HIV-1 Infection of Long-Lived Hematopoietic Precursors In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Cell Isolation and Culture
2.3. Antibodies Used for Flow Cytometry
2.4. Virus Preparation and Transductions
2.4.1. Replication-Incompetent Pseudotyped Reporter Viruses
2.4.2. Replication-Competent Reporter Viruses
2.4.3. Replication-Competent Wild-Type HIV-1 Viruses
2.4.4. Virus Titer Determination
2.5. Inhibition Experiments
2.6. Colony-Formation Assays
2.7. Replating Assay
2.8. Analysis of env Sequences from HIV+ Bone Marrow
2.9. qPCR-Assay
2.10. Confocal Imaging
2.11. Correlation Analysis
3. Results
3.1. In Vitro Infection/Transduction of Cord Blood and Bone Marrow-Derived HSPCs with HIV-1
3.2. Co-Expression of CD4 and HIV Co-Receptors on HSPC Subsets
3.3. HIV (Co-)Receptor Expression and Localization on HSPCs Using Confocal Micrographs
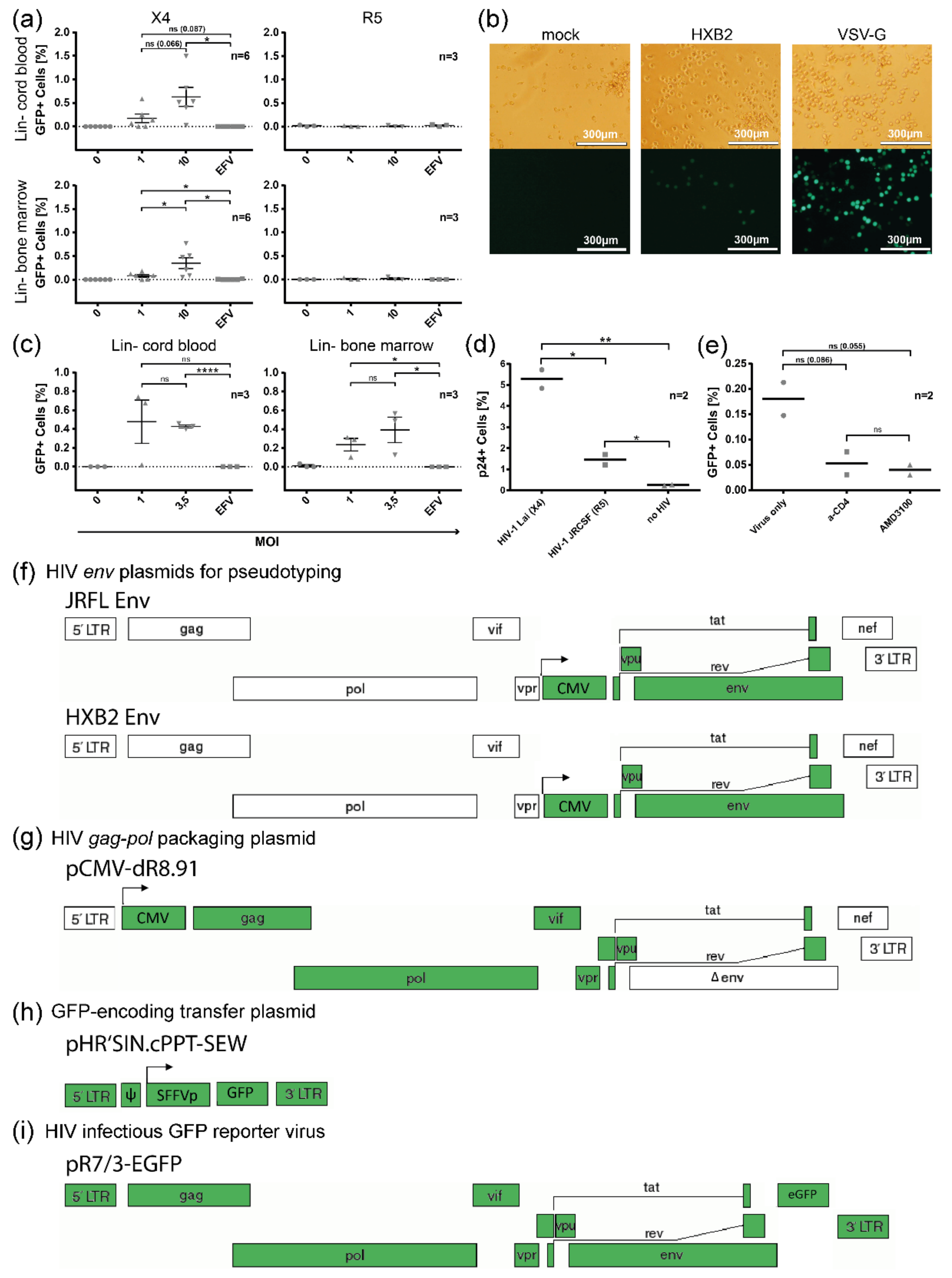
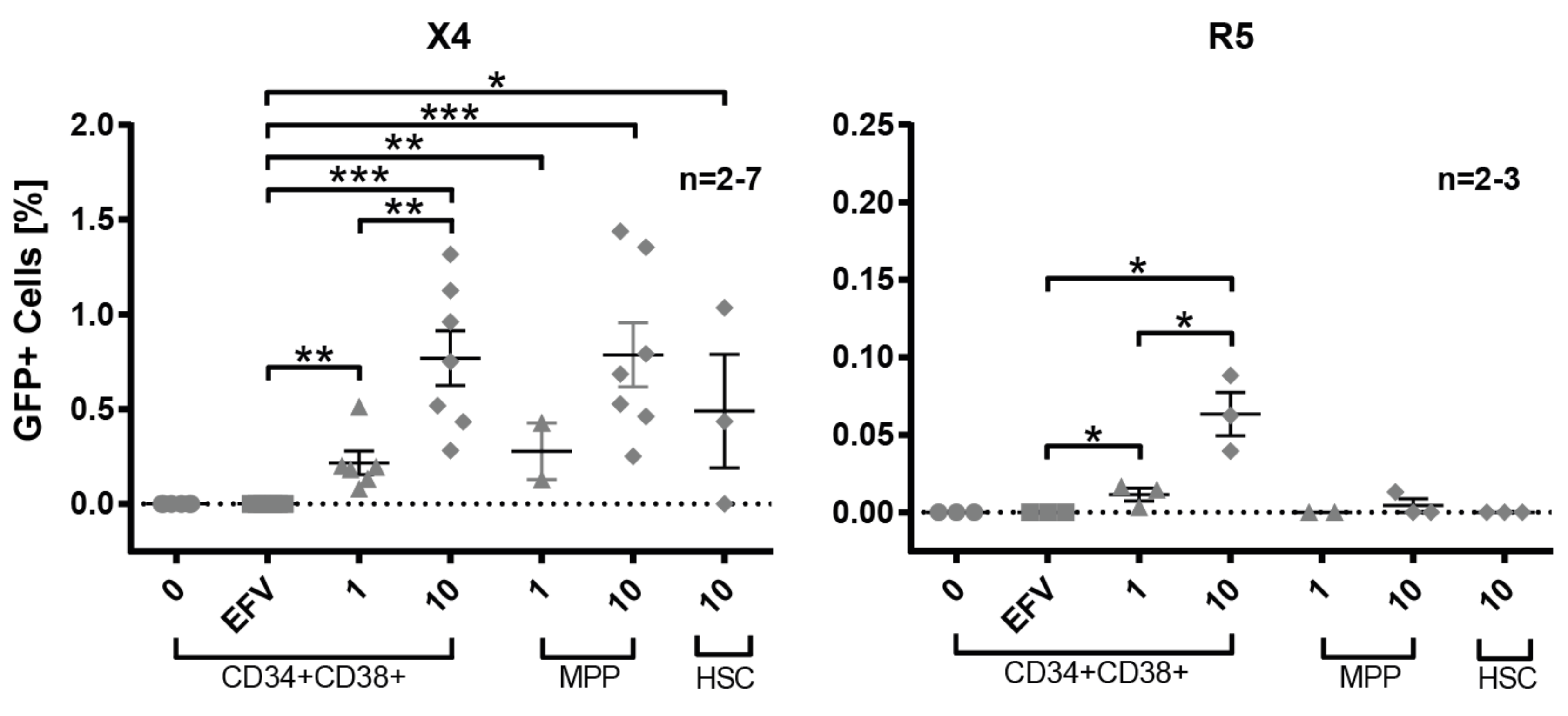
3.4. Susceptibility of Defined HSPC Subsets to X4 and R5-Mediated HIV-1 Entry
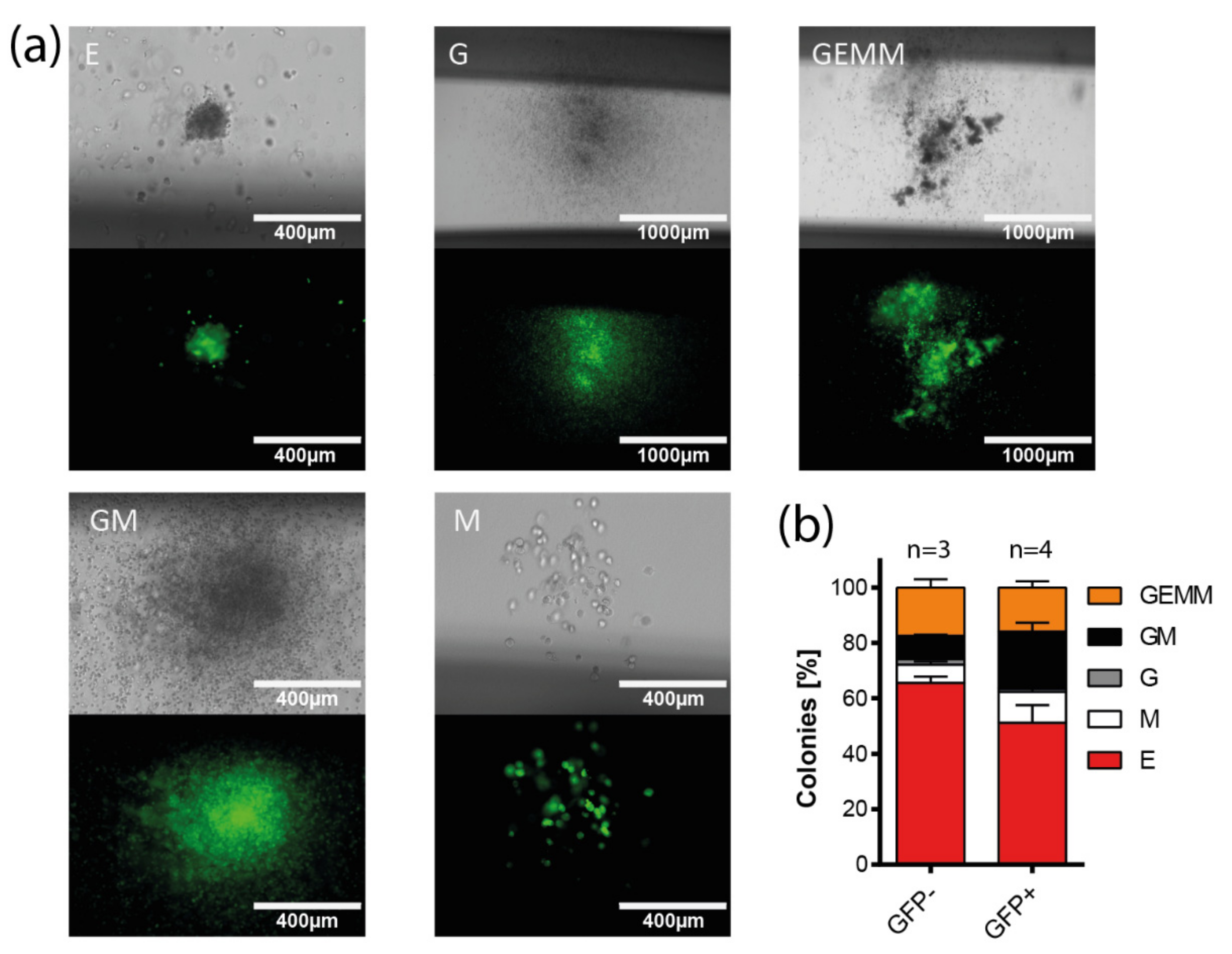
3.5. Multi-Lineage Colony Formation Capacity of HIV-1-Transduced Hematopoietic Progenitors
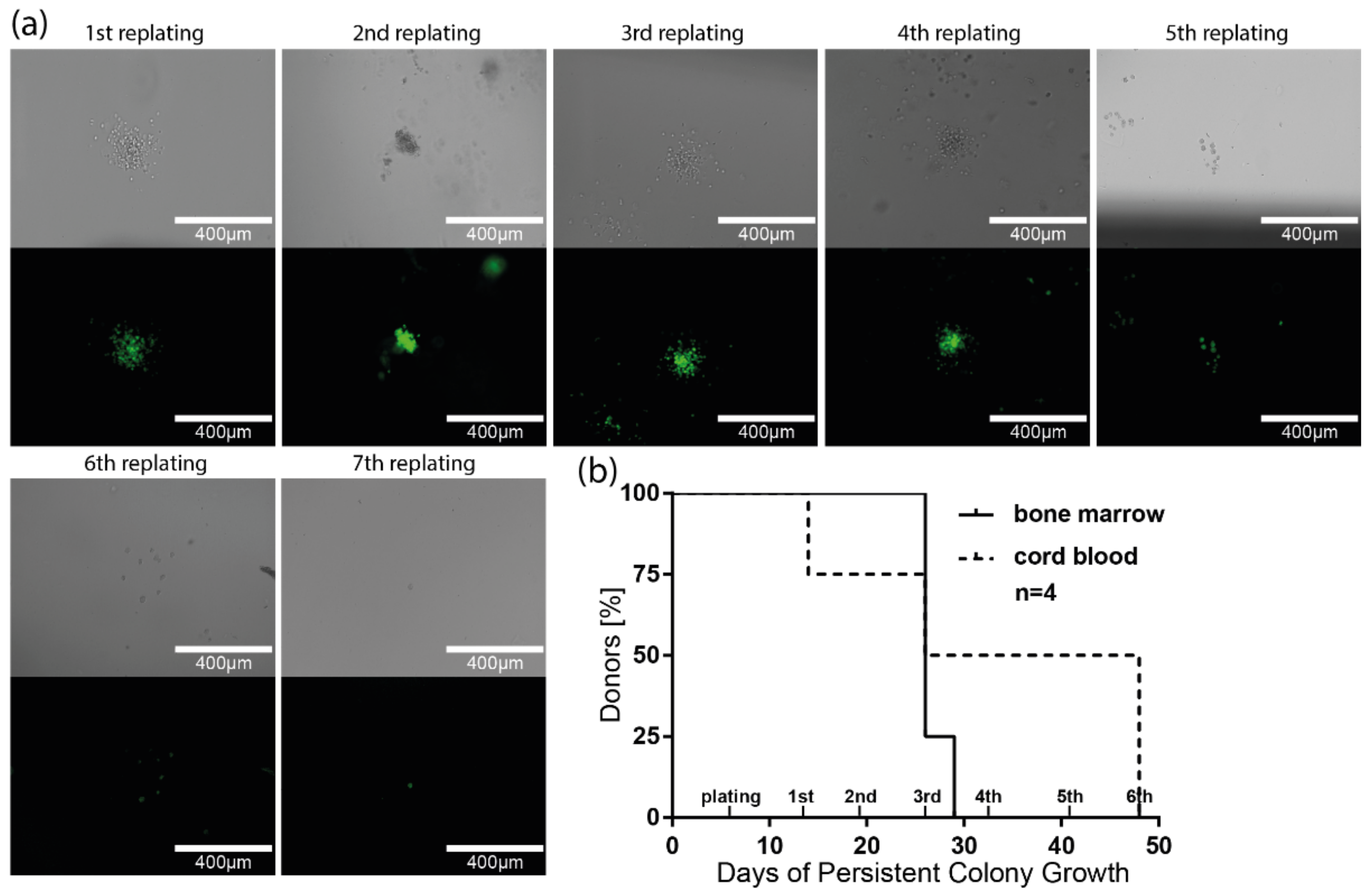
3.6. Long-Term Colony-Forming Capacity of HIV-1-Susceptible HSPCs
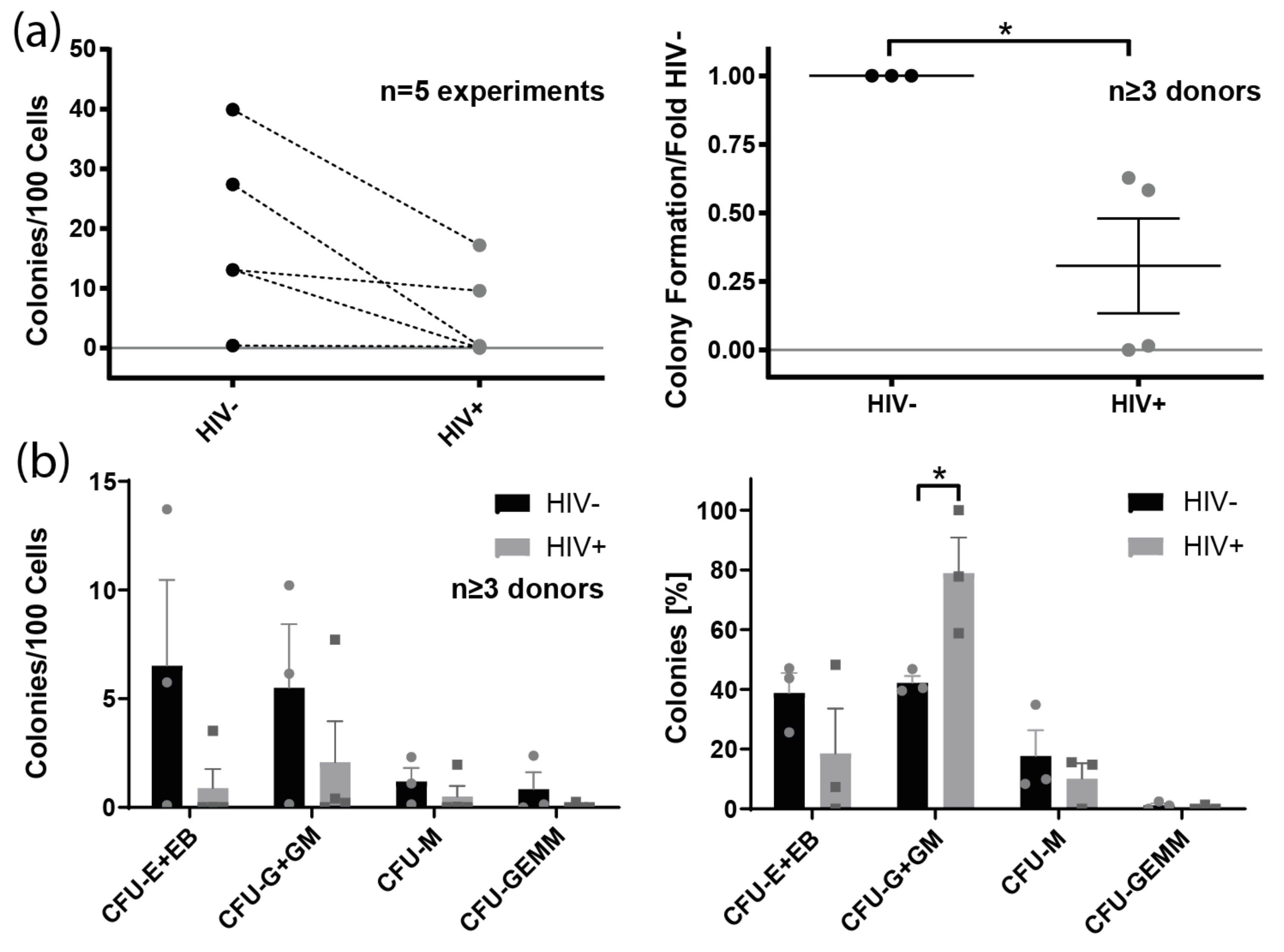
3.7. Colony Formation of Bone Marrow-Derived Lin− Cells from HIV+ Individuals
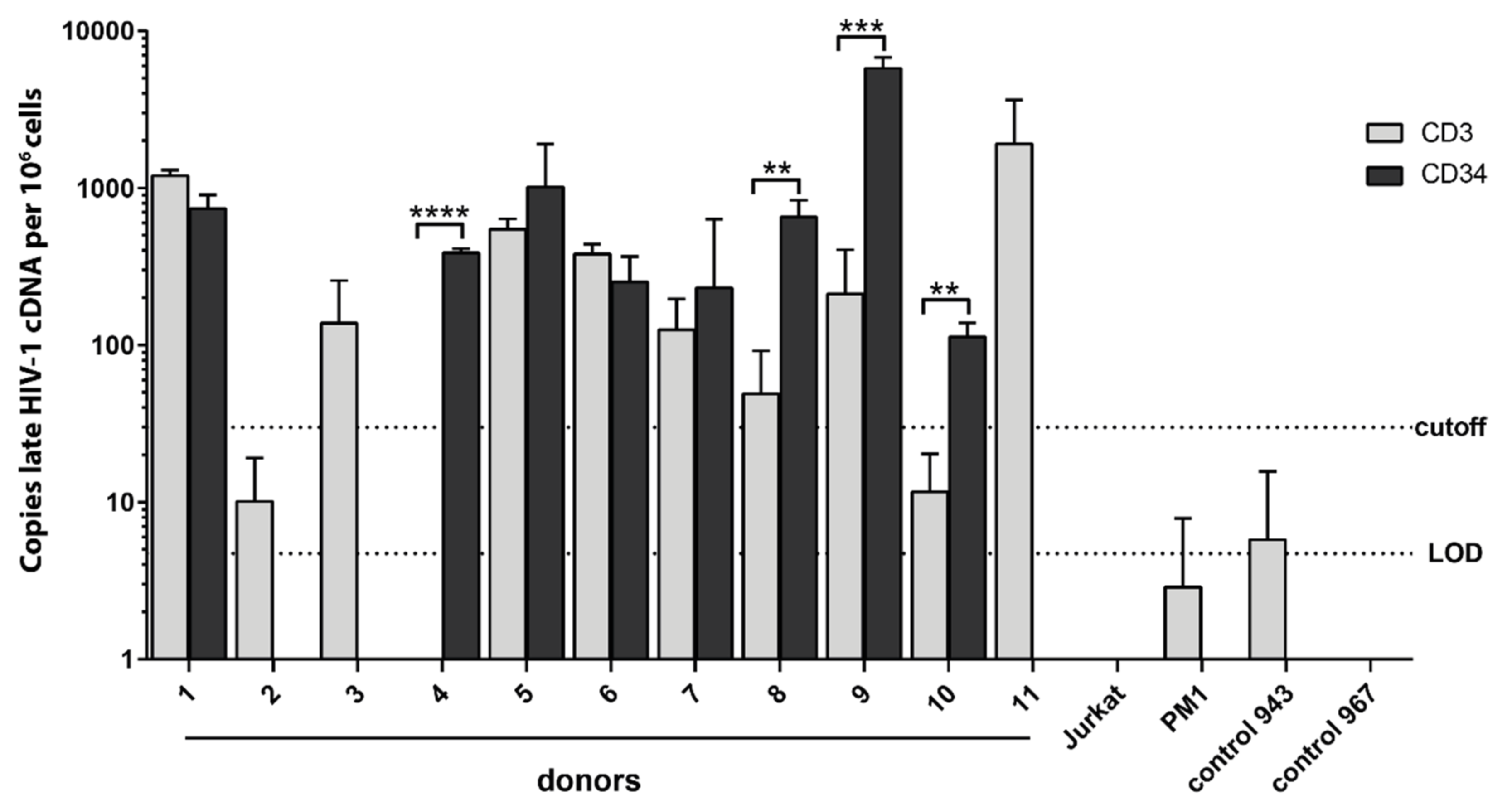
3.8. Clinical Evidence for HIV-1-Infected HSPCs in Bone Marrow from HIV-1-Infected Individuals
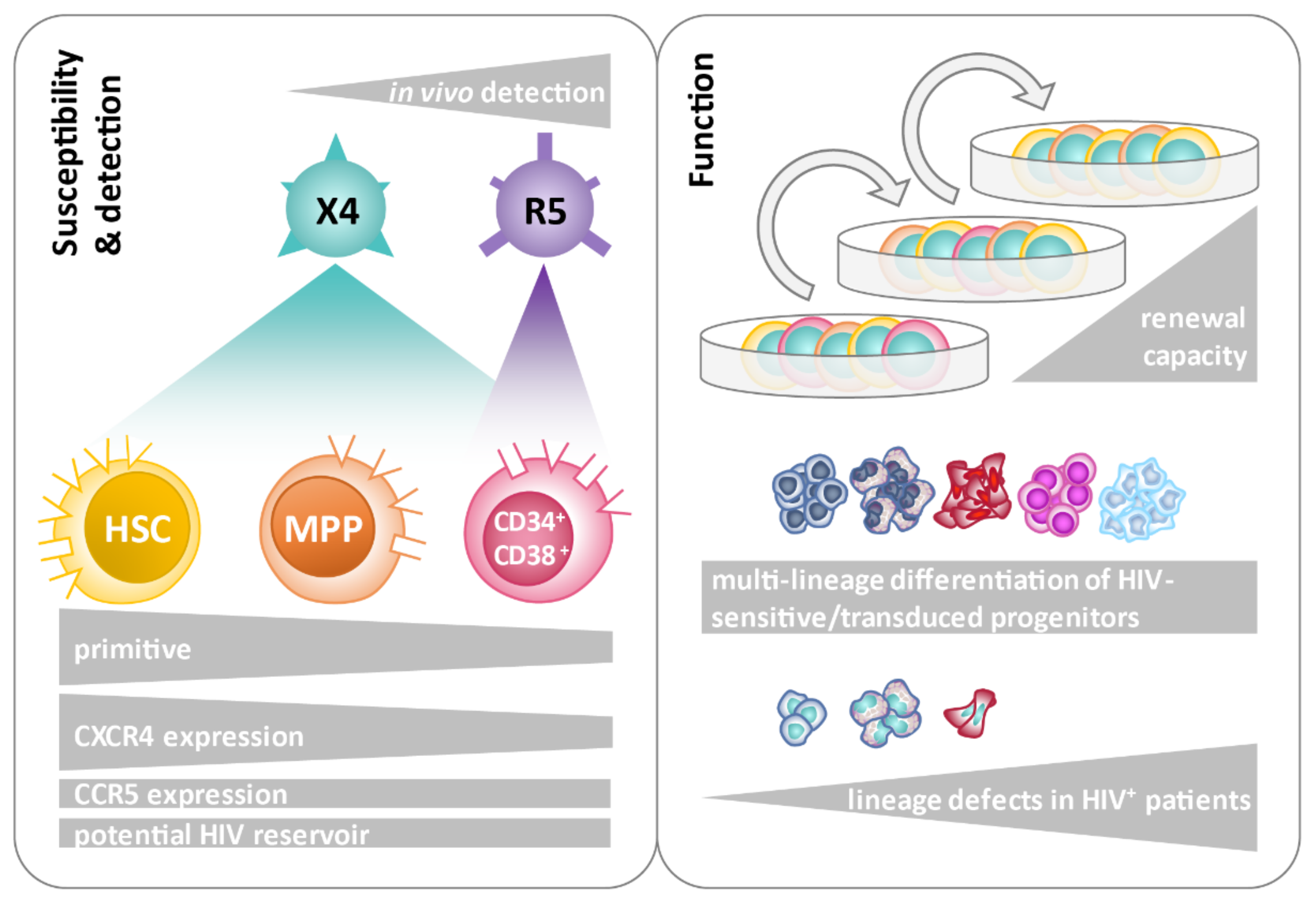
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davey, R.T., Jr.; Bhat, N.; Yoder, C.; Chun, T.W.; Metcalf, J.A.; Dewar, R.; Natarajan, V.; Lempicki, R.A.; Adelsberger, J.W.; Miller, K.D.; et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA 1999, 96, 15109–15114. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.W.; Finzi, D.; Margolick, J.; Chadwick, K.; Schwartz, D.; Siliciano, R.F. In vivo fate of HIV-1-infected T cells: Quantitative analysis of the transition to stable latency. Nat. Med. 1995, 1, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef] [PubMed]
- Kulpa, D.A.; Brehm, J.H.; Fromentin, R.; Cooper, A.; Cooper, C.; Ahlers, J.; Chomont, N.; Sekaly, R.P. The immunological synapse: The gateway to the HIV reservoir. Immunol. Rev. 2013, 254, 305–325. [Google Scholar] [CrossRef]
- Chun, T.W.; Engel, D.; Mizell, S.B.; Ehler, L.A.; Fauci, A.S. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 1998, 188, 83–91. [Google Scholar] [CrossRef]
- Devadas, K.; Hardegen, N.J.; Wahl, L.M.; Hewlett, I.K.; Clouse, K.A.; Yamada, K.M.; Dhawan, S. Mechanisms for macrophage-mediated HIV-1 induction. J. Immunol. 2004, 173, 6735–6744. [Google Scholar] [CrossRef]
- Trono, D.; Van Lint, C.; Rouzioux, C.; Verdin, E.; Barre-Sinoussi, F.; Chun, T.W.; Chomont, N. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 2010, 329, 174–180. [Google Scholar] [CrossRef]
- Blankson, J.N.; Persaud, D.; Siliciano, R.F. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 2002, 53, 557–593. [Google Scholar] [CrossRef]
- Castro-Gonzalez, S.; Colomer-Lluch, M.; Serra-Moreno, R. Barriers for HIV Cure: The Latent Reservoir. AIDS Res. Hum. Retrovir. 2018, 34, 739–759. [Google Scholar] [CrossRef]
- Sadowski, I.; Hashemi, F.B. Strategies to eradicate HIV from infected patients: Elimination of latent provirus reservoirs. Cell Mol. Life Sci. 2019, 76, 3583–3600. [Google Scholar] [CrossRef]
