophet, S.M.; Shao, L.; Zhao, C.; Lusk, C.P.; Schlieker, C. Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J. Cell Biol. 2020, 219, e201910185. [Google Scholar] [CrossRef]
- Elsiena Kuiper, E.F.; Gallardo, P.; Bergsma, T.; Mari, M.; Musskopf, M.K.; Kuipers, J.; Giepmans, B.N.G.; Steen, A.; Veenhoff, L.M.; Kampinga, H.H.; et al. The molecular chaperone DNAJB6 provides surveillance of FG-Nups and is required for interphase nuclear pore complex biogenesis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Prophet, S.M.; Rampello, A.J.; Niescier, R.F.; Shaw, J.E.; Koleske, A.J.; Schlieker, C. MLF2 modulates phase separated nuclear envelope condensates that provoke dual proteotoxicity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kessel, R.G. Annulate lamellae: A last frontier in cellular organelles. Int. Rev. Cytol. 1992, 133, 43–120. [Google Scholar] [CrossRef]
- Iino, H.; Maeshima, K.; Nakatomi, R.; Kose, S.; Hashikawa, T.; Tachibana, T.; Imamoto, N. Live imaging system for visualizing nuclear pore complex (NPC) formation during interphase in mammalian cells. Genes Cells 2010, 15, 647–660. [Google Scholar] [CrossRef]
- Ryan, K.J.; McCaffery, J.M.; Wente, S.R. The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 2003, 160, 1041–1053. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.J.; Zhou, Y.; Wente, S.R. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell 2007, 18, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Bernis, C.; Swift-Taylor, B.; Nord, M.; Carmona, S.; Chook, Y.M.; Forbes, D.J. Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol. Biol. Cell 2014, 25, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Harel, A.; Chan, R.C.; Lachish-Zalait, A.; Zimmerman, E.; Elbaum, M.; Forbes, D.J. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell 2003, 14, 4387–4396. [Google Scholar] [CrossRef]
- Lau, C.K.; Delmar, V.A.; Chan, R.C.; Phung, Q.; Bernis, C.; Fichtman, B.; Rasala, B.A.; Forbes, D.J. Transportin regulates major mitotic assembly events: From spindle to nuclear pore assembly. Mol. Biol. Cell 2009, 20, 4043–4058. [Google Scholar] [CrossRef]
- Rotem, A.; Gruber, R.; Shorer, H.; Shaulov, L.; Klein, E.; Harel, A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol. Biol. Cell 2009, 20, 4031–4042. [Google Scholar] [CrossRef]
- Walther, T.C.; Askjaer, P.; Gentzel, M.; Habermann, A.; Griffiths, G.; Wilm, M.; Mattaj, I.W.; Hetzer, M. RanGTP mediates nuclear pore complex assembly. Nature 2003, 424, 689–694. [Google Scholar] [CrossRef]
- Zhang, C.; Clarke, P.R. Chromatin-independent nuclear envelope assembly induced by Ran GTPase in Xenopus egg extracts. Science 2000, 288, 1429–1432. [Google Scholar] [CrossRef]
- Dasso, M. The Ran GTPase: Theme and variations. Curr. Biol. 2002, 12, R502–R508. [Google Scholar] [CrossRef]
- Kalab, P.; Heald, R. The RanGTP gradient-A GPS for the mitotic spindle. J. Cell Sci. 2008, 121, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, T.; Enninga, J.; Dales, S.; Blobel, G.; Zhong, H.L. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2003, 100, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, A.G.; Piano, F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr. Biol. 2006, 16, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Saldivar, G.; Fernandez, A.; Hirano, Y.; Mauro, M.; Lai, A.; Ayuso, C.; Haraguchi, T.; Hiraoka, Y.; Piano, F.; Askjaer, P. Identification of Conserved MEL-28/ELYS Domains with Essential Roles in Nuclear Assembly and Chromosome Segregation. PLoS Genet. 2016, 12, e1006131. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Zhang, Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 2014, 21, 609–616. [Google Scholar] [CrossRef]
- Kobayashi, W.; Takizawa, Y.; Aihara, M.; Negishi, L.; Ishii, H.; Kurumizaka, H. Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding. Commun. Biol. 2019, 2, 163. [Google Scholar] [CrossRef]
- Zierhut, C.; Jenness, C.; Kimura, H.; Funabiki, H. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 2014, 21, 617–625. [Google Scholar] [CrossRef]
- Hetzer, M.; Bilbao-Cortes, D.; Walther, T.C.; Gruss, O.J.; Mattaj, I.W. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell 2000, 5, 1013–1024. [Google Scholar] [CrossRef]
- Holzer, G.; De Magistris, P.; Gramminger, C.; Sachdev, R.; Magalska, A.; Schooley, A.; Scheufen, A.; Lennartz, B.; Tatarek-Nossol, M.; Lue, H.; et al. The nucleoporin Nup50 activates the Ran guanine nucleotide exchange factor RCC1 to promote NPC assembly at the end of mitosis. EMBO J. 2021, 40, e108788. [Google Scholar] [CrossRef]
