Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model of T1D
2.2. Cell Culture and miRNA Transfection
2.3. RNA Extraction
2.4. Microarray Expression Profiles
2.5. Microarray Data Analyses
2.6. Quantitative Real-Time PCR Analysis (qRT-PCR)
2.7. Luciferase Assay
2.8. Western Blot
2.9. Statistical Analysis
3. Results
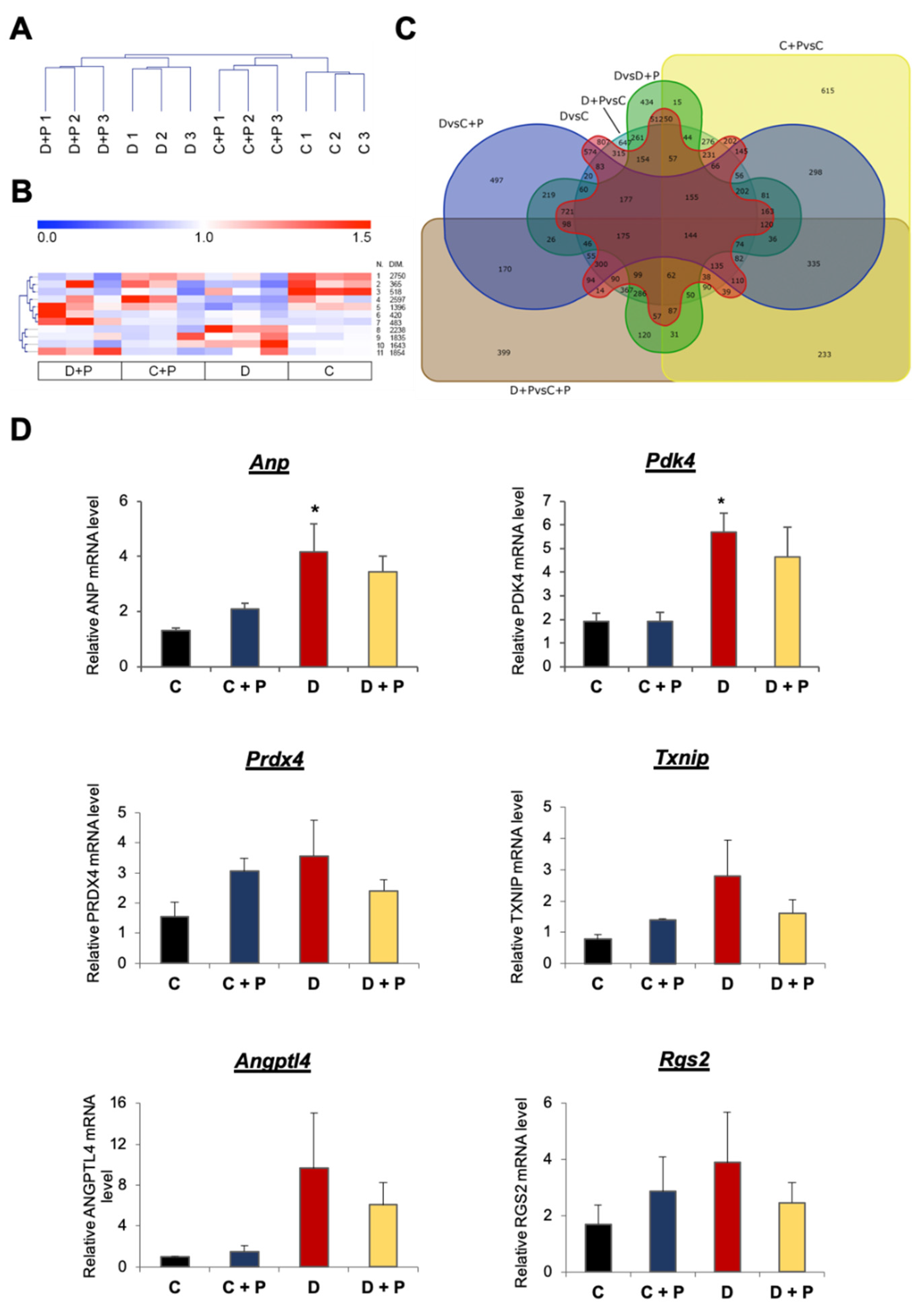
3.1. mRNA Expression Analysis
3.2. Validation of the mRNA Microarray
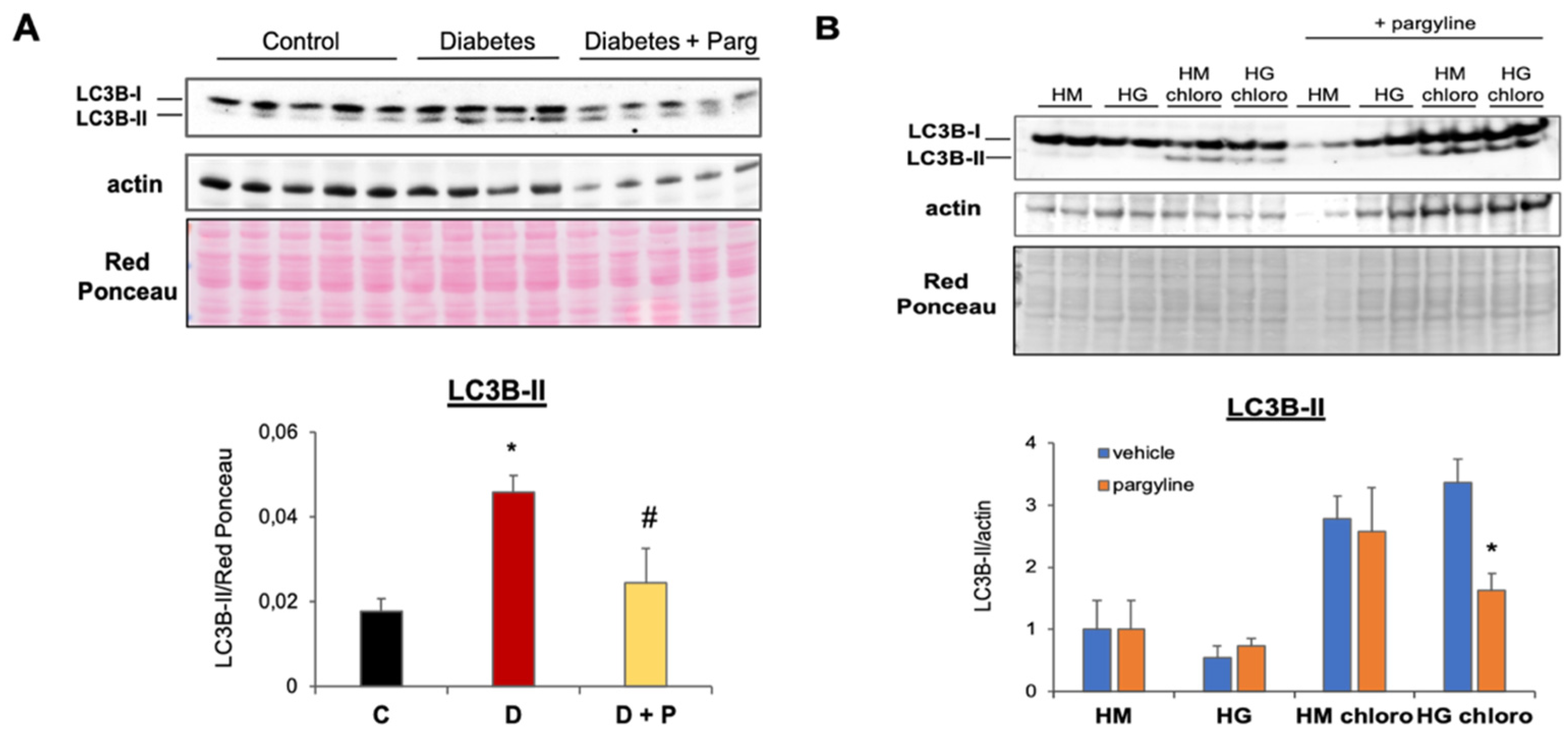
3.3. MAO Inhibition Reduces Aberrant Autophagy Flux Activation in T1D Hearts
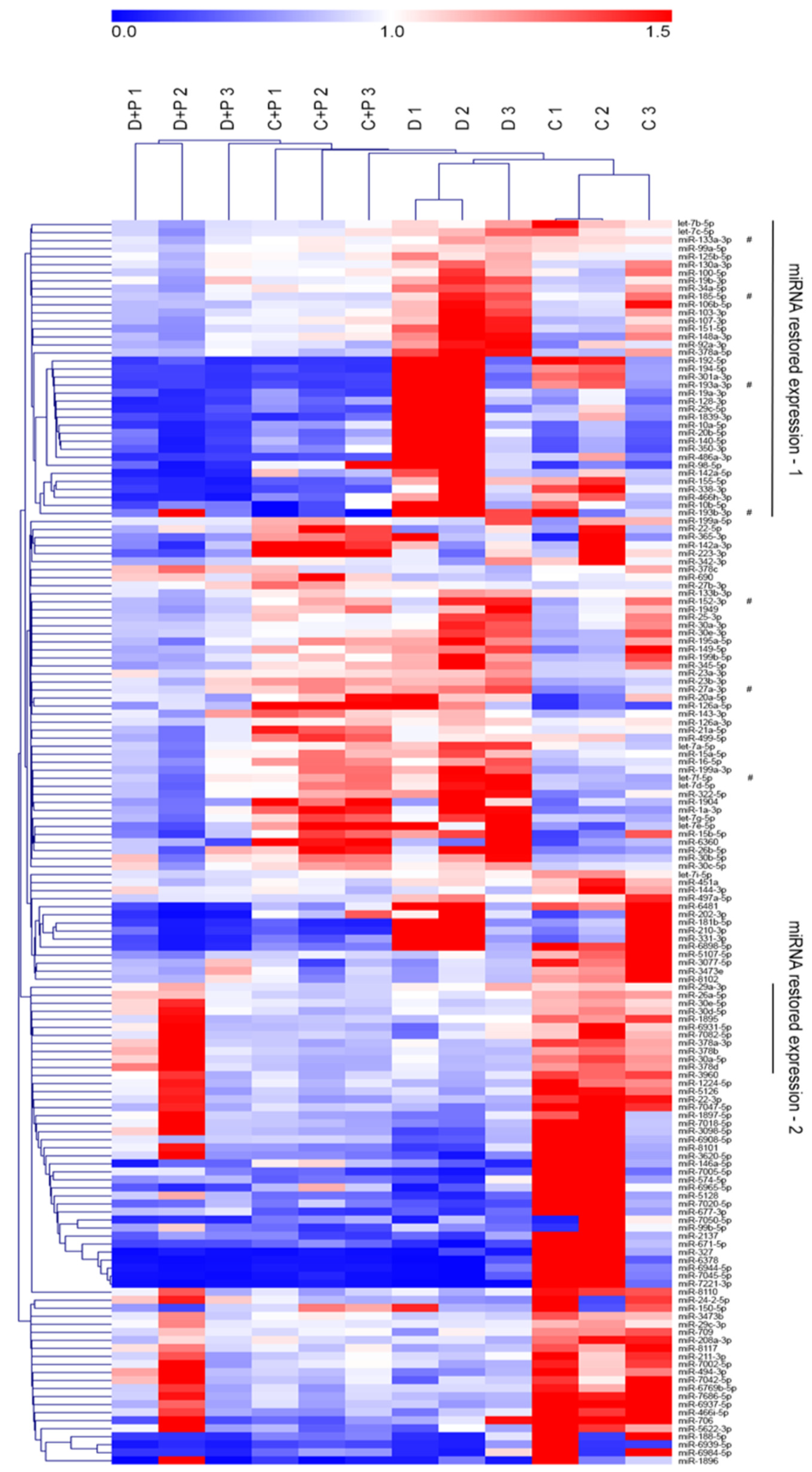
3.4. MiRNA Gene Expression Analysis
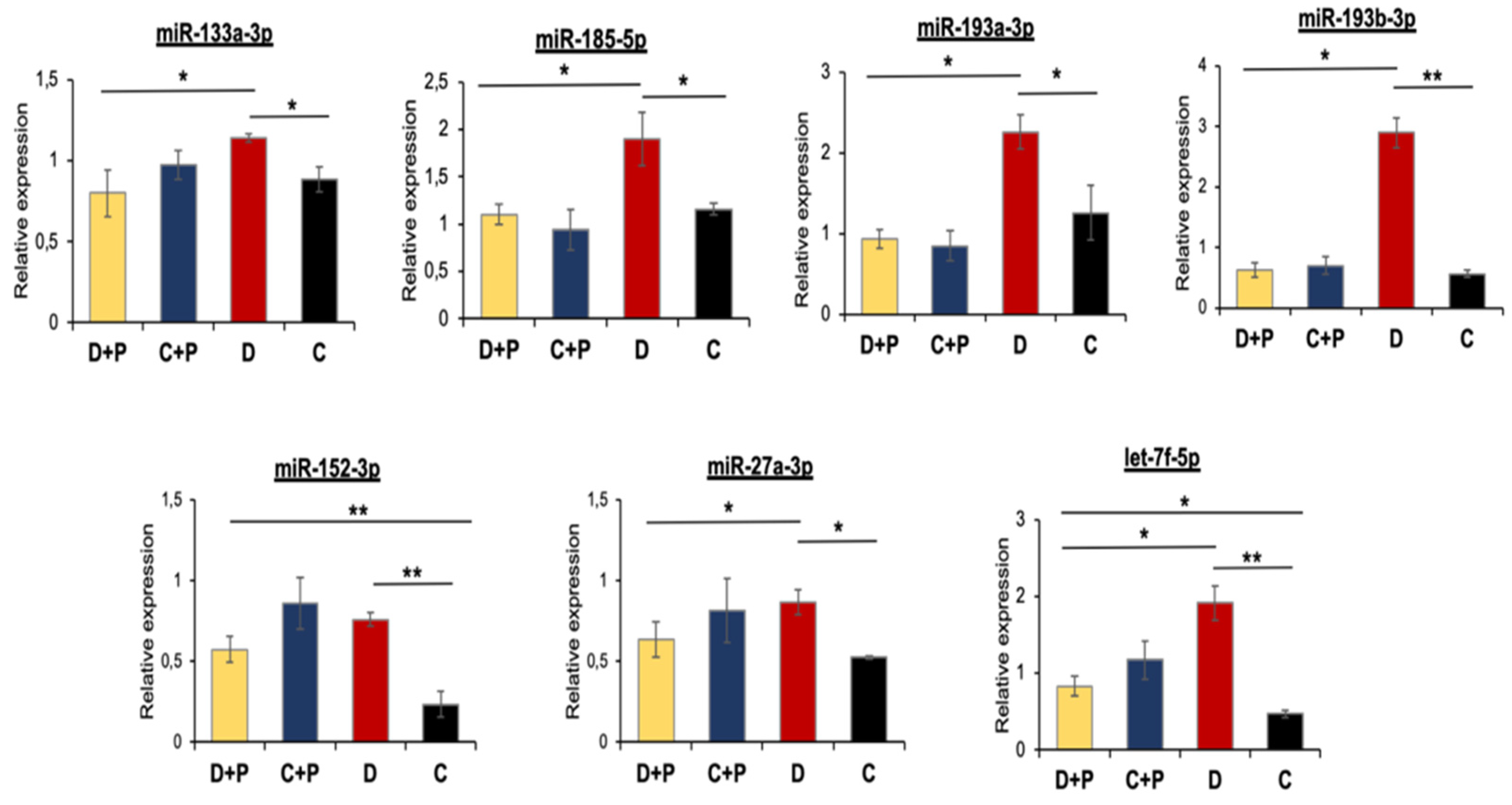
3.5. Validation of miRNA Microarray Data by qRT-PCR
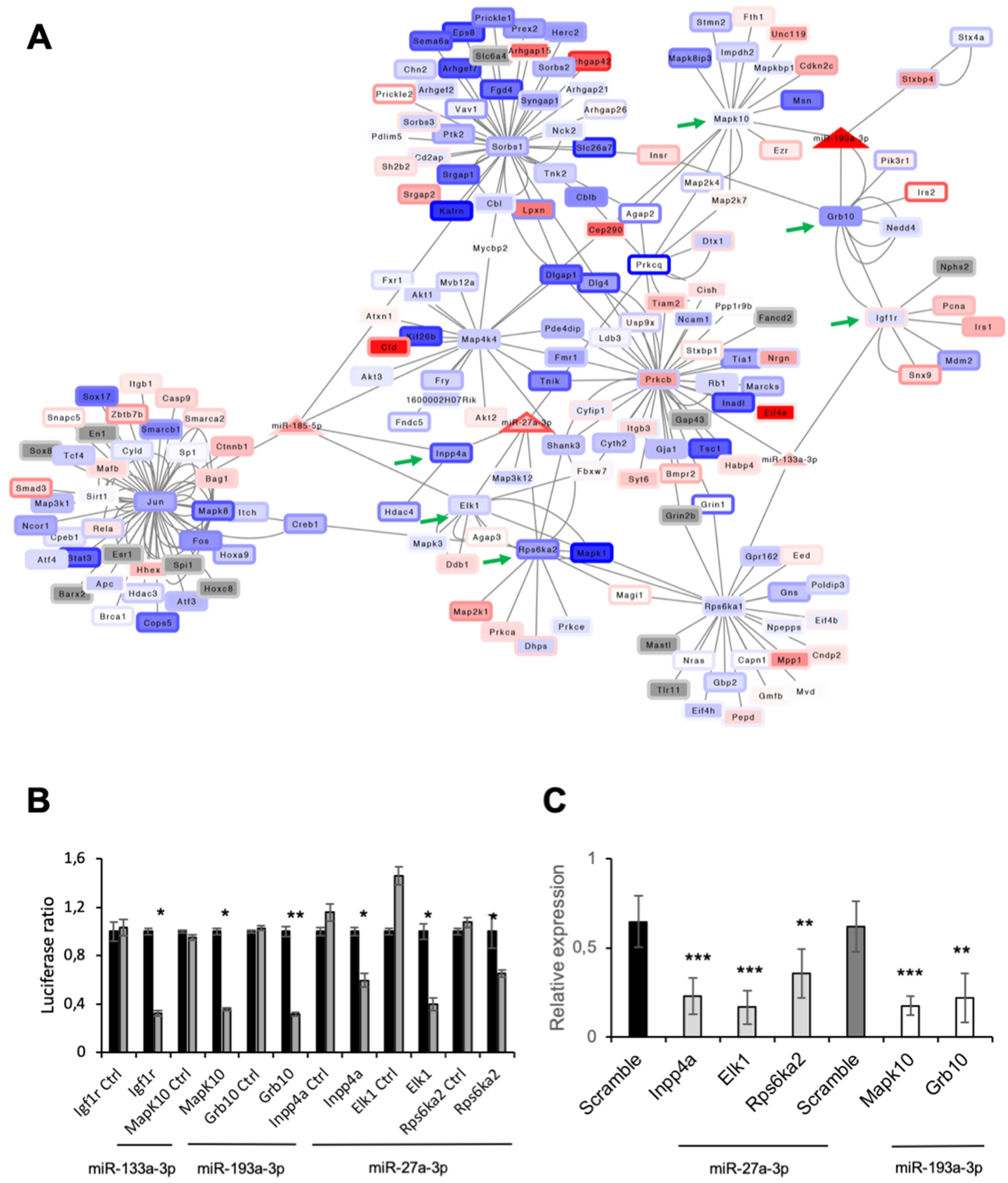
3.6. Function of miRNAs Affected by MAO Inhibition in T1D Hearts
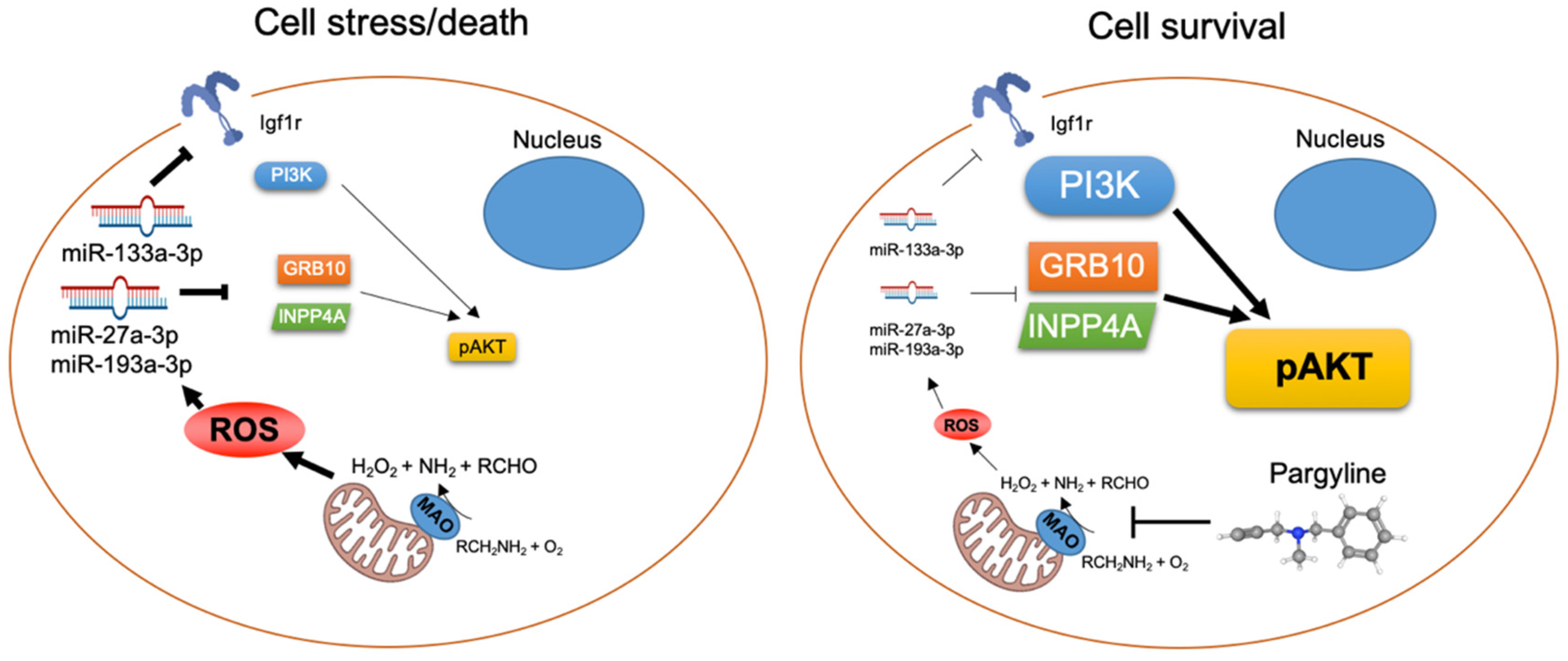
3.7. MAO Inhibition Restored AKT Activation in Diabetic Hearts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaludercic, N.; Di Lisa, F. Mitochondrial ROS Formation in the Pathogenesis of Diabetic Cardiomyopathy. Front. Cardiovasc. Med. 2020, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Rota, M.; Le Capitaine, N.; Hosoda, T.; Boni, A.; De Angelis, A.; Padin-Iruegas, M.E.; Esposito, G.; Vitale, S.; Urbanek, K.; Casarsa, C.; et al. Diabetes promotes cardiac stem cell aging and heart failure, which are prevented by deletion of the p66shc gene. Circ. Res. 2006, 99, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, M.; Migliaccio, E.; Orsini, F.; Paolucci, D.; Moroni, M.; Contursi, C.; Pelliccia, G.; Luzi, L.; Minucci, S.; Marcaccio, M.; et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell 2005, 122, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Carpi, A.; Menabo, R.; Kaludercic, N.; Pelicci, P.; Di Lisa, F.; Giorgio, M. The cardioprotective effects elicited by p66(Shc) ablation demonstrate the crucial role of mitochondrial ROS formation in ischemia/reperfusion injury. Biochim. Biophys. Acta 2009, 1787, 774–780. [Google Scholar] [CrossRef]
- Maalouf, R.M.; Eid, A.A.; Gorin, Y.C.; Block, K.; Escobar, G.P.; Bailey, S.; Abboud, H.E. Nox4-derived reactive oxygen species mediate cardiomyocyte injury in early type 1 diabetes. Am. J. Physiol. Cell Physiol. 2012, 302, C597–C604. [Google Scholar] [CrossRef]
- Kuroda, J.; Ago, T.; Matsushima, S.; Zhai, P.; Schneider, M.D.; Sadoshima, J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc. Natl. Acad. Sci. USA 2010, 107, 15565–15570. [Google Scholar] [CrossRef]
- Beretta, M.; Santos, C.X.; Molenaar, C.; Hafstad, A.D.; Miller, C.C.; Revazian, A.; Betteridge, K.; Schroder, K.; Streckfuss-Bomeke, K.; Doroshow, J.H.; et al. Nox4 regulates InsP3 receptor-dependent Ca(2+) release into mitochondria to promote cell survival. EMBO J. 2020, 39, e103530. [Google Scholar] [CrossRef]
- Deshwal, S.; Di Sante, M.; Di Lisa, F.; Kaludercic, N. Emerging role of monoamine oxidase as a therapeutic target for cardiovascular disease. Curr. Opin. Pharm. 2017, 33, 64–69. [Google Scholar] [CrossRef]