- Anderson, E.M.; Maldarelli, F. The role of integration and clonal expansion in HIV infection: Live long and prosper. Retrovirology 2018, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Chahroudi, A.; Silvestri, G.; Lichterfeld, M. T memory stem cells and HIV: A long-term relationship. Curr. HIV/AIDS Rep. 2015, 12, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Henrich, T.J.; Deeks, S.G.; Pillai, S.K. Measuring the Size of the Latent Human Immunodeficiency Virus Reservoir: The Present and Future of Evaluating Eradication Strategies. J. Infect. Dis. 2017, 215, S134–S141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Borducchi, E.N.; Liu, J.; Nkolola, J.P.; Cadena, A.M.; Yu, W.H.; Fischinger, S.; Broge, T.; Abbink, P.; Mercado, N.B.; Chandrashekar, A.; et al. Publisher Correction: Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 2018, 564, E8. [Google Scholar] [CrossRef]
- Li, P.; Kaiser, P.; Lampiris, H.W.; Kim, P.; Yukl, S.A.; Havlir, D.V.; Greene, W.C.; Wong, J.K. Stimulating the RIG-I pathway to kill cells in the latent HIV reservoir following viral reactivation. Nat. Med. 2016, 22, 807–811. [Google Scholar] [CrossRef]
- Dash, P.K.; Kaminski, R.; Bella, R.; Su, H.; Mathews, S.; Ahooyi, T.M.; Chen, C.; Mancuso, P.; Sariyer, R.; Ferrante, P.; et al. Sequential LASER ART and CRISPR Treatments Eliminate HIV-1 in a Subset of Infected Humanized Mice. Nat. Commun. 2019, 10, 2753. [Google Scholar] [CrossRef]
- Lorenzi, J.C.; Cohen, Y.Z.; Cohn, L.B.; Kreider, E.F.; Barton, J.P.; Learn, G.H.; Oliveira, T.; Lavine, C.L.; Horwitz, J.A.; Settler, A.; et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E7908–E7916. [Google Scholar] [CrossRef]
- Gaebler, C.; Lorenzi, J.C.C.; Oliveira, T.Y.; Nogueira, L.; Ramos, V.; Lu, C.L.; Pai, J.A.; Mendoza, P.; Jankovic, M.; Caskey, M.; et al. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J. Exp. Med. 2019, 216, 2253–2264. [Google Scholar] [CrossRef]
- Pardons, M.; Baxter, A.E.; Massanella, M.; Pagliuzza, A.; Fromentin, R.; Dufour, C.; Leyre, L.; Routy, J.P.; Kaufmann, D.E.; Chomont, N. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog. 2019, 15, e1007619. [Google Scholar] [CrossRef]
- Sanyal, A.; Mailliard, R.B.; Rinaldo, C.R.; Ratner, D.; Ding, M.; Chen, Y.; Zerbato, J.M.; Giacobbi, N.S.; Venkatachari, N.J.; Patterson, B.K.; et al. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4(+) T cells. Nat. Med. 2017, 23, 885–889. [Google Scholar] [CrossRef]
- Whitney, J.B.; Hill, A.L.; Sanisetty, S.; Penaloza-MacMaster, P.; Liu, J.; Shetty, M.; Parenteau, L.; Cabral, C.; Shields, J.; Blackmore, S.; et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014, 512, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.; Gaebler, C.; Olveira, T.; Ramos, V.; Saad, M.; Lorenzi, J.C.C.; Gazumyan, A.; Moir, S.; Caskey, M.; Chun, T.W.; et al. Longitudinal clonal dynamics of HIV-1 latent reservoirs measured by combination quadruplex polymerase chain reaction and sequencing. Proc. Natl. Acad. Sci. USA 2022, 119, e2117630119. [Google Scholar] [CrossRef] [PubMed]
- Einkauf, K.B.; Osborn, M.R.; Gao, C.; Sun, W.; Sun, X.; Lian, X.; Parsons, E.M.; Gladkov, G.T.; Seiger, K.W.; Blackmer, J.E.; et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell 2022, 185, 266–282.e15. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, B.; Jutte, B.B.; Love, L.; Assi, W.; Roux, J.; Sonnerborg, A.; Tezil, T.; Verdin, E.; Svensson, J.P. T cell stimulation remodels the latently HIV-1 infected cell population by differential activation of proviral chromatin. PLoS Pathog. 2022, 18, e1010555. [Google Scholar] [CrossRef]
- Palmer, S.; Maldarelli, F.; Wiegand, A.; Bernstein, B.; Hanna, G.J.; Brun, S.C.; Kempf, D.J.; Mellors, J.W.; Coffin, J.M.; King, M.S. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2008, 105, 3879–3884. [Google Scholar] [CrossRef]
- Eisele, E.; Siliciano, R.F. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012, 37, 377–388. [Google Scholar] [CrossRef]
- Vibholm, L.K.; Lorenzi, J.C.C.; Pai, J.A.; Cohen, Y.Z.; Oliveira, T.Y.; Barton, J.P.; Garcia Noceda, M.; Lu, C.L.; Ablanedo-Terrazas, Y.; Del Rio Estrada, P.M.; et al. Characterization of Intact Proviruses in Blood and Lymph Node from HIV-Infected Individuals Undergoing Analytical Treatment Interruption. J. Virol. 2019, 93, e01920-18. [Google Scholar] [CrossRef]