- Yavuz, S.; Santarella-Mellwig, R.; Koch, B.; Jaedicke, A.; Mattaj, I.W.; Antonin, W. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010, 584, 3292–3298. [Google Scholar] [CrossRef]
- Floch, A.G.; Tareste, D.; Fuchs, P.F.; Chadrin, A.; Naciri, I.; Léger, T.; Schlenstedt, G.; Palancade, B.; Doye, V. Nuclear pore targeting of the yeast Pom33 nucleoporin depends on karyopherin and lipid binding. J. Cell Sci. 2015, 128, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Schrader, N.; Stelter, P.; Flemming, D.; Kunze, R.; Hurt, E.; Vetter, I.R. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell 2008, 29, 46–55. [Google Scholar] [CrossRef]
- Boni, A.; Politi, A.Z.; Strnad, P.; Xiang, W.; Hossain, M.J.; Ellenberg, J. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J. Cell Biol. 2015, 209, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Ungricht, R.; Klann, M.; Horvath, P.; Kutay, U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J. Cell Biol. 2015, 209, 687–703. [Google Scholar] [CrossRef]
- Lusk, C.P.; Makhnevych, T.; Marelli, M.; Aitchison, J.D.; Wozniak, R.W. Karyopherins in nuclear pore biogenesis: A role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 2002, 159, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.; Mykytka, B.; Allen, N.P.; Huang, L.; Al, B.; Rexach, M. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 2001, 154, 937–950. [Google Scholar] [CrossRef] [PubMed]
- De Magistris, P.; Tatarek-Nossol, M.; Dewor, M.; Antonin, W. A self-inhibitory interaction within Nup155 and membrane binding are required for nuclear pore complex formation. J. Cell Sci. 2018, 131, jcs208538. [Google Scholar] [CrossRef]
- De Souza, C.P.; Osmani, A.H.; Hashmi, S.B.; Osmani, S.A. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 2004, 14, 1973–1984. [Google Scholar] [CrossRef]
- Laurell, E.; Beck, K.; Krupina, K.; Theerthagiri, G.; Bodenmiller, B.; Horvath, P.; Aebersold, R.; Antonin, W.; Kutay, U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 2011, 144, 539–550. [Google Scholar] [CrossRef]
- Linder, M.I.; Kohler, M.; Boersema, P.; Weberruss, M.; Wandke, C.; Marino, J.; Ashiono, C.; Picotti, P.; Antonin, W.; Kutay, U. Mitotic Disassembly of Nuclear Pore Complexes Involves CDK1- and PLK1-Mediated Phosphorylation of Key Interconnecting Nucleoporins. Dev. Cell 2017, 43, 141–156.e7. [Google Scholar] [CrossRef]
- Martino, L.; Morchoisne-Bolhy, S.; Cheerambathur, D.K.; Van Hove, L.; Dumont, J.; Joly, N.; Desai, A.; Doye, V.; Pintard, L. Channel Nucleoporins Recruit PLK-1 to Nuclear Pore Complexes to Direct Nuclear Envelope Breakdown in C. elegans. Dev. Cell 2017, 43, 157–171.e7. [Google Scholar] [CrossRef]
- Hattersley, N.; Cheerambathur, D.; Moyle, M.; Stefanutti, M.; Richardson, A.; Lee, K.Y.; Dumont, J.; Oegema, K.; Desai, A. A Nucleoporin Docks Protein Phosphatase 1 to Direct Meiotic Chromosome Segregation and Nuclear Assembly. Dev. Cell 2016, 38, 463–477. [Google Scholar] [CrossRef]
- Moorhead, G.B.; Trinkle-Mulcahy, L.; Nimick, M.; De Wever, V.; Campbell, D.G.; Gourlay, R.; Lam, Y.W.; Lamond, A.I. Displacement affinity chromatography of protein phosphatase one (PP1) complexes. BMC Biochem. 2008, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- de Castro, I.J.; Budzak, J.; Di Giacinto, M.L.; Ligammari, L.; Gokhan, E.; Spanos, C.; Moralli, D.; Richardson, C.; de Las Heras, J.I.; Salatino, S.; et al. Repo-Man/PP1 regulates heterochromatin formation in interphase. Nat. Commun. 2017, 8, 14048. [Google Scholar] [CrossRef] [PubMed]
- Vagnarelli, P.; Ribeiro, S.; Sennels, L.; Sanchez-Pulido, L.; de Lima Alves, F.; Verheyen, T.; Kelly, D.A.; Ponting, C.P.; Rappsilber, J.; Earnshaw, W.C. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell 2011, 21, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Makio, T.; Stanton, L.H.; Lin, C.C.; Goldfarb, D.S.; Weis, K.; Wozniak, R.W. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 2009, 185, 459–473. [Google Scholar] [CrossRef]
- Bailer, S.M.; Siniossoglou, S.; Podtelejnikov, A.; Hellwig, A.; Mann, M.; Hurt, E. Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 1998, 17, 1107–1119. [Google Scholar] [CrossRef]