- Kaludercic, N.; Mialet-Perez, J.; Paolocci, N.; Parini, A.; Di Lisa, F. Monoamine oxidases as sources of oxidants in the heart. J. Mol. Cell. Cardiol. 2014, 73, 34–42. [Google Scholar] [CrossRef]
- Antonucci, S.; Di Sante, M.; Tonolo, F.; Pontarollo, L.; Scalcon, V.; Alanova, P.; Menabo, R.; Carpi, A.; Bindoli, A.; Rigobello, M.P.; et al. The Determining Role of Mitochondrial Reactive Oxygen Species Generation and Monoamine Oxidase Activity in Doxorubicin-Induced Cardiotoxicity. Antioxid. Redox Signal. 2021, 34, 531–550. [Google Scholar] [CrossRef]
- Kaludercic, N.; Carpi, A.; Nagayama, T.; Sivakumaran, V.; Zhu, G.; Lai, E.W.; Bedja, D.; De Mario, A.; Chen, K.; Gabrielson, K.L.; et al. Monoamine oxidase B prompts mitochondrial and cardiac dysfunction in pressure overloaded hearts. Antioxid. Redox Signal. 2014, 20, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Kaludercic, N.; Takimoto, E.; Nagayama, T.; Feng, N.; Lai, E.W.; Bedja, D.; Chen, K.; Gabrielson, K.L.; Blakely, R.D.; Shih, J.C.; et al. Monoamine oxidase A-mediated enhanced catabolism of norepinephrine contributes to adverse remodeling and pump failure in hearts with pressure overload. Circ. Res. 2010, 106, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, P.; Kunduzova, O.; Masini, E.; Cambon, C.; Bani, D.; Raimondi, L.; Seguelas, M.H.; Nistri, S.; Colucci, W.; Leducq, N.; et al. Oxidative stress by monoamine oxidase mediates receptor-independent cardiomyocyte apoptosis by serotonin and postischemic myocardial injury. Circulation 2005, 112, 3297–3305. [Google Scholar] [CrossRef]
- Villeneuve, C.; Guilbeau-Frugier, C.; Sicard, P.; Lairez, O.; Ordener, C.; Duparc, T.; De Paulis, D.; Couderc, B.; Spreux-Varoquaux, O.; Tortosa, F.; et al. p53-PGC-1alpha pathway mediates oxidative mitochondrial damage and cardiomyocyte necrosis induced by monoamine oxidase-A upregulation: Role in chronic left ventricular dysfunction in mice. Antioxid. Redox Signal. 2013, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Deshwal, S.; Forkink, M.; Hu, C.H.; Buonincontri, G.; Antonucci, S.; Di Sante, M.; Murphy, M.P.; Paolocci, N.; Mochly-Rosen, D.; Krieg, T.; et al. Monoamine oxidase-dependent endoplasmic reticulum-mitochondria dysfunction and mast cell degranulation lead to adverse cardiac remodeling in diabetes. Cell Death Differ. 2018, 25, 1671–1685. [Google Scholar] [CrossRef]
- Umbarkar, P.; Singh, S.; Arkat, S.; Bodhankar, S.L.; Lohidasan, S.; Sitasawad, S.L. Monoamine oxidase-A is an important source of oxidative stress and promotes cardiac dysfunction, apoptosis, and fibrosis in diabetic cardiomyopathy. Free Radic. Biol. Med. 2015, 87, 263–273. [Google Scholar] [CrossRef]
- Thum, T.; Galuppo, P.; Wolf, C.; Fiedler, J.; Kneitz, S.; van Laake, L.W.; Doevendans, P.A.; Mummery, C.L.; Borlak, J.; Haverich, A.; et al. MicroRNAs in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation 2007, 116, 258–267. [Google Scholar] [CrossRef]
- Yildirim, S.S.; Akman, D.; Catalucci, D.; Turan, B. Relationship between Downregulation of miRNAs and Increase of Oxidative Stress in the Development of Diabetic Cardiac Dysfunction: Junctin as a Target Protein of miR-1. Cell Biochem.Biophys. 2013, 67, 1397–1408. [Google Scholar] [CrossRef]