- Ruiz, A.; Blanch-Lombarte, O.; Jimenez-Moyano, E.; Ouchi, D.; Mothe, B.; Pena, R.; Galvez, C.; Genesca, M.; Martinez-Picado, J.; Goulder, P.; et al. Antigen Production After Latency Reversal and Expression of Inhibitory Receptors in CD8+ T Cells Limit the Killing of HIV-1 Reactivated Cells. Front. Immunol. 2018, 9, 3162. [Google Scholar] [CrossRef]
- Darcis, G.; Das, A.T.; Berkhout, B. Tackling HIV Persistence: Pharmacological versus CRISPR-Based Shock Strategies. Viruses 2018, 10, 157. [Google Scholar] [CrossRef]
- Lichterfeld, M. Reactivation of latent HIV moves shock-and-kill treatments forward. Nature 2020, 578, 42–43. [Google Scholar] [CrossRef]
- McDougal, J.S.; Kennedy, M.S.; Sligh, J.M.; Cort, S.P.; Mawle, A.; Nicholson, J.K. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science 1986, 231, 382–385. [Google Scholar] [CrossRef] [PubMed]
- Kazazi, F.; Mathijs, J.M.; Foley, P.; Cunningham, A.L. Variations in CD4 expression by human monocytes and macrophages and their relationships to infection with the human immunodeficiency virus. J. Gen. Virol. 1989, 70 Pt 10, 2661–2672. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Broder, C.C.; Kennedy, P.E.; Berger, E.A. HIV-1 entry cofactor: Functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996, 272, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Liu, R.; Ellmeier, W.; Choe, S.; Unutmaz, D.; Burkhart, M.; Di Marzio, P.; Marmon, S.; Sutton, R.E.; Hill, C.M.; et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996, 381, 661–666. [Google Scholar] [CrossRef]
- Schuitemaker, H.; Koot, M.; Kootstra, N.A.; Dercksen, M.W.; de Goede, R.E.; van Steenwijk, R.P.; Lange, J.M.; Schattenkerk, J.K.; Miedema, F.; Tersmette, M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: Progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 1992, 66, 1354–1360. [Google Scholar] [CrossRef]
- Connor, R.I.; Sheridan, K.E.; Ceradini, D.; Choe, S.; Landau, N.R. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J. Exp. Med. 1997, 185, 621–628. [Google Scholar] [CrossRef]
- Li, X.; Xue, Y.; Zhou, L.; Lin, Y.; Yu, X.; Wang, X.; Zhen, X.; Zhang, W.; Ning, Z.; Yue, Q.; et al. Evidence that HIV-1 CRF01_AE is associated with low CD4+T cell count and CXCR4 co-receptor usage in recently infected young men who have sex with men (MSM) in Shanghai, China. PLoS ONE 2014, 9, e89462. [Google Scholar] [CrossRef] [PubMed]
- Kouri, V.; Khouri, R.; Aleman, Y.; Abrahantes, Y.; Vercauteren, J.; Pineda-Pena, A.C.; Theys, K.; Megens, S.; Moutschen, M.; Pfeifer, N.; et al. CRF19_cpx is an Evolutionary fit HIV-1 Variant Strongly Associated With Rapid Progression to AIDS in Cuba. EBioMedicine 2015, 2, 244–254. [Google Scholar] [CrossRef]
- Fouchier, R.A.; Groenink, M.; Kootstra, N.A.; Tersmette, M.; Huisman, H.G.; Miedema, F.; Schuitemaker, H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 1992, 66, 3183–3187. [Google Scholar] [CrossRef]
- Lengauer, T.; Sander, O.; Sierra, S.; Thielen, A.; Kaiser, R. Bioinformatics prediction of HIV coreceptor usage. Nat. Biotechnol. 2007, 25, 1407–1410. [Google Scholar] [CrossRef]
- Delgado, E.; Fernandez-Garcia, A.; Vega, Y.; Cuevas, T.; Pinilla, M.; Garcia, V.; Sanchez, M.; Gonzalez, M.; Sanchez, A.M.; Thomson, M.M.; et al. Evaluation of genotypic tropism prediction tests compared with in vitro co-receptor usage in HIV-1 primary isolates of diverse subtypes. J. Antimicrob. Chemother. 2012, 67, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Mbondji-Wonje, C.; Ragupathy, V.; Zhao, J.; Nanfack, A.; Lee, S.; Torimiro, J.; Nyambi, P.; Hewlett, I.K. Genotypic prediction of tropism of highly diverse HIV-1 strains from Cameroon. PLoS ONE 2014, 9, e112434. [Google Scholar] [CrossRef]
