Epigenetic Mechanisms of Senescence in Plants
Abstract
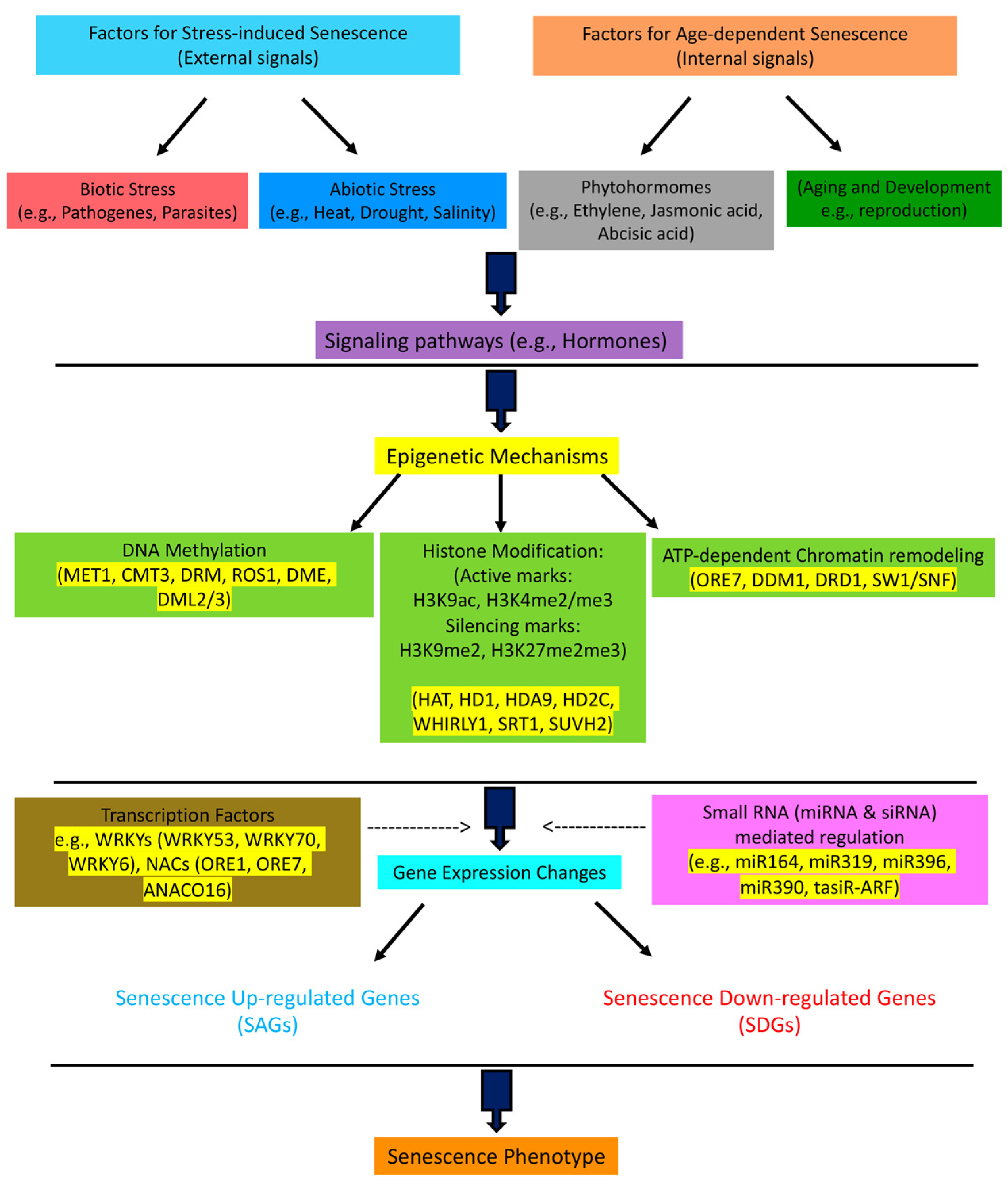
:1. Overview
2. DNA Methylation and Plant Senescence
3. Histone Modifications and Plant Senescence
4. ATP-Dependent Chromatin Remodeling and Senescence
5. Stress-Induced vs. Age-Dependent Senescence
6. DNA Damage, Aging, and Epigenetics
7. Small RNAs and Senescence
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar]
- Yolcu, S.; Li, X.; Li, S.; Kim, Y.J. Beyond the genetic code in leaf senescence. J. Exp. Bot. 2018, 69, 801–810. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death—Regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef] [Green Version]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef] [Green Version]
- Woo, H.R.; Koo, H.J.; Kim, J.; Jeong, H.; Yang, J.O.; Lee, I.H.; Jun, J.H.; Choi, S.H.; Park, S.J.; Kang, B.; et al. Programming of Plant Leaf Senescence with Temporal and Inter-Organellar Coordination of Transcriptome in Arabidopsis. Plant Physiol. 2016, 171, 452–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinderhofer, K.; Zentgraf, U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 2001, 213, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Humbeck, K. Epigenetic and small RNA regulation of senescence. Plant Mol. Biol. 2013, 82, 529–537. [Google Scholar] [CrossRef]
- Stroud, H.; Greenberg, M.V.; Feng, S.; Bernatavichute, Y.V.; Jacobsen, S.E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 2013, 152, 352–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ay, N.; Janack, B.; Humbeck, K. Epigenetic control of plant senescence and linked processes. J. Exp. Bot. 2014, 65, 3875–3887. [Google Scholar] [CrossRef] [Green Version]
- Miryeganeh, M. Senescence: The Compromised Time of Death That Plants May Call on Themselves. Genes 2021, 12, 143. [Google Scholar] [CrossRef] [PubMed]
- Vanyushin, B.F.; Ashapkin, V.V. DNA methylation in higher plants: Past, present and future. Biochim. Biophys. Acta 2011, 1809, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Gehring, M.; Reik, W.; Henikoff, S. DNA demethylation by DNA repair. Trends Genet. 2009, 25, 82–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inamdar, N.M.; Ehrlich, K.C.; Ehrlich, M. CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol. Biol. 1991, 17, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Henderson, I.R.; Jacobsen, S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005, 6, 351–360. [Google Scholar] [CrossRef]
- Monteuuis, O.; Doulbeau, S.; Verdeil, J.-L. DNA methylation in different origin clonal offspring from a mature Sequoiadendron giganteum genotype. Trees 2008, 22, 779. [Google Scholar] [CrossRef]
- Ostrowska-Mazurek, A.; Kasprzak, P.; Kubala, S.; Zaborowska, M.; Sobieszczuk-Nowicka, E. Epigenetic Landmarks of Leaf Senescence and Crop Improvement. Int. J. Mol. Sci. 2020, 21, 5125. [Google Scholar] [CrossRef]
- Li, H.; Freeling, M.; Lisch, D. Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 2010, 107, 22184. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Ohr, H.; Lee, J.W.; Hyun, Y.; Fischer, R.L.; Choi, Y. Temporal and spatial downregulation of Arabidopsis MET1 activity results in global DNA hypomethylation and developmental defects. Mol. Cells 2008, 26, 611–615. [Google Scholar] [PubMed]
- Horváth, V.; Merenciano, M.; González, J. Revisiting the Relationship between Transposable Elements and the Eukaryotic Stress Response. Trends Genet. 2017, 33, 832–841. [Google Scholar] [CrossRef]