- Kura, B.; Szeiffova Bacova, B.; Kalocayova, B.; Sykora, M.; Slezak, J. Oxidative Stress-Responsive MicroRNAs in Heart Injury. Int. J. Mol. Sci. 2020, 21, 358. [Google Scholar] [CrossRef]
- Shantikumar, S.; Caporali, A.; Emanueli, C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc.Res. 2012, 93, 583–593. [Google Scholar] [CrossRef]
- Lew, J.K.; Pearson, J.T.; Saw, E.; Tsuchimochi, H.; Wei, M.; Ghosh, N.; Du, C.K.; Zhan, D.Y.; Jin, M.; Umetani, K.; et al. Exercise Regulates MicroRNAs to Preserve Coronary and Cardiac Function in the Diabetic Heart. Circ. Res. 2020, 127, 1384–1400. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Treguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Orekhov, A.N.; Bobryshev, Y.V. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction). J. Mol. Cell. Cardiol. 2016, 94, 107–121. [Google Scholar] [CrossRef] [PubMed]
- van Rooij, E.; Sutherland, L.B.; Liu, N.; Williams, A.H.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 2006, 103, 18255–18260. [Google Scholar] [CrossRef]
- van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef]
- Seok, H.; Lee, H.; Lee, S.; Ahn, S.H.; Lee, H.S.; Kim, G.D.; Peak, J.; Park, J.; Cho, Y.K.; Jeong, Y.; et al. Position-specific oxidation of miR-1 encodes cardiac hypertrophy. Nature 2020, 584, 279–285. [Google Scholar] [CrossRef]
- Chemello, F.; Grespi, F.; Zulian, A.; Cancellara, P.; Hebert-Chatelain, E.; Martini, P.; Bean, C.; Alessio, E.; Buson, L.; Bazzega, M.; et al. Transcriptomic Analysis of Single Isolated Myofibers Identifies miR-27a-3p and miR-142-3p as Regulators of Metabolism in Skeletal Muscle. Cell Rep. 2019, 26, 3784–3797.e3788. [Google Scholar] [CrossRef] [PubMed]
- Alessio, E.; Buson, L.; Chemello, F.; Peggion, C.; Grespi, F.; Martini, P.; Massimino, M.L.; Pacchioni, B.; Millino, C.; Romualdi, C.; et al. Single cell analysis reveals the involvement of the long non-coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Res. 2019, 47, 1653–1670. [Google Scholar] [CrossRef] [PubMed]
- Herrero, J.; Valencia, A.; Dopazo, J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 2001, 17, 126–136. [Google Scholar] [CrossRef]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef]
- Cheng, X.; Yan, J.; Liu, Y.; Wang, J.; Taubert, S. eVITTA: A web-based visualization and inference toolbox for transcriptome analysis. Nucleic Acids Res. 2021, 49, W207–W215. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pagliari, M.; Munari, F.; Toffoletto, M.; Lonardi, S.; Chemello, F.; Codolo, G.; Millino, C.; Della Bella, C.; Pacchioni, B.; Vermi, W.; et al. Helicobacter pylori Affects the Antigen Presentation Activity of Macrophages Modulating the Expression of the Immune Receptor CD300E through miR-4270. Front. Immunol. 2017, 8, 1288. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, X.; Wan, Y.; Zhang, S.; Zhao, Y.; Fan, R.; Cui, Q.; Zhou, Y. TAM 2.0: Tool for MicroRNA set analysis. Nucleic Acids Res. 2018, 46, W180–W185. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef]
- Jung, H.J.; Suh, Y. Regulation of IGF -1 signaling by microRNAs. Front. Genet. 2014, 5, 472. [Google Scholar] [CrossRef]