- Gupta, S.; Neogi, U.; Srinivasa, H.; Shet, A. Performance of genotypic tools for prediction of tropism in HIV-1 subtype C V3 loop sequences. Intervirology 2015, 58, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Riemenschneider, M.; Cashin, K.Y.; Budeus, B.; Sierra, S.; Shirvani-Dastgerdi, E.; Bayanolhagh, S.; Kaiser, R.; Gorry, P.R.; Heider, D. Genotypic Prediction of Co-receptor Tropism of HIV-1 Subtypes A and C. Sci. Rep. 2016, 6, 24883. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.R.; Sedaghat, A.R.; Kieffer, T.; Brennan, T.; Lee, P.K.; Wind-Rotolo, M.; Haggerty, C.M.; Kamireddi, A.R.; Liu, Y.; Lee, J.; et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 2006, 80, 6441–6457. [Google Scholar] [CrossRef] [PubMed]
- Kruize, Z.; Kootstra, N.A. The Role of Macrophages in HIV-1 Persistence and Pathogenesis. Front. Microbiol. 2019, 10, 2828. [Google Scholar] [CrossRef]
- Mullins, J.I.; Frenkel, L.M. Clonal Expansion of Human Immunodeficiency Virus-Infected Cells and Human Immunodeficiency Virus Persistence During Antiretroviral Therapy. J. Infect. Dis. 2017, 215, S119–S127. [Google Scholar] [CrossRef]
- Satou, Y.; Katsuya, H.; Fukuda, A.; Misawa, N.; Ito, J.; Uchiyama, Y.; Miyazato, P.; Islam, S.; Fassati, A.; Melamed, A.; et al. Dynamics and mechanisms of clonal expansion of HIV-1-infected cells in a humanized mouse model. Sci. Rep. 2017, 7, 6913. [Google Scholar] [CrossRef]
- Simonetti, F.R.; Sobolewski, M.D.; Fyne, E.; Shao, W.; Spindler, J.; Hattori, J.; Anderson, E.M.; Watters, S.A.; Hill, S.; Wu, X.; et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 1883–1888. [Google Scholar] [CrossRef]
- Buzon, M.J.; Sun, H.; Li, C.; Shaw, A.; Seiss, K.; Ouyang, Z.; Martin-Gayo, E.; Leng, J.; Henrich, T.J.; Li, J.Z.; et al. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med. 2014, 20, 139–142. [Google Scholar] [CrossRef]
- Brennan, T.P.; Woods, J.O.; Sedaghat, A.R.; Siliciano, J.D.; Siliciano, R.F.; Wilke, C.O. Analysis of Human Immunodeficiency Virus Type 1 Viremia and Provirus in Resting Cd4+ T Cells Reveals a Novel Source of Residual Viremia in Patients on Antiretroviral Therapy. J. Virol. 2009, 83, 8470–8481. [Google Scholar] [CrossRef] [PubMed]
- Sahu, G.K.; Paar, D.; Frost, S.D.; Smith, M.M.; Weaver, S.; Cloyd, M.W. Low-Level Plasma Hivs in Patients on Prolonged Suppressive Highly Active Antiretroviral Therapy Are Produced Mostly by Cells Other Than Cd4 T-Cells. J. Med. Virol. 2009, 81, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zalar, A.; Figueroa, M.I.; Ruibal-Ares, B.; Bare, P.; Cahn, P.; de Bracco, M.M.; Belmonte, L. Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antivir. Res. 2010, 87, 269–271. [Google Scholar] [CrossRef]
- Cribbs, S.K.; Lennox, J.; Caliendo, A.M.; Brown, L.A.; Guidot, D.M. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res. Hum. Retrovir. 2015, 31, 64–70. [Google Scholar] [CrossRef] [PubMed]
- McIlroy, D.; Autran, B.; Cheynier, R.; Wain-Hobson, S.; Clauvel, J.P.; Oksenhendler, E.; Debre, P.; Hosmalin, A. Infection frequency of dendritic cells and CD4+ T lymphocytes in spleens of human immunodeficiency virus-positive patients. J. Virol. 1995, 69, 4737–4745. [Google Scholar] [CrossRef] [PubMed]
- Popov, S.; Chenine, A.L.; Gruber, A.; Li, P.L.; Ruprecht, R.M. Long-term productive human immunodeficiency virus infection of CD1a-sorted myeloid dendritic cells. J. Virol. 2005, 79, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, H.; Herbst, H.; Niedobitek, G.; Foss, H.D.; Stein, H. Follicular dendritic cells are a major reservoir for human immunodeficiency virus type 1 in lymphoid tissues facilitating infection of CD4+ T-helper cells. Am. J. Pathol. 1992, 140, 15–22. [Google Scholar]
- Zhang, J.; Perelson, A.S. Contribution of follicular dendritic cells to persistent HIV viremia. J. Virol. 2013, 87, 7893–7901. [Google Scholar] [CrossRef]