- Groth, M.; Moissiard, G.; Wirtz, M.; Wang, H.; Garcia-Salinas, C.; Ramos-Parra, P.A.; Bischof, S.; Feng, S.; Cokus, S.J.; John, A.; et al. MTHFD1 controls DNA methylation in Arabidopsis. Nat. Commun. 2016, 7, 11640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Gan, S.-S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef]
- Ay, N.; Clauß, K.; Barth, O.; Humbeck, K. Identification and characterization of novel senescence-associated genes from barley (Hordeum vulgare) primary leaves. Plant Biol. 2008, 10, 121–135. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wu, W.; Zinta, G.; Yang, L.; Wang, D.; Liu, R.; Zhang, H.; Zheng, Z.; Huang, H.; Zhang, Q.; et al. A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nat. Commun. 2018, 9, 460. [Google Scholar] [CrossRef] [PubMed]
- Lambé, P.; Mutambel, H.S.N.; Fouché, J.-G.; Deltour, R.; Foidart, J.-M.; Gaspar, T. DNA methylation as a key process in regulation of organogenic totipotency and plant neoplastic progression? In Vitro Cell. Dev. Biol.—Plant 1997, 33, 155–162. [Google Scholar] [CrossRef]
- Trejo-Arellano, M.S.; Mehdi, S.; de Jonge, J.; Dvorák Tomastíková, E.; Köhler, C.; Hennig, L. Dark-Induced Senescence Causes Localized Changes in DNA Methylation1. Plant Physiol. 2020, 182, 949–961. [Google Scholar] [CrossRef]
- Vatov, E.; Zentgraf, U.; Ludewig, U. Moderate DNA methylation changes associated with nitrogen remobilization and leaf senescence in Arabidopsis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Greenwood, M.S. Juvenility and maturation in conifers: Current concepts. Tree Physiol. 1995, 15, 433–438. [Google Scholar] [CrossRef] [Green Version]
- Galaud, J.-P.; Gaspar, T.; Boyer, N. Effect of anti-DNA methylation drugs on growth, level of methylated DNA, peroxidase activity and ethylene production of Bryonia dioica internodes. Physiol. Plant. 1993, 87, 528–534. [Google Scholar] [CrossRef]
- Fraga, M.F.; Rodríguez, R.; Cañal, M.J. Genomic DNA methylation-demethylation during aging and reinvigoration of Pinus radiata. Tree Physiol. 2002, 22, 813–816. [Google Scholar] [CrossRef] [Green Version]
- Ogneva, Z.V.; Dubrovina, A.S.; Kiselev, K.V. Age-associated alterations in DNA methylation and expression of methyltransferase and demethylase genes in Arabidopsis thaliana. Biol. Plant. 2016, 60, 628–634. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, D.; Cao, L.; Yu, N.; Liu, K.; Guo, Y.; Gan, S.; Chen, L. Regulation of Leaf Longevity by DML3-Mediated DNA Demethylation. Mol. Plant 2020, 13, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [Green Version]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 5. [Google Scholar] [CrossRef]
- Ay, N.; Irmler, K.; Fischer, A.; Uhlemann, R.; Reuter, G.; Humbeck, K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009, 58, 333–346. [Google Scholar] [CrossRef]
- Brusslan, J.A.; Bonora, G.; Rus-Canterbury, A.M.; Tariq, F.; Jaroszewicz, A.; Pellegrini, M. A Genome-Wide Chronological Study of Gene Expression and Two Histone Modifications, H3K4me3 and H3K9ac, during Developmental Leaf Senescence. Plant Physiol. 2015, 168, 1246–1261. [Google Scholar] [CrossRef] [Green Version]
- Naumann, K.; Fischer, A.; Hofmann, I.; Krauss, V.; Phalke, S.; Irmler, K.; Hause, G.; Aurich, A.C.; Dorn, R.; Jenuwein, T.; et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005, 24, 1418–1429. [Google Scholar] [CrossRef]