- Casas, A.I.; Dao, V.T.; Daiber, A.; Maghzal, G.J.; Di Lisa, F.; Kaludercic, N.; Leach, S.; Cuadrado, A.; Jaquet, V.; Seredenina, T.; et al. Reactive Oxygen-Related Diseases: Therapeutic Targets and Emerging Clinical Indications. Antioxid. Redox Signal. 2015, 23, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Kaludercic, N.; Deshwal, S.; Di Lisa, F. Reactive oxygen species and redox compartmentalization. Front. Physiol. 2014, 5, 285. [Google Scholar] [CrossRef] [PubMed]
- Sturza, A.; Duicu, O.M.; Vaduva, A.; Danila, M.D.; Noveanu, L.; Varro, A.; Muntean, D.M. Monoamine oxidases are novel sources of cardiovascular oxidative stress in experimental diabetes. Can. J. Physiol. Pharm. 2015, 93, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Maurel, A.; Hernandez, C.; Kunduzova, O.; Bompart, G.; Cambon, C.; Parini, A.; Frances, B. Age-dependent increase in hydrogen peroxide production by cardiac monoamine oxidase A in rats. Am. J. Physiol Heart Circ. Physiol 2003, 284, H1460–H1467. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef]
- Santin, Y.; Sicard, P.; Vigneron, F.; Guilbeau-Frugier, C.; Dutaur, M.; Lairez, O.; Couderc, B.; Manni, D.; Korolchuk, V.I.; Lezoualc’h, F.; et al. Oxidative Stress by Monoamine Oxidase-A Impairs Transcription Factor EB Activation and Autophagosome Clearance, Leading to Cardiomyocyte Necrosis and Heart Failure. Antioxid. Redox Signal. 2016, 25, 10–27. [Google Scholar] [CrossRef]
- Ugun-Klusek, A.; Theodosi, T.S.; Fitzgerald, J.C.; Burte, F.; Ufer, C.; Boocock, D.J.; Yu-Wai-Man, P.; Bedford, L.; Billett, E.E. Monoamine oxidase-A promotes protective autophagy in human SH-SY5Y neuroblastoma cells through Bcl-2 phosphorylation. Redox Biol. 2019, 20, 167–181. [Google Scholar] [CrossRef]
- Kaludercic, N.; Maiuri, M.C.; Kaushik, S.; Fernandez, A.F.; de Bruijn, J.; Castoldi, F.; Chen, Y.; Ito, J.; Mukai, R.; Murakawa, T.; et al. Comprehensive autophagy evaluation in cardiac diseases models. Cardiovasc. Res. 2019, 116, 483–504. [Google Scholar] [CrossRef]
- Xu, X.; Kobayashi, S.; Chen, K.; Timm, D.; Volden, P.; Huang, Y.; Gulick, J.; Yue, Z.; Robbins, J.; Epstein, P.N.; et al. Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J. Biol. Chem. 2013, 288, 18077–18092. [Google Scholar] [CrossRef]
- Tong, M.; Saito, T.; Zhai, P.; Oka, S.I.; Mizushima, W.; Nakamura, M.; Ikeda, S.; Shirakabe, A.; Sadoshima, J. Mitophagy Is Essential for Maintaining Cardiac Function During High Fat Diet-Induced Diabetic Cardiomyopathy. Circ. Res. 2019, 124, 1360–1371. [Google Scholar] [CrossRef]
- Tong, M.; Saito, T.; Zhai, P.; Oka, S.I.; Mizushima, W.; Nakamura, M.; Ikeda, S.; Shirakabe, A.; Sadoshima, J. Alternative Mitophagy Protects the Heart Against Obesity-Associated Cardiomyopathy. Circ. Res. 2021, 129, 1105–1121. [Google Scholar] [CrossRef]
- Gao, S.; Wassler, M.; Zhang, L.; Li, Y.; Wang, J.; Zhang, Y.; Shelat, H.; Williams, J.; Geng, Y.J. MicroRNA-133a regulates insulin-like growth factor-1 receptor expression and vascular smooth muscle cell proliferation in murine atherosclerosis. Atherosclerosis 2014, 232, 171–179. [Google Scholar] [CrossRef]