- Neal, T.F.; Holland, H.K.; Baum, C.M.; Villinger, F.; Ansari, A.A.; Saral, R.; Wingard, J.R.; Fleming, W.H. CD34+ progenitor cells from asymptomatic patients are not a major reservoir for human immunodeficiency virus-1. Blood 1995, 86, 1749–1756. [Google Scholar] [CrossRef]
- Sebastian, N.T.; Collins, K.L. Targeting HIV latency: Resting memory T cells, hematopoietic progenitor cells and future directions. Expert Rev. Anti-Infect. Ther. 2014, 12, 1187–1201. [Google Scholar] [CrossRef][Green Version]
- Jenkins, M.; Hanley, M.B.; Moreno, M.B.; Wieder, E.; McCune, J.M. Human immunodeficiency virus-1 infection interrupts thymopoiesis and multilineage hematopoiesis in vivo. Blood 1998, 91, 2672–2678. [Google Scholar] [CrossRef] [PubMed]
- Moses, A.; Nelson, J.; Bagby, G.C., Jr. The influence of human immunodeficiency virus-1 on hematopoiesis. Blood 1998, 91, 1479–1495. [Google Scholar] [CrossRef] [PubMed]
- Koka, P.S.; Fraser, J.K.; Bryson, Y.; Bristol, G.C.; Aldrovandi, G.M.; Daar, E.S.; Zack, J.A. Human immunodeficiency virus inhibits multilineage hematopoiesis in vivo. J. Virol. 1998, 72, 5121–5127. [Google Scholar] [CrossRef]
- Vishnu, P.; Aboulafia, D.M. Haematological manifestations of human immune deficiency virus infection. Br. J. Haematol. 2015, 171, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Crumpacker, C. Hematopoietic Stem and Immune Cells in Chronic HIV Infection. Stem Cells Int. 2015, 2015, 148064. [Google Scholar] [CrossRef]
- Li, G.; Zhao, J.; Cheng, L.; Jiang, Q.; Kan, S.; Qin, E.; Tu, B.; Zhang, X.; Zhang, L.; Su, L.; et al. HIV-1 infection depletes human CD34+CD38- hematopoietic progenitor cells via pDC-dependent mechanisms. PLoS Pathog. 2017, 13, e1006505. [Google Scholar] [CrossRef]
- Guo, X.; He, S.; Lv, X.; Ding, H.; Li, S.; Kang, J.; Liu, J.; Qin, C.; Geng, W.; Jiang, Y.; et al. The Role of HIV-1 in Affecting the Proliferation Ability of HPCs Derived From BM. J. Acquir. Immune. Defic. Syndr. 2016, 71, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Xing, J.; Wang, F.; Chen, X.; Liu, Q.; Wang, J.; Zou, S.; Chen, L.; Fu, X.; Zhou, Z.; et al. HIV-1LAI Nef blocks the development of hematopoietic stem/progenitor cells into T lymphoid cells. AIDS 2021, 35, 851–860. [Google Scholar] [CrossRef]
- Zou, W.; Xing, J.; Zou, S.; Jiang, M.; Chen, X.; Chen, Q.; Liu, D.; Zhang, X.; Fu, X. HIV-1LAI Nef blocks the development of hematopoietic stem/progenitor cells into myeloid-erythroid lineage cells. Biol. Direct 2021, 16, 27. [Google Scholar] [CrossRef]
- Slobod, K.S.; Bennett, T.A.; Freiden, P.J.; Kechli, A.M.; Howlett, N.; Flynn, P.M.; Head, D.R.; Srivastava, D.K.; Boyett, J.M.; Brenner, M.K.; et al. Mobilization of CD34+ progenitor cells by granulocyte colony-stimulating factor in human immunodeficiency virus type 1-infected adults. Blood 1996, 88, 3329–3335. [Google Scholar] [CrossRef]
- Von Laer, D.; Hufert, F.T.; Fenner, T.E.; Schwander, S.; Dietrich, M.; Schmitz, H.; Kern, P. CD34+ hematopoietic progenitor cells are not a major reservoir of the human immunodeficiency virus. Blood 1990, 76, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Zauli, G.; Re, M.C.; Davis, B.; Sen, L.; Visani, G.; Gugliotta, L.; Furlini, G.; La Placa, M. Impaired in vitro growth of purified (CD34+) hematopoietic progenitors in human immunodeficiency virus-1 seropositive thrombocytopenic individuals. Blood 1992, 79, 2680–2687. [Google Scholar] [CrossRef] [PubMed]
- Stanley, S.K.; Kessler, S.W.; Justement, J.S.; Schnittman, S.M.; Greenhouse, J.J.; Brown, C.C.; Musongela, L.; Musey, K.; Kapita, B.; Fauci, A.S. CD34+ bone marrow cells are infected with HIV in a subset of seropositive individuals. J. Immunol. 1992, 149, 689–697. [Google Scholar]