- Jing, Y.; Sun, H.; Yuan, W.; Wang, Y.; Li, Q.; Liu, Y.; Li, Y.; Qian, W. SUVH2 and SUVH9 Couple Two Essential Steps for Transcriptional Gene Silencing in Arabidopsis. Mol. Plant 2016, 9, 1156–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brusslan, J.A.; Rus Alvarez-Canterbury, A.M.; Nair, N.U.; Rice, J.C.; Hitchler, M.J.; Pellegrini, M. Genome-wide evaluation of histone methylation changes associated with leaf senescence in Arabidopsis. PLoS ONE 2012, 7, e33151. [Google Scholar] [CrossRef] [PubMed]
- Weaver, L.M.; Gan, S.; Quirino, B.; Amasino, R.M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 1998, 37, 455–469. [Google Scholar] [CrossRef]
- Yan, H.; Liu, Y.; Zhang, K.; Song, J.; Xu, W.; Su, Z. Chromatin State-Based Analysis of Epigenetic H3K4me3 Marks of Arabidopsis in Response to Dark Stress. Front. Genet. 2019, 10, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebsch, D.; Keech, O. Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytol. 2016, 212, 563–570. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, S.; Zhou, B.; Luo, X.; Zhou, X.F.; Cai, B.; Jin, Y.H.; Niu, D.; Lin, J.; Cao, X.; et al. The Histone H3K4 Demethylase JMJ16 Represses Leaf Senescence in Arabidopsis. Plant Cell 2019, 31, 430–443. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Gao, J.; Gao, S.; Song, Y.; Yang, Z.; Kuai, B. The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet. 2019, 15, e1008068. [Google Scholar] [CrossRef]
- Lu, F.; Cui, X.; Zhang, S.; Jenuwein, T.; Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011, 43, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015, 16, 258–264. [Google Scholar] [CrossRef]
- Chen, X.; Lu, L.; Mayer, K.S.; Scalf, M.; Qian, S.; Lomax, A.; Smith, L.M.; Zhong, X. POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 2016, 5, e17214. [Google Scholar] [CrossRef]
- Zhu, M.; Li, Y.; Chen, G.; Ren, L.; Xie, Q.; Zhao, Z.; Hu, Z. Silencing SlELP2L, a tomato Elongator complex protein 2-like gene, inhibits leaf growth, accelerates leaf, sepal senescence, and produces dark-green fruit. Sci. Rep. 2015, 5, 7693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Lan, W.; Li, D.; Deng, B.; Lin, W.; Ren, Y.; Miao, Y. WHIRLY1 Occupancy Affects Histone Lysine Modification and WRKY53 Transcription in Arabidopsis Developmental Manner. Front. Plant Sci. 2018, 9, 1503. [Google Scholar] [CrossRef]
- Hinckley, W.E.; Keymanesh, K.; Cordova, J.A.; Brusslan, J.A. The HAC1 histone acetyltransferase promotes leaf senescence and regulates the expression of ERF022. Plant Direct. 2019, 3, e00159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Lu, Y.; Zhao, Y.; Zhou, D.X. Histone Acetylation Dynamics Integrates Metabolic Activity to Regulate Plant Response to Stress. Front. Plant Sci. 2019, 10, 1236. [Google Scholar] [CrossRef]
- Ueda, M.; Seki, M. Histone Modifications Form Epigenetic Regulatory Networks to Regulate Abiotic Stress Response. Plant Physiol. 2020, 182, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Chen, Z.J. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 2001, 98, 200–205. [Google Scholar] [CrossRef]
- Wu, K.; Zhang, L.; Zhou, C.; Yu, C.W.; Chaikam, V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 2008, 59, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef] [Green Version]