- Hodge, C.A.; Choudhary, V.; Wolyniak, M.J.; Scarcelli, J.J.; Schneiter, R.; Cole, C.N. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 2010, 123, 141–151. [Google Scholar] [CrossRef]
- Kralt, A.; Wojtynek, M.; Fischer, J.S.; Agote-Aran, A.; Mancini, R.; Dultz, E.; Noor, E.; Uliana, F.; Tatarek-Nossol, M.; Antonin, W.; et al. An amphipathic helix in Brl1 is required for membrane fusion during nuclear pore complex biogenesis in S. cerevisiae. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lone, M.A.; Atkinson, A.E.; Hodge, C.A.; Cottier, S.; Martinez-Montanes, F.; Maithel, S.; Mene-Saffrane, L.; Cole, C.N.; Schneiter, R. Yeast Integral Membrane Proteins Apq12, Brl1, and Brr6 Form a Complex Important for Regulation of Membrane Homeostasis and Nuclear Pore Complex Biogenesis. Eukaryot. Cell 2015, 14, 1217–1227. [Google Scholar] [CrossRef]
- Scarcelli, J.J.; Hodge, C.A.; Cole, C.N. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol. 2007, 178, 799–812. [Google Scholar] [CrossRef]
- Vitale, J.; Khan, A.; Neuner, A.; Schiebel, E. A perinuclear alpha-helix with amphipathic features in Brl1 promotes NPC assembly. Mol. Biol. Cell 2022, 33, mbcE21120616. [Google Scholar] [CrossRef]
- Wente, S.R.; Blobel, G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J. Cell Biol. 1993, 123, 275–284. [Google Scholar] [CrossRef]
- Laudermilch, E.; Tsai, P.L.; Graham, M.; Turner, E.; Zhao, C.; Schlieker, C. Dissecting Torsin/cofactor function at the nuclear envelope: A genetic study. Mol. Biol. Cell 2016, 27, 3964–3971. [Google Scholar] [CrossRef]
- Drin, G.; Antonny, B. Amphipathic helices and membrane curvature. FEBS Lett. 2010, 584, 1840–1847. [Google Scholar] [CrossRef]
- Casey, A.K.; Chen, S.; Novick, P.; Ferro-Novick, S.; Wente, S.R. Nuclear pore complex integrity requires Lnp1, a regulator of cortical endoplasmic reticulum. Mol. Biol. Cell 2015, 26, 2833–2844. [Google Scholar] [CrossRef]
- Dawson, T.R.; Lazarus, M.D.; Hetzer, M.W.; Wente, S.R. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 2009, 184, 659–675. [Google Scholar] [CrossRef]
- Peeters, B.W.A.; Piet, A.C.A.; Fornerod, M. Generating Membrane Curvature at the Nuclear Pore: A Lipid Point of View. Cells 2022, 11, 469. [Google Scholar] [CrossRef]
- Thaller, D.J.; Tong, D.; Marklew, C.J.; Ader, N.R.; Mannino, P.J.; Borah, S.; King, M.C.; Ciani, B.; Lusk, C.P. Direct binding of ESCRT protein Chm7 to phosphatidic acid-rich membranes at nuclear envelope herniations. J. Cell Biol. 2021, 220. [Google Scholar] [CrossRef]
- Zhang, W.; Khan, A.; Vitale, J.; Neuner, A.; Rink, K.; Luchtenborg, C.; Brugger, B.; Sollner, T.H.; Schiebel, E. A short perinuclear amphipathic alpha-helix in Apq12 promotes nuclear pore complex biogenesis. Open Biol. 2021, 11, 210250. [Google Scholar] [CrossRef]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019, 593, 2428–2451. [Google Scholar] [CrossRef]
- Kusumaatmaja, H.; May, A.I.; Feeney, M.; McKenna, J.F.; Mizushima, N.; Frigerio, L.; Knorr, R.L. Wetting of phase-separated droplets on plant vacuole membranes leads to a competition between tonoplast budding and nanotube formation. Proc. Natl. Acad. Sci. USA 2021, 118, e2024109118. [Google Scholar] [CrossRef]
- Yuan, F.; Alimohamadi, H.; Bakka, B.; Trementozzi, A.N.; Day, K.J.; Fawzi, N.L.; Rangamani, P.; Stachowiak, J.C. Membrane bending by protein phase separation. Proc. Natl. Acad. Sci. USA 2021, 118, e2017435118. [Google Scholar] [CrossRef]
- Kozlov, M.M.; McMahon, H.T.; Chernomordik, L.V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010, 35, 699–706. [Google Scholar] [CrossRef]
- Zhang, W.; Neuner, A.; Ruthnick, D.; Sachsenheimer, T.; Luchtenborg, C.; Brugger, B.; Schiebel, E. Brr6 and Brl1 locate to nuclear pore complex assembly sites to promote their biogenesis. J. Cell Biol. 2018, 217, 877–894. [Google Scholar] [CrossRef]
- Ciechonska, M.; Duncan, R. Reovirus FAST proteins: Virus-encoded cellular fusogens. Trends Microbiol. 2014, 22, 715–724. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef]
- Saitoh, Y.H.; Ogawa, K.; Nishimoto, T. Brl1p—A novel nuclear envelope protein required for nuclear transport. Traffic 2005, 6, 502–517. [Google Scholar] [CrossRef]
- Tanabe, L.M.; Liang, C.C.; Dauer, W.T. Neuronal Nuclear Membrane Budding Occurs during a Developmental Window Modulated by Torsin Paralogs. Cell Rep. 2016, 16, 3322–3333. [Google Scholar] [CrossRef]
- VanGompel, M.J.; Nguyen, K.C.; Hall, D.H.; Dauer, W.T.; Rose, L.S. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol. Biol. Cell 2015, 26, 1752–1763. [Google Scholar] [CrossRef]