- Izarra, A.; Moscoso, I.; Levent, E.; Canon, S.; Cerrada, I.; Diez-Juan, A.; Blanca, V.; Nunez-Gil, I.J.; Valiente, I.; Ruiz-Sauri, A.; et al. miR-133a enhances the protective capacity of cardiac progenitors cells after myocardial infarction. Stem Cell Rep. 2014, 3, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Meng, X.P.; Delafontaine, P. Transcriptional regulation of the insulin-like growth factor-I receptor gene: Evidence for protein kinase C-dependent and -independent pathways. Endocrinology 1996, 137, 1378–1384. [Google Scholar] [CrossRef][Green Version]
- Scheidegger, K.J.; Du, J.; Delafontaine, P. Distinct and common pathways in the regulation of insulin-like growth factor-1 receptor gene expression by angiotensin II and basic fibroblast growth factor. J. Biol. Chem. 1999, 274, 3522–3530. [Google Scholar] [CrossRef]
- Troncoso, R.; Ibarra, C.; Vicencio, J.M.; Jaimovich, E.; Lavandero, S. New insights into IGF-1 signaling in the heart. Trends Endocrinol. Metab. TEM 2014, 25, 128–137. [Google Scholar] [CrossRef]
- Li, Q.; Li, B.; Wang, X.; Leri, A.; Jana, K.P.; Liu, Y.; Kajstura, J.; Baserga, R.; Anversa, P. Overexpression of insulin-like growth factor-1 in mice protects from myocyte death after infarction, attenuating ventricular dilation, wall stress, and cardiac hypertrophy. J. Clin. Investig. 1997, 100, 1991–1999. [Google Scholar] [CrossRef]
- Lay, I.S.; Kuo, W.W.; Shibu, M.A.; Ho, T.J.; Cheng, S.M.; Day, C.H.; Ban, B.; Wang, S.; Li, Q.; Huang, C.Y. Exercise training restores IGFIR survival signaling in d-galactose induced-aging rats to suppress cardiac apoptosis. J. Adv. Res. 2021, 28, 35–41. [Google Scholar] [CrossRef]
- McMullen, J.R.; Shioi, T.; Huang, W.Y.; Zhang, L.; Tarnavski, O.; Bisping, E.; Schinke, M.; Kong, S.; Sherwood, M.C.; Brown, J.; et al. The insulin-like growth factor 1 receptor induces physiological heart growth via the phosphoinositide 3-kinase(p110alpha) pathway. J. Biol. Chem. 2004, 279, 4782–4793. [Google Scholar] [CrossRef] [PubMed]
- Moellendorf, S.; Kessels, C.; Peiseler, L.; Raupach, A.; Jacoby, C.; Vogt, N.; Lindecke, A.; Koch, L.; Bruning, J.; Heger, J.; et al. IGF-IR signaling attenuates the age-related decline of diastolic cardiac function. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E213–E222. [Google Scholar] [CrossRef] [PubMed]
- Ock, S.; Lee, W.S.; Ahn, J.; Kim, H.M.; Kang, H.; Kim, H.S.; Jo, D.; Abel, E.D.; Lee, T.J.; Kim, J. Deletion of IGF-1 Receptors in Cardiomyocytes Attenuates Cardiac Aging in Male Mice. Endocrinology 2016, 157, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Quipildor, G.F.; Tabrizian, T.; Novaj, A.; Guan, F.; Walters, R.O.; Delahaye, F.; Hubbard, G.B.; Ikeno, Y.; Ejima, K.; et al. Late-life targeting of the IGF-1 receptor improves healthspan and lifespan in female mice. Nat. Commun. 2018, 9, 2394. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, M.; Trummer-Herbst, V.; Heberle, A.M.; Humnig, A.; Pendl, T.; Durand, S.; Cerrato, G.; Hofer, S.J.; Islam, M.; Voglhuber, J.; et al. Fine-Tuning Cardiac Insulin-Like Growth Factor 1 Receptor Signaling to Promote Health and Longevity. Circulation 2022, 145, 1853–1866. [Google Scholar] [CrossRef] [PubMed]