- Kuang, J.F.; Chen, J.Y.; Luo, M.; Wu, K.Q.; Sun, W.; Jiang, Y.M.; Lu, W.J. Histone deacetylase HD2 interacts with ERF1 and is involved in longan fruit senescence. J. Exp. Bot. 2012, 63, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Han, S.K.; Wu, M.F.; Cui, S.; Wagner, D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015, 83, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Kusch, T.; Workman, J.L. Histone variants and complexes involved in their exchange. Subcell Biochem. 2007, 41, 91–109. [Google Scholar] [PubMed]
- Chodavarapu, R.K.; Feng, S.; Bernatavichute, Y.V.; Chen, P.Y.; Stroud, H.; Yu, Y.; Hetzel, J.A.; Kuo, F.; Kim, J.; Cokus, S.J.; et al. Relationship between nucleosome positioning and DNA methylation. Nature 2010, 466, 388–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berr, A.; Shafiq, S.; Shen, W.-H. Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2011, 1809, 567–576. [Google Scholar] [CrossRef]
- Miryeganeh, M. Plants’ Epigenetic Mechanisms and Abiotic Stress. Genes 2021, 12, 1106. [Google Scholar] [CrossRef]
- Pradhan, B.; Jangid, K.K.; Sarwat, M.; Bishi, S.K. Chapter 11—Role of Histones During Leaf Senescence. In Senescence Signalling and Control in Plants; Sarwat, M., Tuteja, N., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 187–197. [Google Scholar]
- Lim, P.O.; Kim, Y.; Breeze, E.; Koo, J.C.; Woo, H.R.; Ryu, J.S.; Park, D.H.; Beynon, J.; Tabrett, A.; Buchanan-Wollaston, V.; et al. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 2007, 52, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gu, L.; Gao, L.; Chen, C.; Wei, C.-Q.; Qiu, Q.; Chien, C.-W.; Wang, S.; Jiang, L.; Ai, L.-F.; et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016, 48, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Han, S.K.; Kim, H.J.; Wu, M.F.; Steiner, E.; Birnbaum, K.D.; Hong, J.C.; Eshed, Y.; Wagner, D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 2013, 24, 438–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archacki, R.; Yatusevich, R.; Buszewicz, D.; Krzyczmonik, K.; Patryn, J.; Iwanicka-Nowicka, R.; Biecek, P.; Wilczynski, B.; Koblowska, M.; Jerzmanowski, A.; et al. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res. 2017, 45, 3116–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.J.; Choi, S.H.; Kim, J.H.; Kim, J.E.; Lee, M.H.; Chung, B.Y.; Woo, H.R.; Kim, J.H. A Mutation in Plant-Specific SWI2/SNF2-Like Chromatin-Remodeling Proteins, DRD1 and DDM1, Delays Leaf Senescence in Arabidopsis thaliana. PLoS ONE 2016, 11, e0146826. [Google Scholar] [CrossRef]
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flügge, U.-I.; Kunze, R. Transcription Analysis of Arabidopsis Membrane Transporters and Hormone Pathways during Developmental and Induced Leaf Senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef] [Green Version]
- Schippers, J.H.M.; Nunes-Nesi, A.; Apetrei, R.; Hille, J.; Fernie, A.R.; Dijkwel, P.P. The Arabidopsis onset of leaf death5 Mutation of Quinolinate Synthase Affects Nicotinamide Adenine Dinucleotide Biosynthesis and Causes Early Ageing. Plant Cell 2008, 20, 2909–2925. [Google Scholar] [CrossRef] [Green Version]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Köhler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvino, L.; Guzmán-Benito, I.; Del Toro, F.J.; Donaire, L.; Castro-Sanz, A.B.; Ruíz-Ferrer, V.; Llave, C. Activation of senescence-associated Dark-inducible (DIN) genes during infection contributes to enhanced susceptibility to plant viruses. Mol. Plant Pathol. 2016, 17, 3–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner, N.; Mittelsten Scheid, O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 2010, 22, 3118–3129. [Google Scholar] [CrossRef] [Green Version]