- Goodchild, R.E.; Kim, C.E.; Dauer, W.T. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 2005, 48, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wu, S.; Zhou, Q.; Vivona, S.; Cipriano, D.J.; Cheng, Y.; Brunger, A.T. Mechanistic insights into the recycling machine of the SNARE complex. Nature 2015, 518, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Thaller, D.J.; Allegretti, M.; Borah, S.; Ronchi, P.; Beck, M.; Lusk, C.P. An ESCRT-LEM protein surveillance system is poised to directly monitor the nuclear envelope and nuclear transport system. eLife 2019, 8, e45284. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.M.; Colombi, P.; Jager, J.; Lusk, C.P. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 2014, 159, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Hakhverdyan, Z.; Molloy, K.R.; Keegan, S.; Herricks, T.; Lepore, D.M.; Munson, M.; Subbotin, R.I.; Fenyo, D.; Aitchison, J.D.; Fernandez-Martinez, J.; et al. Dissecting the Structural Dynamics of the Nuclear Pore Complex. Mol. Cell 2021, 81, 153–165.e7. [Google Scholar] [CrossRef] [PubMed]
- Derrer, C.P.; Mancini, R.; Vallotton, P.; Huet, S.; Weis, K.; Dultz, E. The RNA export factor Mex67 functions as a mobile nucleoporin. J. Cell Biol. 2019, 218, 3967–3976. [Google Scholar] [CrossRef]
- Fernandez-Martinez, J.; Rout, M.P. One ring to rule them all? Structural and functional diversity in the nuclear pore complex. Trends Biochem. Sci. 2021, 46, 595–607. [Google Scholar] [CrossRef]
- Varberg, J.M.; Unruh, J.R.; Bestul, A.J.; Khan, A.A.; Jaspersen, S.L. Quantitative analysis of nuclear pore complex organization in Schizosaccharomyces pombe. Life Sci. Alliance 2022, 5. [Google Scholar] [CrossRef]
- Bensidoun, P.; Reiter, T.; Montpetit, B.; Zenklusen, D.; Oeffinger, M. Nuclear mRNA metabolism drives selective basket assembly on a subset of nuclear pores in budding yeast. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dey, G.; Culley, S.; Curran, S.; Schmidt, U.; Henriques, R.; Kukulski, W.; Baum, B. Closed mitosis requires local disassembly of the nuclear envelope. Nature 2020, 585, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Exposito-Serrano, M.; Sanchez-Molina, A.; Gallardo, P.; Salas-Pino, S.; Daga, R.R. Selective Nuclear Pore Complex Removal Drives Nuclear Envelope Division in Fission Yeast. Curr. Biol. 2020, 30, 3212–3222.e2. [Google Scholar] [CrossRef]
- Galy, V.; Gadal, O.; Fromont-Racine, M.; Romano, A.; Jacquier, A.; Nehrbass, U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 2004, 116, 63–73. [Google Scholar] [CrossRef]
- Denoth-Lippuner, A.; Krzyzanowski, M.K.; Stober, C.; Barral, Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife 2014, 3, e03790. [Google Scholar] [CrossRef] [PubMed]
- Meinema, A.C.; Marzelliusardottir, A.; Mirkovic, M.; Aspert, T.; Lee, S.S.; Charvin, G.; Barral, Y. DNA circles promote yeast ageing in part through stimulating the reorganization of nuclear pore complexes. eLife 2022, 11, e71196. [Google Scholar] [CrossRef]
- Souquet, B.; Freed, E.; Berto, A.; Andric, V.; Audugé, N.; Reina-San-Martin, B.; Lacy, E.; Doye, V. Nup133 is required for proper nuclear pore basket assembly and dynamics in embryonic stem cells. Cell Rep. 2018, 23, 2443–2454. [Google Scholar] [CrossRef]
- Capelson, M.; Hetzer, M.W. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009, 10, 697–705. [Google Scholar] [CrossRef]
- Gomez-Cavazos, J.S.; Hetzer, M.W. Outfits for different occasions: Tissue-specific roles of Nuclear Envelope proteins. Curr. Opin. Cell Biol. 2012, 24, 775–783. [Google Scholar] [CrossRef][Green Version]
- Kane, M.; Rebensburg, S.V.; Takata, M.A.; Zang, T.M.; Yamashita, M.; Kvaratskhelia, M.; Bieniasz, P.D. Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. eLife 2018, 7, e35738. [Google Scholar] [CrossRef]
- Carmody, S.R.; Tran, E.J.; Apponi, L.H.; Corbett, A.H.; Wente, S.R. The mitogen-activated protein kinase Slt2 regulates nuclear retention of non-heat shock mRNAs during heat shock-induced stress. Mol. Cell Biol. 2010, 30, 5168–5179. [Google Scholar] [CrossRef]
- Cho, U.H.; Hetzer, M.W. Caspase-mediated nuclear pore complex trimming in cell differentiation and endoplasmic reticulum stress. bioRxiv 2022. [Google Scholar] [CrossRef]