- Fabbi, P.; Spallarossa, P.; Garibaldi, S.; Barisione, C.; Mura, M.; Altieri, P.; Rebesco, B.; Monti, M.G.; Canepa, M.; Ghigliotti, G.; et al. Doxorubicin impairs the insulin-like growth factor-1 system and causes insulin-like growth factor-1 resistance in cardiomyocytes. PLoS ONE 2015, 10, e0124643. [Google Scholar] [CrossRef]
- Urbanek, K.; Rota, M.; Cascapera, S.; Bearzi, C.; Nascimbene, A.; De Angelis, A.; Hosoda, T.; Chimenti, S.; Baker, M.; Limana, F.; et al. Cardiac stem cells possess growth factor-receptor systems that after activation regenerate the infarcted myocardium, improving ventricular function and long-term survival. Circ. Res. 2005, 97, 663–673. [Google Scholar] [CrossRef]
- D’Amario, D.; Cabral-Da-Silva, M.C.; Zheng, H.; Fiorini, C.; Goichberg, P.; Steadman, E.; Ferreira-Martins, J.; Sanada, F.; Piccoli, M.; Cappetta, D.; et al. Insulin-like growth factor-1 receptor identifies a pool of human cardiac stem cells with superior therapeutic potential for myocardial regeneration. Circ. Res. 2011, 108, 1467–1481. [Google Scholar] [CrossRef]
- Leslie, N.R. The redox regulation of PI 3-kinase-dependent signaling. Antioxid. Redox Signal. 2006, 8, 1765–1774. [Google Scholar] [CrossRef]
- Yasukawa, T.; Tokunaga, E.; Ota, H.; Sugita, H.; Martyn, J.A.; Kaneki, M. S-nitrosylation-dependent inactivation of Akt/protein kinase B in insulin resistance. J. Biol. Chem. 2005, 280, 7511–7518. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, D.; Reece, E.A.; Wang, A.R.; Yang, P. Oxidative stress-induced miR-27a targets the redox gene nuclear factor erythroid 2-related factor 2 in diabetic embryopathy. Am. J. Obstet. Gynecol. 2018, 218, 136.e1–136.e10. [Google Scholar] [CrossRef] [PubMed]
- Costantino, S.; Paneni, F.; Luscher, T.F.; Cosentino, F. MicroRNA profiling unveils hyperglycaemic memory in the diabetic heart. Eur. Heart J. 2016, 37, 572–576. [Google Scholar] [CrossRef]
- Li, D.; Shen, M.; Deng, X.; Bai, Y. MicroRNA miR-27a-3p accelerates cardiac hypertrophy by targeting neuro-oncological ventral antigen 1. Bioengineered 2022, 13, 8982–8993. [Google Scholar] [CrossRef] [PubMed]
- Vyas, P.; Norris, F.A.; Joseph, R.; Majerus, P.W.; Orkin, S.H. Inositol polyphosphate 4-phosphatase type I regulates cell growth downstream of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 2000, 97, 13696–13701. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, J.; Kofuji, S.; Itoh, R.; Momiyama, T.; Takayama, K.; Murakami, H.; Chida, S.; Tsuya, Y.; Takasuga, S.; Eguchi, S.; et al. The PtdIns(3,4)P(2) phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature 2010, 465, 497–501. [Google Scholar] [CrossRef]
- Kwon, J.E.; Kim, B.Y.; Kwak, S.Y.; Bae, I.H.; Han, Y.H. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis Int. J. Program. Cell Death 2013, 18, 896–909. [Google Scholar] [CrossRef]
- Huang, J.; Wang, F.; Sun, X.; Chu, X.; Jiang, R.; Wang, Y.; Pang, L. Myocardial infarction cardiomyocytes-derived exosomal miR-328-3p promote apoptosis via Caspase signaling. Am. J. Transl. Res. 2021, 13, 2365–2378. [Google Scholar]