- Knoth, C.; Ringler, J.; Dangl, J.L.; Eulgem, T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 2007, 20, 120–128. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Venegas, R.; Abdallat, A.A.; Guo, M.; Alfano, J.R.; Avramova, Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2007, 2, 106–113. [Google Scholar] [CrossRef] [Green Version]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Hu, Y.; Dong, Q.; Yu, D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012, 185–186, 288–297. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Avramova, Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005, 33, 5199–5207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.M.; To, T.K.; Ishida, J.; Matsui, A.; Kimura, H.; Seki, M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 847–856. [Google Scholar] [CrossRef]
- Janack, B.; Sosoi, P.; Krupinska, K.; Humbeck, K. Knockdown of WHIRLY1 Affects Drought Stress-Induced Leaf Senescence and Histone Modifications of the Senescence-Associated Gene HvS40. Plants 2016, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- Mengel, A.; Ageeva, A.; Georgii, E.; Bernhardt, J.; Wu, K.; Durner, J.; Lindermayr, C. Nitric Oxide Modulates Histone Acetylation at Stress Genes by Inhibition of Histone Deacetylases. Plant Physiol. 2017, 173, 1434–1452. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.T.; Luo, M.; Wang, Y.Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.T.; Wu, K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 2010, 5, 1318–1320. [Google Scholar] [CrossRef] [Green Version]
- Becker, W.; Apel, K. Differences in gene expression between natural and artificially induced leaf senescence. Planta 1993, 189, 74–79. [Google Scholar] [CrossRef]
- Mehta, A.; Haber, J.E. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 2014, 6, a016428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuteja, N.; Singh, M.B.; Misra, M.K.; Bhalla, P.L.; Tuteja, R. Molecular mechanisms of DNA damage and repair: Progress in plants. Crit. Rev. Biochem. Mol. Biol. 2001, 36, 337–397. [Google Scholar] [CrossRef] [PubMed]
- White, R.R.; Vijg, J. Do DNA Double-Strand Breaks Drive Aging? Mol. Cell 2016, 63, 729–738. [Google Scholar] [CrossRef] [Green Version]
- Waterworth, W.M.; Footitt, S.; Bray, C.M.; Finch-Savage, W.E.; West, C.E. DNA damage checkpoint kinase ATM regulates germination and maintains genome stability in seeds. Proc. Natl. Acad. Sci. USA 2016, 113, 9647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britt, A.B. DNA damage and repair in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 75–100. [Google Scholar] [CrossRef]
- Culligan, K.M.; Robertson, C.E.; Foreman, J.; Doerner, P.; Britt, A.B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006, 48, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.; Conklin, P.A.; Huefner, N.D.; Britt, A.B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef] [Green Version]