- Heinrich, S.; Hondele, M.; Marchand, D.; Derrer, C.P.; Zedan, M.; Oswald, A.; Uliana, F.; Mancini, R.; Grunwald, D.; Weis, K. Condensation of a nuclear mRNA export factor regulates mRNA transport during stress. bioRxiv 2022. [Google Scholar] [CrossRef]
- Takemura, R.; Inoue, Y.; Izawa, S. Stress response in yeast mRNA export factor: Reversible changes in Rat8p localization are caused by ethanol stress but not heat shock. J. Cell Sci. 2004, 117, 4189–4197. [Google Scholar] [CrossRef]
- Gomar-Alba, M.; Pozharskaia, V.; Schaal, C.; Kumar, A.; Jacquel, B.; Charvin, G.; Igual, J.C.; Mendoza, M. Nuclear Pore Complex Acetylation Regulates mRNA Export and Cell Cycle Commitment in Budding Yeast. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, P.; Gomar-Alba, M.; Shcheprova, Z.; Daulny, A.; Sanmartin, T.; Matucci, I.; Funaya, C.; Beato, M.; Mendoza, M. Daughter-cell-specific modulation of nuclear pore complexes controls cell cycle entry during asymmetric division. Nat. Cell Biol. 2018, 20, 432–442. [Google Scholar] [CrossRef]
- Folz, H.; Nino, C.A.; Taranum, S.; Caesar, S.; Latta, L.; Waharte, F.; Salamero, J.; Schlenstedt, G.; Dargemont, C. SUMOylation of the nuclear pore complex basket is involved in sensing cellular stresses. J. Cell Sci. 2019, 132, jcs224279. [Google Scholar] [CrossRef]
- Nino, C.A.; Guet, D.; Gay, A.; Brutus, S.; Jourquin, F.; Mendiratta, S.; Salamero, J.; Geli, V.; Dargemont, C. Posttranslational marks control architectural and functional plasticity of the nuclear pore complex basket. J. Cell Biol. 2016, 212, 167–180. [Google Scholar] [CrossRef]
- Regot, S.; de Nadal, E.; Rodriguez-Navarro, S.; Gonzalez-Novo, A.; Perez-Fernandez, J.; Gadal, O.; Seisenbacher, G.; Ammerer, G.; Posas, F. The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J. Biol. Chem. 2013, 288, 17384–17398. [Google Scholar] [CrossRef]
- Hayakawa, A.; Babour, A.; Sengmanivong, L.; Dargemont, C. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J. Cell Biol. 2012, 196, 19–27. [Google Scholar] [CrossRef]
- Buendia, B.; Santa-Maria, A.; Courvalin, J.C. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci. 1999, 112 Pt 11, 1743–1753. [Google Scholar] [CrossRef]
- Ferrando-May, E.; Cordes, V.; Biller-Ckovric, I.; Mirkovic, J.; Görlich, D.; Nicotera, P. Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 2001, 8, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Kihlmark, M.; Imreh, G.; Hallberg, E. Sequential degradation of proteins from the nuclear envelope during apoptosis. J. Cell Sci. 2001, 114, 3643–3653. [Google Scholar] [CrossRef] [PubMed]
- Kihlmark, M.; Rustum, C.; Eriksson, C.; Beckman, M.; Iverfeldt, K.; Hallberg, E. Correlation between nucleocytoplasmic transport and caspase-3-dependent dismantling of nuclear pores during apoptosis. Exp. Cell Res. 2004, 293, 346–356. [Google Scholar] [CrossRef]
- Patre, M.; Tabbert, A.; Hermann, D.; Walczak, H.; Rackwitz, H.R.; Cordes, V.C.; Ferrando-May, E. Caspases target only two architectural components within the core structure of the nuclear pore complex. J. Biol. Chem. 2006, 281, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Liashkovich, I.; Oberleithner, H.; Ludwig, S.; Mazur, I.; Shahin, V. Apoptosis leads to a degradation of vital components of active nuclear transport and a dissociation of the nuclear lamina. Proc. Natl. Acad. Sci. USA 2008, 105, 11236–11241. [Google Scholar] [CrossRef]
- Kramer, A.; Liashkovich, I.; Oberleithner, H.; Shahin, V. Caspase-9-dependent decrease of nuclear pore channel hydrophobicity is accompanied by nuclear envelope leakiness. Nanomedicine 2010, 6, 605–611. [Google Scholar] [CrossRef]
- Juhlen, R.; Fahrenkrog, B. Moonlighting nuclear pore proteins: Tissue-specific nucleoporin function in health and disease. Histochem. Cell Biol. 2018, 150, 593–605. [Google Scholar] [CrossRef]
- Dultz, E.; Tjong, H.; Weider, E.; Herzog, M.; Young, B.; Brune, C.; Mullner, D.; Loewen, C.; Alber, F.; Weis, K. Global reorganization of budding yeast chromosome conformation in different physiological conditions. J. Cell Biol. 2016, 212, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Luthra, R.; Kerr, S.C.; Harreman, M.T.; Apponi, L.H.; Fasken, M.B.; Ramineni, S.; Chaurasia, S.; Valentini, S.R.; Corbett, A.H. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 2007, 282, 3042–3049. [Google Scholar] [CrossRef]