- Sun, Y.; Xu, R.; Huang, J.; Yao, Y.; Pan, X.; Chen, Z.; Ma, G. Insulin-like growth factor-1-mediated regulation of miR-193a expression promotes the migration and proliferation of c-kit-positive mouse cardiac stem cells. Stem Cell Res. Ther. 2018, 9, 41. [Google Scholar] [CrossRef]
- Manni, M.E.; Rigacci, S.; Borchi, E.; Bargelli, V.; Miceli, C.; Giordano, C.; Raimondi, L.; Nediani, C. Monoamine Oxidase Is Overactivated in Left and Right Ventricles from Ischemic Hearts: An Intriguing Therapeutic Target. Oxid Med Cell Longev 2016, 2016, 4375418. [Google Scholar] [CrossRef]
- Anderson, E.J.; Efird, J.T.; Davies, S.W.; O’Neal, W.T.; Darden, T.M.; Thayne, K.A.; Katunga, L.A.; Kindell, L.C.; Ferguson, T.B.; Anderson, C.A.; et al. Monoamine oxidase is a major determinant of redox balance in human atrial myocardium and is associated with postoperative atrial fibrillation. J. Am. Heart Assoc. 2014, 3, e000713. [Google Scholar] [CrossRef]
- Nelson, M.M.; Efird, J.T.; Kew, K.A.; Katunga, L.A.; Monroe, T.B.; Doorn, J.A.; Beatty, C.N.; Shi, Q.; Akhter, S.A.; Alwair, H.; et al. Enhanced Catecholamine Flux and Impaired Carbonyl Metabolism Disrupt Cardiac Mitochondrial Oxidative Phosphorylation in Diabetes Patients. Antioxid. Redox Signal. 2021, 35, 235–251. [Google Scholar] [CrossRef]


| Cluster Number | Description | p-Value |
|---|---|---|
| 8 | GO:0005783 Endoplasmic reticulum | 0.000027 |
| KW-0496 Mitochondrion | 0.017 | |
| GO:0097352 Autophagosome maturation | 0.005 | |
| 10 | KW-0832 Ubl conjugation | 0.00049 |
| KW-0496 Mitochondrion | 0.000017 | |
| GO:0005783 Endoplasmic reticulum | 0.00087 | |
| GO:0004842 Ubiquitin-protein transferase activity | 0.02 | |
| 5 | KW-0832 Ubl conjugation | 1.9 × 10−8 |
| GO:0005794 Golgi apparatus | 0.0000012 | |
| KW-0805 Transcription regulation | 4.07 × 10−6 | |
| GO:0003281 Ventricular septum development; GO:0060976 Coronary vasculature development | 0.03 |
| Cluster Number | Pathway | p-Value |
|---|---|---|
| KEGG Ubiquitin-mediated proteolysis | 1.74 × 10−8 | |
| KEGG Fluid shear stress and atherosclerosis | 4.13 × 10−6 | |
| WP Oxidative Stress and Redox Pathway | 8.47 × 10−6 | |
| KEGG Peroxisome | 0.000029 | |
| WP Glycolysis and Gluconeogenesis | 0.00010 | |
| WP Calcium Regulation in the Cardiac Cell | 0.00010 | |
| 10 | KEGG Protein processing in endoplasmic reticulum | 2.57 × 10-15 |
| KEGG Phagosome | 3.14 × 10−11 | |
| KEGG Ubiquitin-mediated proteolysis | 3.60 × 10−7 | |
| KEGG Tight junction | 4.90 × 10−7 | |
| WP Calcium Regulation in the Cardiac Cell | 6.80 × 10−7 | |
| KEGG mTOR signaling pathway | 8.04 × 10−7 | |
| WP MicroRNAs in Cardiomyocyte Hypertrophy | 1.10 × 10−6 | |
| WP Regulation of Actin Cytoskeleton | 2.92 × 10−6 | |
| KEGG Peroxisome | 4.77 × 10−6 | |
| 5 | KEGG Cellular senescence | 2.69 × 10−10 |
| WP EGFR1 Signaling Pathway | 6.92 × 10−10 | |
| WP Insulin Signaling | 1.14 × 10−6 |
| miRNA Expression Cluster | Term | FDR | miRNAs |
|---|---|---|---|
| 1 | Cluster | ||
| miR-17 | 1.54 × 10−3 | miR-19a, miR-19b-1, miR-92a-1 | |
| miR-106a | 1.54 × 10−3 | miR-20b, miR-19b-2, miR-92a-2 | |
| miR-99a | 3.71 × 10−3 | miR-99a, let-7c | |