- Du, L.L.; Nakamura, T.M.; Russell, P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006, 20, 1583–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drury, G.E.; Dowle, A.A.; Ashford, D.A.; Waterworth, W.M.; Thomas, J.; West, C.E. Dynamics of plant histone modifications in response to DNA damage. Biochem. J. 2012, 445, 393–401. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Kim, J.H.; Kim, J.; Lyu, J.I.; Zhang, Y.; Guo, H.; Nam, H.G.; Woo, H.R. ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. New Phytol. 2020, 227, 473–484. [Google Scholar] [CrossRef]
- Freitas, A.A.; de Magalhães, J.P. A review and appraisal of the DNA damage theory of ageing. Mutat. Res. 2011, 728, 12–22. [Google Scholar] [CrossRef]
- Pulido, A.; Laufs, P. Co-ordination of developmental processes by small RNAs during leaf development. J. Exp. Bot. 2010, 61, 1277–1291. [Google Scholar] [CrossRef] [Green Version]
- Rubio-Somoza, I.; Weigel, D. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 2011, 16, 258–264. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Ito, H. Small RNAs and transposon silencing in plants. Dev. Growth Differ. 2012, 54, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of Jasmonate Biosynthesis and Senescence by miR319 Targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [Green Version]
- Debernardi, J.M.; Mecchia, M.A.; Vercruyssen, L.; Smaczniak, C.; Kaufmann, K.; Inze, D.; Rodriguez, R.E.; Palatnik, J.F. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014, 79, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Lee, I.C.; Kim, J.; Kim, H.J.; Ryu, J.S.; Woo, H.R.; Nam, H.G. Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 2010, 61, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef] [Green Version]
- Arikit, S.; Xia, R.; Kakrana, A.; Huang, K.; Zhai, J.; Yan, Z.; Valdés-López, O.; Prince, S.; Musket, T.A.; Nguyen, H.T.; et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 2014, 26, 4584–4601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [Green Version]
- Luan, W.; Dai, Y.; Li, X.-Y.; Wang, Y.; Tao, X.; Li, C.-X.; Mao, P.; Ma, X.-R. Identification of tRFs and phasiRNAs in tomato (Solanum lycopersicum) and their responses to exogenous abscisic acid. BMC Plant Biol. 2020, 20, 320. [Google Scholar] [CrossRef] [PubMed]
- Miryeganeh, M. Synchronization of senescence and desynchronization of flowering in Arabidopsis thaliana. AoB Plants 2020, 12, plaa018. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Paszkowski, J. DNA and histone methylation in plants. Trends Genet. 2004, 20, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Tsugane, K.; Kanno, T.; Nishimura, T. DNA methylation in plants: Relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 2012, 53, 766–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miryeganeh, M.; Saze, H. Epigenetic inheritance and plant evolution. Popul. Ecol. 2020, 62, 17–27. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miryeganeh, M. Epigenetic Mechanisms of Senescence in Plants. Cells 2022, 11, 251. https://doi.org/10.3390/cells11020251
Miryeganeh M. Epigenetic Mechanisms of Senescence in Plants. Cells. 2022; 11(2):251. https://doi.org/10.3390/cells11020251
Chicago/Turabian StyleMiryeganeh, Matin. 2022. "Epigenetic Mechanisms of Senescence in Plants" Cells 11, no. 2: 251. https://doi.org/10.3390/cells11020251
APA StyleMiryeganeh, M. (2022). Epigenetic Mechanisms of Senescence in Plants. Cells, 11(2), 251. https://doi.org/10.3390/cells11020251