- Redd, A.D.; Avalos, A.; Essex, M. Infection of hematopoietic progenitor cells by HIV-1 subtype C, and its association with anemia in southern Africa. Blood 2007, 110, 3143–3149. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.C.; Onafuwa-Nuga, A.; McNamara, L.A.; Riddell, J.t.; Bixby, D.; Savona, M.R.; Collins, K.L. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat. Med. 2010, 16, 446–451. [Google Scholar] [CrossRef]
- Carter, C.C.; McNamara, L.A.; Onafuwa-Nuga, A.; Shackleton, M.; Riddell, J.t.; Bixby, D.; Savona, M.R.; Morrison, S.J.; Collins, K.L. HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe 2011, 9, 223–234. [Google Scholar] [CrossRef]
- Josefsson, L.; Eriksson, S.; Sinclair, E.; Ho, T.; Killian, M.; Epling, L.; Shao, W.; Lewis, B.; Bacchetti, P.; Loeb, L.; et al. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J. Infect. Dis. 2012, 206, 28–34. [Google Scholar] [CrossRef]
- Durand, C.M.; Ghiaur, G.; Siliciano, J.D.; Rabi, S.A.; Eisele, E.E.; Salgado, M.; Shan, L.; Lai, J.F.; Zhang, H.; Margolick, J.; et al. HIV-1 DNA is detected in bone marrow populations containing CD4+ T cells but is not found in purified CD34+ hematopoietic progenitor cells in most patients on antiretroviral therapy. J. Infect. Dis. 2012, 205, 1014–1018. [Google Scholar] [CrossRef]
- McNamara, L.A.; Ganesh, J.A.; Collins, K.L. Latent HIV-1 infection occurs in multiple subsets of hematopoietic progenitor cells and is reversed by NF-kappaB activation. J. Virol. 2012, 86, 9337–9350. [Google Scholar] [CrossRef]
- McNamara, L.A.; Onafuwa-Nuga, A.; Sebastian, N.T.; Riddell, J.t.; Bixby, D.; Collins, K.L. CD133+ hematopoietic progenitor cells harbor HIV genomes in a subset of optimally treated people with long-term viral suppression. J. Infect. Dis. 2013, 207, 1807–1816. [Google Scholar] [CrossRef]
- Cheung, A.K.L.; Huang, Y.; Kwok, H.Y.; Chen, M.; Chen, Z. Latent human cytomegalovirus enhances HIV-1 infection in CD34(+) progenitor cells. Blood Adv. 2017, 1, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, T.; Okada, S. The use of RetroNectin in studies requiring in vitro HIV-1 infection of human hematopoietic stem/progenitor cells. J. Virol. Methods 2017, 248, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Laurenti, E.; Dick, J.E. Hematopoiesis: A human perspective. Cell Stem Cell 2012, 10, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Louache, F.; Debili, N.; Marandin, A.; Coulombel, L.; Vainchenker, W. Expression of CD4 by human hematopoietic progenitors. Blood 1994, 84, 3344–3355. [Google Scholar] [CrossRef] [PubMed]
- Zauli, G.; Furlini, G.; Vitale, M.; Re, M.C.; Gibellini, D.; Zamai, L.; Visani, G.; Borgatti, P.; Capitani, S.; La Placa, M. A subset of human CD34+ hematopoietic progenitors express low levels of CD4, the high-affinity receptor for human immunodeficiency virus-type 1. Blood 1994, 84, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Banda, N.K.; Simon, G.R.; Sipple, J.D.; Terrell, K.L.; Archer, P.; Shpall, E.J.; Akkina, R.K.; Myers, A.M.; Harrison, G.S. Depletion of CD34+ CD4+ cells in bone marrow from HIV-1-infected individuals. Biol Blood Marrow Transpl. 1999, 5, 162–172. [Google Scholar] [CrossRef]
- Shen, H.; Cheng, T.; Preffer, F.I.; Dombkowski, D.; Tomasson, M.H.; Golan, D.E.; Yang, O.; Hofmann, W.; Sodroski, J.G.; Luster, A.D.; et al. Intrinsic human immunodeficiency virus type 1 resistance of hematopoietic stem cells despite coreceptor expression. J. Virol. 1999, 73, 728–737. [Google Scholar] [CrossRef]
- Muench, M.O.; Roncarolo, M.G.; Namikawa, R. Phenotypic and functional evidence for the expression of CD4 by hematopoietic stem cells isolated from human fetal liver. Blood 1997, 89, 1364–1375. [Google Scholar] [CrossRef]
- Zhang, Y.; Foudi, A.; Geay, J.F.; Berthebaud, M.; Buet, D.; Jarrier, P.; Jalil, A.; Vainchenker, W.; Louache, F. Intracellular localization and constitutive endocytosis of CXCR4 in human CD34+ hematopoietic progenitor cells. Stem Cells 2004, 22, 1015–1029. [Google Scholar] [CrossRef]