- Iglesias, N.; Paulo, J.A.; Tatarakis, A.; Wang, X.; Edwards, A.L.; Bhanu, N.V.; Garcia, B.A.; Haas, W.; Gygi, S.P.; Moazed, D. Native Chromatin Proteomics Reveals a Role for Specific Nucleoporins in Heterochromatin Organization and Maintenance. Mol. Cell 2020, 77, 51–66.e8. [Google Scholar] [CrossRef]
- Lapetina, D.L.; Ptak, C.; Roesner, U.K.; Wozniak, R.W. Yeast silencing factor Sir4 and a subset of nucleoporins form a complex distinct from nuclear pore complexes. J. Cell Biol. 2017, 216, 3145–3159. [Google Scholar] [CrossRef] [PubMed]
- Gozalo, A.; Duke, A.; Lan, Y.; Pascual-Garcia, P.; Talamas, J.A.; Nguyen, S.C.; Shah, P.P.; Jain, R.; Joyce, E.F.; Capelson, M. Core Components of the Nuclear Pore Bind Distinct States of Chromatin and Contribute to Polycomb Repression. Mol. Cell 2020, 77, 67–81.e7. [Google Scholar] [CrossRef] [PubMed]
- Osmani, A.H.; Davies, J.; Liu, H.L.; Nile, A.; Osmani, S.A. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 2006, 17, 4946–4961. [Google Scholar] [CrossRef]
- Arai, K.; Sato, M.; Tanaka, K.; Yamamoto, M. Nuclear compartmentalization is abolished during fission yeast meiosis. Curr. Biol. 2010, 20, 1913–1918. [Google Scholar] [CrossRef]
- Asakawa, H.; Kojidani, T.; Mori, C.; Osakada, H.; Sato, M.; Ding, D.Q.; Hiraoka, Y.; Haraguchi, T. Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr. Biol. 2010, 20, 1919–1925. [Google Scholar] [CrossRef]
- Denoth Lippuner, A.; Julou, T.; Barral, Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol. Rev. 2014, 38, 300–325. [Google Scholar] [CrossRef] [PubMed]
- Khmelinskii, A.; Keller, P.J.; Lorenz, H.; Schiebel, E.; Knop, M. Segregation of yeast nuclear pores. Nature 2010, 466, E1. [Google Scholar] [CrossRef]
- Makio, T.; Lapetina, D.L.; Wozniak, R.W. Inheritance of yeast nuclear pore complexes requires the Nsp1p subcomplex. J. Cell Biol. 2013, 203, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Shcheprova, Z.; Baldi, S.; Frei, S.B.; Gonnet, G.; Barral, Y. A mechanism for asymmetric segregation of age during yeast budding. Nature 2008, 454, 728–734. [Google Scholar] [CrossRef]
- Colombi, P.; Webster, B.M.; Frohlich, F.; Lusk, C.P. The transmission of nuclear pore complexes to daughter cells requires a cytoplasmic pool of Nsp1. J. Cell Biol. 2013, 203, 215–232. [Google Scholar] [CrossRef]
- King, G.A.; Goodman, J.S.; Schick, J.G.; Chetlapalli, K.; Jorgens, D.M.; McDonald, K.L.; Unal, E. Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast. eLife 2019, 8, e47156. [Google Scholar] [CrossRef] [PubMed]
- Koch, B.A.; Staley, E.; Jin, H.; Yu, H.G. The ESCRT-III complex is required for nuclear pore complex sequestration and regulates gamete replicative lifespan in budding yeast meiosis. Nucleus 2020, 11, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Regmi, S.G.; Lee, H.; Kaufhold, R.; Fichtman, B.; Chen, S.; Aksenova, V.; Turcotte, E.; Harel, A.; Arnaoutov, A.; Dasso, M. The Nuclear Pore Complex consists of two independent scaffolds. bioRxiv 2020. [Google Scholar] [CrossRef]
- Aksenova, V.; Smith, A.; Lee, H.; Bhat, P.; Esnault, C.; Chen, S.; Iben, J.; Kaufhold, R.; Yau, K.C.; Echeverria, C.; et al. Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat. Commun. 2020, 11, 4577. [Google Scholar] [CrossRef]
- Salas-Pino, S.; Gallardo, P.; Barrales, R.R.; Braun, S.; Daga, R.R. The fission yeast nucleoporin Alm1 is required for proteasomal degradation of kinetochore components. J. Cell Biol. 2017, 216, 3591–3608. [Google Scholar] [CrossRef]
- Webster, B.M.; Thaller, D.J.; Jager, J.; Ochmann, S.E.; Borah, S.; Lusk, C.P. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 2016, 35, 2447–2467. [Google Scholar] [CrossRef]
- Capella, M.; Martin Caballero, L.; Pfander, B.; Braun, S.; Jentsch, S. ESCRT recruitment by the S. cerevisiae inner nuclear membrane protein Heh1 is regulated by Hub1-mediated alternative splicing. J. Cell Sci. 2020, 133, jcs250688. [Google Scholar] [CrossRef]
- Vietri, M.; Schultz, S.W.; Bellanger, A.; Jones, C.M.; Petersen, L.I.; Raiborg, C.; Skarpen, E.; Pedurupillay, C.R.J.; Kjos, I.; Kip, E.; et al. Unrestrained ESCRT-III drives micronuclear catastrophe and chromosome fragmentation. Nat. Cell Biol. 2020, 22, 856–867. [Google Scholar] [CrossRef]
- Borah, S.; Thaller, D.J.; Hakhverdyan, Z.; Rodriguez, E.C.; Isenhour, A.W.; Rout, M.P.; King, M.C.; Lusk, C.P. Heh2/Man1 may be an evolutionarily conserved sensor of NPC assembly state. Mol. Biol. Cell 2021, 32, 1359–1373. [Google Scholar] [CrossRef]