| miR-100 | 9.10 × 10−3 | miR-100, miR-125b-1 | |
| miR-6749 | 0.0163 | miR-194-2, miR-192 | |
| Family | |||
| miR-10 | 5.71 × 10−8 | miR-100, miR-10a, miR-10b, miR-125b-1, miR-125b-2, miR-99a | |
| miR-19 | 1.18 × 10−4 | miR-19a, miR-19b-1, miR-19b-2 | |
| miR-194 | 3.71 × 10−3 | miR-194-1, miR-194-2 | |
| miR-193 | 3.71 × 10−3 | miR-193a, miR-193b | |
| miR-128 | 3.71 × 10−3 | miR-128-1, miR-128-2 | |
| miR-133 | 9.10 × 10−3 | miR-133a-1, miR-133a-2 | |
| let-7 | 0.0109 | let-7b, let-7c, miR-98 | |
| miR-130 | 0.0163 | miR-130a, miR-301a | |
| miR-25 | 0.0163 | miR-92a-1, miR-92a-2 | |
| 2 | Family | ||
| miR-30 | 1.66 × 10−4 | miR-30a, miR-30d, miR-30e | |
| miR-378 | 1.25 × 10−7 | miR-378a, miR-378b, miR-378d | |
| miRNA Expression Cluster | Pathway | p-Value | Overlap Genes |
|---|---|---|---|
| 1 | WP Insulin Signaling | 3.95 × 10−19 | JUN, SNAP25, MAP2K2, IGF1R, INPP4A, SNAP23, PIK3CA, PIK3CD, CBLB, MAP3K8, SORBS1, FLOT2, MAPK4, MAP3K7, GRB10, MAP4K4, CRK, PIK3R3, RPS6KA2, RPS6KA1, MINK1, GSK3B, RAF1, MAPK12, SLC2A4 |
| WP EGFR1 Signaling | 7.72 × 10−17 | JUN, WNK1, MAP2K2, PIK3CA, PIK3CD, CBLB, ASAP1, WASL, STAT3, STAT1, VAV3, GJA1, CTNND1, CAV1, CREB1, HTT, GRB10, EPS15, CRK, PIK3R3, RALBP1, RPS6KA2, RPS6KA1, RAF1 | |
| WP Regulation of Actin Cytoskeleton | 3.45 × 10−15 | MAP2K2, MSN, SSH2, SSH1, ARHGEF7, PIK3CA, LIMK1, PIK3CD, ITGA1, ARHGEF6, MAPK4, APC, MYLK, PAK2, IQGAP1, CRK, WASF2, PIK3R3, ARPC5, PTK2, RAF1 | |
| BP actin cytoskeleton organization | 3.52 × 10−16 | ABLIM3, ACTR2, ARFIP2, ARHGEF18, ATXN3, CDC42BPA, CORO1C, CORO2B, CRK, CSRP1, FLNB, PIK3CA, SDAD1, SSH1, SSH2, TAOK2, WASF2, WASL, ACTN4, LIMK1, SPTAN1, DMD, UTRN, SPTBN1 | |
| BP endocytosis | 4.66 × 10−15 | AAK1, ANK2, ANKFY1, AP2A2, CAV1, CLCN5, DNM1L, DNM2, DNM3, FCHSD2, FKBP15, FNBP1L, MICALL1, MYO6, PSTPIP1, RABEP2, RALBP1, SGIP1, SYNRG, WASF2, NCKIPSD, ITSN1, EPS15 | |
| 2 | WP Insulin Signaling | 0.005 | PRKAA2, CBLB, PRKCA, RPS6KB1 |
| BP negative regulation of insulin receptor signaling pathway | 0.016 | PRKCA, PTPN2, RPS6KB1 | |
| BP positive regulation of autophagy | 0.04 | MTDH, PRKAA2, DEPDC5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cagnin, S.; Brugnaro, M.; Millino, C.; Pacchioni, B.; Troiano, C.; Di Sante, M.; Kaludercic, N. Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs. Cells 2022, 11, 2697. https://doi.org/10.3390/cells11172697
Cagnin S, Brugnaro M, Millino C, Pacchioni B, Troiano C, Di Sante M, Kaludercic N. Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs. Cells. 2022; 11(17):2697. https://doi.org/10.3390/cells11172697
Chicago/Turabian StyleCagnin, Stefano, Marco Brugnaro, Caterina Millino, Beniamina Pacchioni, Carmen Troiano, Moises Di Sante, and Nina Kaludercic. 2022. "Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs" Cells 11, no. 17: 2697. https://doi.org/10.3390/cells11172697
APA StyleCagnin, S., Brugnaro, M., Millino, C., Pacchioni, B., Troiano, C., Di Sante, M., & Kaludercic, N. (2022). Monoamine Oxidase-Dependent Pro-Survival Signaling in Diabetic Hearts Is Mediated by miRNAs. Cells, 11(17), 2697. https://doi.org/10.3390/cells11172697