- Aiuti, A.; Turchetto, L.; Cota, M.; Cipponi, A.; Brambilla, A.; Arcelloni, C.; Paroni, R.; Vicenzi, E.; Bordignon, C.; Poli, G. Human CD34(+) cells express CXCR4 and its ligand stromal cell-derived factor-1. Implications for infection by T-cell tropic human immunodeficiency virus. Blood 1999, 94, 62–73. [Google Scholar] [CrossRef]
- Nixon, C.C.; Vatakis, D.N.; Reichelderfer, S.N.; Dixit, D.; Kim, S.G.; Uittenbogaart, C.H.; Zack, J.A. HIV-1 infection of hematopoietic progenitor cells in vivo in humanized mice. Blood 2013, 122, 2195–2204. [Google Scholar] [CrossRef] [PubMed]
- Bordoni, V.; Bibas, M.; Abbate, I.; Viola, D.; Rozera, G.; Agrati, C.; Rinaldi, A.; Amendola, A.; Ammassari, A.; Capobianchi, M.R.; et al. Bone marrow CD34+ progenitor cells may harbour HIV-DNA even in successfully treated patients. Clin. Microbiol. Infect. 2015, 21, 290.e5–290.e8. [Google Scholar] [CrossRef]
- Sebastian, N.T.; Zaikos, T.D.; Terry, V.; Taschuk, F.; McNamara, L.A.; Onafuwa-Nuga, A.; Yucha, R.; Signer, R.A.J.; Riddell, J.I.; Bixby, D.; et al. CD4 is expressed on a heterogeneous subset of hematopoietic progenitors, which persistently harbor CXCR4 and CCR5-tropic HIV proviral genomes in vivo. PLoS Pathog. 2017, 13, e1006509. [Google Scholar] [CrossRef]
- Painter, M.M.; Zaikos, T.D.; Collins, K.L. Quiescence Promotes Latent HIV Infection and Resistance to Reactivation from Latency with Histone Deacetylase Inhibitors. J. Virol. 2017, 91, e01080-17. [Google Scholar] [CrossRef] [PubMed]
- Zaikos, T.D.; Terry, V.H.; Sebastian Kettinger, N.T.; Lubow, J.; Painter, M.M.; Virgilio, M.C.; Neevel, A.; Taschuk, F.; Onafuwa-Nuga, A.; McNamara, L.A.; et al. Hematopoietic Stem and Progenitor Cells Are a Distinct HIV Reservoir that Contributes to Persistent Viremia in Suppressed Patients. Cell Rep. 2018, 25, 3759–3773.e9. [Google Scholar] [CrossRef] [PubMed]
- Dervillez, X.; Huther, A.; Schuhmacher, J.; Griesinger, C.; Cohen, J.H.; von Laer, D.; Dietrich, U. Stable expression of soluble therapeutic peptides in eukaryotic cells by multimerisation: Application to the HIV-1 fusion inhibitory peptide C46. ChemMedChem 2006, 1, 330–339. [Google Scholar] [CrossRef]
- Naldini, L.; Blomer, U.; Gallay, P.; Ory, D.; Mulligan, R.; Gage, F.H.; Verma, I.M.; Trono, D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996, 272, 263–267. [Google Scholar] [CrossRef]
- Demaison, C.; Parsley, K.; Brouns, G.; Scherr, M.; Battmer, K.; Kinnon, C.; Grez, M.; Thrasher, A.J. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 2002, 13, 803–813. [Google Scholar] [CrossRef]
- Lohrengel, S.; Hermann, F.; Hagmann, I.; Oberwinkler, H.; Scrivano, L.; Hoffmann, C.; von Laer, D.; Dittmar, M.T. Determinants of human immunodeficiency virus type 1 resistance to membrane-anchored gp41-derived peptides. J. Virol. 2005, 79, 10237–10246. [Google Scholar] [CrossRef]
- Baldauf, H.M.; Stegmann, L.; Schwarz, S.M.; Ambiel, I.; Trotard, M.; Martin, M.; Burggraf, M.; Lenzi, G.M.; Lejk, H.; Pan, X.; et al. Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc. Natl. Acad. Sci. USA 2017, 114, 2729–2734. [Google Scholar] [CrossRef]
- Stephan, C.; Baldauf, H.M.; Barry, J.; Giordano, F.A.; Bartholomae, C.C.; Haberl, A.; Bickel, M.; Schmidt, M.; Laufs, S.; Kaderali, L.; et al. Impact of raltegravir on HIV-1 RNA and DNA forms following initiation of antiretroviral therapy in treatment-naive patients. J. Antimicrob. Chemother. 2014, 69, 2809–2818. [Google Scholar] [CrossRef] [PubMed]
- Endres, M.J.; Clapham, P.R.; Marsh, M.; Ahuja, M.; Turner, J.D.; McKnight, A.; Thomas, J.F.; Stoebenau-Haggarty, B.; Choe, S.; Vance, P.J.; et al. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 1996, 87, 745–756. [Google Scholar] [CrossRef]
- R_Core_Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2014. Available online: http://www.R-project.org/ (accessed on 15 December 2021).