- Foresti, O.; Rodriguez-Vaello, V.; Funaya, C.; Carvalho, P. Quality control of inner nuclear membrane proteins by the Asi complex. Science 2014, 346, 751–755. [Google Scholar] [CrossRef]
- Khmelinskii, A.; Blaszczak, E.; Pantazopoulou, M.; Fischer, B.; Omnus, D.J.; Le Dez, G.; Brossard, A.; Gunnarsson, A.; Barry, J.D.; Meurer, M.; et al. Protein quality control at the inner nuclear membrane. Nature 2014, 516, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Smoyer, C.J.; Smith, S.E.; Gardner, J.M.; McCroskey, S.; Unruh, J.R.; Jaspersen, S.L. Distribution of Proteins at the Inner Nuclear Membrane Is Regulated by the Asi1 E3 Ligase in Saccharomyces cerevisiae. Genetics 2019, 211, 1269–1282. [Google Scholar] [CrossRef] [PubMed]
- Shulga, N.; Roberts, P.; Gu, Z.; Spitz, L.; Tabb, M.M.; Nomura, M.; Goldfarb, D.S. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: A role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996, 135, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.K.; Raczniak, G.A.; Ives, E.B.; Wente, S.R. The integral membrane protein snl1p is genetically linked to yeast nuclear pore complex function. Mol. Biol. Cell 1998, 9, 355–373. [Google Scholar] [CrossRef]
- Frey, S.; Gorlich, D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Celetti, G.; Paci, G.; Caria, J.; VanDelinder, V.; Bachand, G.; Lemke, E.A. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J. Cell Biol. 2020, 219, e201907157. [Google Scholar] [CrossRef]
- Mizuguchi-Hata, C.; Ogawa, Y.; Oka, M.; Yoneda, Y. Quantitative regulation of nuclear pore complex proteins by O-GlcNAcylation. Biochim. Biophys. Acta 2013, 1833, 2682–2689. [Google Scholar] [CrossRef][Green Version]
- Zhu, Y.; Liu, T.W.; Madden, Z.; Yuzwa, S.A.; Murray, K.; Cecioni, S.; Zachara, N.; Vocadlo, D.J. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J. Mol. Cell Biol. 2016, 8, 2–16. [Google Scholar] [CrossRef]
- Nosella, M.L.; Tereshchenko, M.; Pritišanac, I.; Chong, P.A.; Toretsky, J.A.; Lee, H.O.; Forman-Kay, J.D. O-GlcNAcylation reduces phase separation and aggregation of the EWS N-terminal low complexity region. bioRxiv 2021. [Google Scholar] [CrossRef]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D.; et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 2016, 353, aaf1420. [Google Scholar] [CrossRef]
- Lee, C.W.; Wilfling, F.; Ronchi, P.; Allegretti, M.; Mosalaganti, S.; Jentsch, S.; Beck, M.; Pfander, B. Selective autophagy degrades nuclear pore complexes. Nat. Cell Biol. 2020, 22, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Tomioka, Y.; Kotani, T.; Kirisako, H.; Oikawa, Y.; Kimura, Y.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes. J. Cell Biol. 2020, 219, e201910063. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Mannino, P.J.; Thaller, D.J.; Ader, N.R.; King, M.C.; Melia, T.J.; Lusk, C.P. Atg39 selectively captures inner nuclear membrane into lumenal vesicles for delivery to the autophagosome. J. Cell Biol. 2021, 220, e202103030. [Google Scholar] [CrossRef]
- Rempel, I.L.; Steen, A.; Veenhoff, L.M. Poor old pores-The challenge of making and maintaining nuclear pore complexes in aging. FEBS J. 2020, 287, 1058–1075. [Google Scholar] [CrossRef]
- Liu, J.; Hetzer, M.W. Nuclear pore complex maintenance and implications for age-related diseases. Trends Cell Biol. 2022, 32, 216–227. [Google Scholar] [CrossRef]
- Janssens, G.E.; Meinema, A.C.; Gonzalez, J.; Wolters, J.C.; Schmidt, A.; Guryev, V.; Bischoff, R.; Wit, E.C.; Veenhoff, L.M.; Heinemann, M. Protein biogenesis machinery is a driver of replicative aging in yeast. eLife 2015, 4, e08527. [Google Scholar] [CrossRef]
- Rempel, I.L.; Crane, M.M.; Thaller, D.J.; Mishra, A.; Jansen, D.P.; Janssens, G.; Popken, P.; Aksit, A.; Kaeberlein, M.; van der Giessen, E.; et al. Age-dependent deterioration of nuclear pore assembly in mitotic cells decreases transport dynamics. eLife 2019, 8, e48186. [Google Scholar] [CrossRef]
- Ori, A.; Toyama, B.H.; Harris, M.S.; Bock, T.; Iskar, M.; Bork, P.; Ingolia, N.T.; Hetzer, M.W.; Beck, M. Integrated Transcriptome and Proteome Analyses Reveal Organ-Specific Proteome Deterioration in Old Rats. Cell Syst. 2015, 1, 224–237. [Google Scholar] [CrossRef]
- Bitetto, G.; Di Fonzo, A. Nucleo-cytoplasmic transport defects and protein aggregates in neurodegeneration. Transl. Neurodegener. 2020, 9, 25. [Google Scholar] [CrossRef]