- RStudio. RStudio Team: A Bundle of RStudio’s Popular Professional Software for Statistical Data Analysis, Package Management, and Sharing Data Products; RStudio, Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Wain-Hobson, S.; Vartanian, J.P.; Henry, M.; Chenciner, N.; Cheynier, R.; Delassus, S.; Martins, L.P.; Sala, M.; Nugeyre, M.T.; Guetard, D.; et al. LAV revisited: Origins of the early HIV-1 isolates from Institut Pasteur. Science 1991, 252, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Bozzano, F.; Marras, F.; Ascierto, M.L.; Cantoni, C.; Cenderello, G.; Dentone, C.; Di Biagio, A.; Orofino, G.; Mantia, E.; Boni, S.; et al. ‘Emergency exit’ of bone-marrow-resident CD34(+)DNAM-1(bright)CXCR4(+)-committed lymphoid precursors during chronic infection and inflammation. Nat. Commun. 2015, 6, 8109. [Google Scholar] [CrossRef]
- Griffin, D.O.; Goff, S.P. HIV-1 Is Restricted prior to Integration of Viral DNA in Primary Cord-Derived Human CD34+ Cells. J. Virol. 2015, 89, 8096–8100. [Google Scholar] [CrossRef] [PubMed]
- Heinzelmann, A.; Stein, S.; Kohl, U.; Dietrich, U.; von Briesen, H. Regulation of HIV-1 infection in cells derived from purified CD34+ cells through manipulation of APOBEC3G expression. Curr. HIV Res. 2010, 8, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Dao Thi, V.L.; Huang, Y.; Billerbeck, E.; Saha, D.; Hoffmann, H.H.; Wang, Y.; Silva, L.A.V.; Sarbanes, S.; Sun, T.; et al. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 2018, 172, 423–438.e25. [Google Scholar] [CrossRef]
- Brummendorf, T.H.; Dragowska, W.; Zijlmans, J.; Thornbury, G.; Lansdorp, P.M. Asymmetric cell divisions sustain long-term hematopoiesis from single-sorted human fetal liver cells. J. Exp. Med. 1998, 188, 1117–1124. [Google Scholar] [CrossRef]
- Cho, R.H.; Muller-Sieburg, C.E. High frequency of long-term culture-initiating cells retain in vivo repopulation and self-renewal capacity. Exp. Hematol. 2000, 28, 1080–1086. [Google Scholar] [CrossRef]
- Hazenberg, M.D.; Stuart, J.W.; Otto, S.A.; Borleffs, J.C.; Boucher, C.A.; de Boer, R.J.; Miedema, F.; Hamann, D. T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: A longitudinal analysis in patients before and during highly active antiretroviral therapy (HAART). Blood 2000, 95, 249–255. [Google Scholar] [CrossRef]
- Mohri, H.; Perelson, A.S.; Tung, K.; Ribeiro, R.M.; Ramratnam, B.; Markowitz, M.; Kost, R.; Hurley, A.; Weinberger, L.; Cesar, D.; et al. Increased turnover of T lymphocytes in HIV-1 infection and its reduction by antiretroviral therapy. J. Exp. Med. 2001, 194, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Toggas, S.M.; Masliah, E.; Rockenstein, E.M.; Rall, G.F.; Abraham, C.R.; Mucke, L. Central nervous system damage produced by expression of the HIV-1 coat protein gp120 in transgenic mice. Nature 1994, 367, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Luke, B.T.; Henry, A.R.; Darko, S.; Brandt, L.D.; Su, L.; Sun, D.; Wells, D.; Joseph, K.W.; Demirov, D.; et al. HIV infected CD4+ T cell clones are more stable than uninfected clones during long-term antiretroviral therapy. PLoS Pathog. 2022, 18, e1010726. [Google Scholar] [CrossRef] [PubMed]
- Banin, A.N.; Tuen, M.; Bimela, J.S.; Tongo, M.; Zappile, P.; Khodadadi-Jamayran, A.; Nanfack, A.J.; Meli, J.; Wang, X.; Mbanya, D.; et al. Development of a Versatile, Near Full Genome Amplification and Sequencing Approach for a Broad Variety of HIV-1 Group M Variants. Viruses 2019, 11, 317. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.; Chackerian, B. Why HIV virions have low numbers of envelope spikes: Implications for vaccine development. PLoS Pathog. 2014, 10, e1004254. [Google Scholar] [CrossRef]
- Louder, M.K.; Sambor, A.; Chertova, E.; Hunte, T.; Barrett, S.; Ojong, F.; Sanders-Buell, E.; Zolla-Pazner, S.; McCutchan, F.E.; Roser, J.D.; et al. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 2005, 339, 226–238. [Google Scholar] [CrossRef]
- Stella, C.C.; Ganser, A.; Hoelzer, D. Defective in vitro growth of the hemopoietic progenitor cells in the acquired immunodeficiency syndrome. J. Clin. Invest. 1987, 80, 286–293. [Google Scholar] [CrossRef]
- Louache, F.; Henri, A.; Bettaieb, A.; Oksenhendler, E.; Raguin, G.; Tulliez, M.; Vainchenker, W. Role of human immunodeficiency virus replication in defective in vitro growth of hematopoietic progenitors. Blood 1992, 80, 2991–2999. [Google Scholar] [CrossRef]
- Costantini, A.; Giuliodoro, S.; Mancini, S.; Butini, L.; Regnery, C.M.; Silvestri, G.; Greco, F.; Leoni, P.; Montroni, M. Impaired in-vitro growth of megakaryocytic colonies derived from CD34 cells of HIV-1-infected patients with active viral replication. AIDS 2006, 20, 1713–1720. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renelt, S.; Schult-Dietrich, P.; Baldauf, H.-M.; Stein, S.; Kann, G.; Bickel, M.; Kielland-Kaisen, U.; Bonig, H.; Marschalek, R.; Rieger, M.A.; et al. HIV-1 Infection of Long-Lived Hematopoietic Precursors In Vitro and In Vivo. Cells 2022, 11, 2968. https://doi.org/10.3390/cells11192968
Renelt S, Schult-Dietrich P, Baldauf H-M, Stein S, Kann G, Bickel M, Kielland-Kaisen U, Bonig H, Marschalek R, Rieger MA, et al. HIV-1 Infection of Long-Lived Hematopoietic Precursors In Vitro and In Vivo. Cells. 2022; 11(19):2968. https://doi.org/10.3390/cells11192968
Chicago/Turabian StyleRenelt, Sebastian, Patrizia Schult-Dietrich, Hanna-Mari Baldauf, Stefan Stein, Gerrit Kann, Markus Bickel, Ulrikke Kielland-Kaisen, Halvard Bonig, Rolf Marschalek, Michael A. Rieger, and et al. 2022. "HIV-1 Infection of Long-Lived Hematopoietic Precursors In Vitro and In Vivo" Cells 11, no. 19: 2968. https://doi.org/10.3390/cells11192968
APA StyleRenelt, S., Schult-Dietrich, P., Baldauf, H.-M., Stein, S., Kann, G., Bickel, M., Kielland-Kaisen, U., Bonig, H., Marschalek, R., Rieger, M. A., Dietrich, U., & Duerr, R. (2022). HIV-1 Infection of Long-Lived Hematopoietic Precursors In Vitro and In Vivo. Cells, 11(19), 2968. https://doi.org/10.3390/cells11192968