- Chou, C.C.; Zhang, Y.; Umoh, M.E.; Vaughan, S.W.; Lorenzini, I.; Liu, F.; Sayegh, M.; Donlin-Asp, P.G.; Chen, Y.H.; Duong, D.M.; et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018, 21, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Eftekharzadeh, B.; Daigle, J.G.; Kapinos, L.E.; Coyne, A.; Schiantarelli, J.; Carlomagno, Y.; Cook, C.; Miller, S.J.; Dujardin, S.; Amaral, A.S.; et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 2018, 99, 925–940.e7. [Google Scholar] [CrossRef] [PubMed]
- Gasset-Rosa, F.; Chillon-Marinas, C.; Goginashvili, A.; Atwal, R.S.; Artates, J.W.; Tabet, R.; Wheeler, V.C.; Bang, A.G.; Cleveland, D.W.; Lagier-Tourenne, C. Polyglutamine-Expanded Huntingtin Exacerbates Age-Related Disruption of Nuclear Integrity and Nucleocytoplasmic Transport. Neuron 2017, 94, 48–57.e4. [Google Scholar] [CrossRef] [PubMed]
- Grima, J.C.; Daigle, J.G.; Arbez, N.; Cunningham, K.C.; Zhang, K.; Ochaba, J.; Geater, C.; Morozko, E.; Stocksdale, J.; Glatzer, J.C.; et al. Mutant Huntingtin Disrupts the Nuclear Pore Complex. Neuron 2017, 94, 93–107.e6. [Google Scholar] [CrossRef]
- Lin, Y.C.; Kumar, M.S.; Ramesh, N.; Anderson, E.N.; Nguyen, A.T.; Kim, B.; Cheung, S.; McDonough, J.A.; Skarnes, W.C.; Lopez-Gonzalez, R.; et al. Interactions between ALS-linked FUS and nucleoporins are associated with defects in the nucleocytoplasmic transport pathway. Nat. Neurosci. 2021, 24, 1077–1088. [Google Scholar] [CrossRef]
- Milles, S.; Huy Bui, K.; Koehler, C.; Eltsov, M.; Beck, M.; Lemke, E.A. Facilitated aggregation of FG nucleoporins under molecular crowding conditions. EMBO Rep. 2013, 14, 178–183. [Google Scholar] [CrossRef]
- Ader, C.; Frey, S.; Maas, W.; Schmidt, H.B.; Gorlich, D.; Baldus, M. Amyloid-like interactions within nucleoporin FG hydrogels. Proc. Natl. Acad. Sci. USA 2010, 107, 6281–6285. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Schmidt, H.B.; Gorlich, D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife 2015, 4, e04251. [Google Scholar] [CrossRef]
- Gonzalez, A.; Mannen, T.; Cagatay, T.; Fujiwara, A.; Matsumura, H.; Niesman, A.B.; Brautigam, C.A.; Chook, Y.M.; Yoshizawa, T. Mechanism of karyopherin-beta2 binding and nuclear import of ALS variants FUS(P525L) and FUS(R495X). Sci. Rep. 2021, 11, 3754. [Google Scholar] [CrossRef]
- Guo, L.; Kim, H.J.; Wang, H.; Monaghan, J.; Freyermuth, F.; Sung, J.C.; O’Donovan, K.; Fare, C.M.; Diaz, Z.; Singh, N.; et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 2018, 173, 677–692.e20. [Google Scholar] [CrossRef] [PubMed]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Strohl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 2018, 173, 720–734.e15. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, T.; Ali, R.; Jiou, J.; Fung, H.Y.J.; Burke, K.A.; Kim, S.J.; Lin, Y.; Peeples, W.B.; Saltzberg, D.; Soniat, M.; et al. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 2018, 173, 693–705.e22. [Google Scholar] [CrossRef]
- Woerner, A.C.; Frottin, F.; Hornburg, D.; Feng, L.R.; Meissner, F.; Patra, M.; Tatzelt, J.; Mann, M.; Winklhofer, K.F.; Hartl, F.U.; et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science 2016, 351, 173–176. [Google Scholar] [CrossRef]
- Coyne, A.N.; Zaepfel, B.L.; Hayes, L.; Fitchman, B.; Salzberg, Y.; Luo, E.C.; Bowen, K.; Trost, H.; Aigner, S.; Rigo, F.; et al. G4C2 Repeat RNA Initiates a POM121-Mediated Reduction in Specific Nucleoporins in C9orf72 ALS/FTD. Neuron 2020, 107, 1124–1140.e11. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.N.; Baskerville, V.; Zaepfel, B.L.; Dickson, D.W.; Rigo, F.; Bennett, F.; Lusk, C.P.; Rothstein, J.D. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med. 2021, 13, eabe1923. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dultz, E.; Wojtynek, M.; Medalia, O.; Onischenko, E. The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth. Cells 2022, 11, 1456. https://doi.org/10.3390/cells11091456
Dultz E, Wojtynek M, Medalia O, Onischenko E. The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth. Cells. 2022; 11(9):1456. https://doi.org/10.3390/cells11091456
Chicago/Turabian StyleDultz, Elisa, Matthias Wojtynek, Ohad Medalia, and Evgeny Onischenko. 2022. "The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth" Cells 11, no. 9: 1456. https://doi.org/10.3390/cells11091456
APA StyleDultz, E., Wojtynek, M., Medalia, O., & Onischenko, E. (2022). The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth. Cells, 11(9), 1456. https://doi.org/10.3390/cells11091456






