Abstract
Betacoronaviruses, responsible for the “Severe Acute Respiratory Syndrome” (SARS) and the “Middle East Respiratory Syndrome” (MERS), use the spikes protruding from the virion envelope to attach and subsequently infect the host cells. The coronavirus spike (S) proteins contain receptor binding domains (RBD), allowing the specific recognition of either the dipeptidyl peptidase CD23 (MERS-CoV) or the angiotensin-converting enzyme ACE2 (SARS-Cov, SARS-CoV-2) host cell receptors. The heavily glycosylated S protein includes both complex and high-mannose type N-glycans that are well exposed at the surface of the spikes. A detailed analysis of the carbohydrate-binding specificity of mannose-binding lectins from plants, algae, fungi, and bacteria, revealed that, depending on their origin, they preferentially recognize either complex type N-glycans, or high-mannose type N-glycans. Since both complex and high-mannose glycans substantially decorate the S proteins, mannose-specific lectins are potentially useful glycan probes for targeting the SARS-CoV, MERS-CoV, and SARS-CoV-2 virions. Mannose-binding legume lectins, like pea lectin, and monocot mannose-binding lectins, like snowdrop lectin or the algal lectin griffithsin, which specifically recognize complex N-glycans and high-mannose glycans, respectively, are particularly adapted for targeting coronaviruses. The biomedical prospects of targeting coronaviruses with mannose-specific lectins are wide-ranging including detection, immobilization, prevention, and control of coronavirus infection.
1. Introduction
The human Betacoronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 responsible for the pandemic COVID 19 that still rages worldwide, share common structural characteristics that account for their transmissibility and infectivity among sensitive people. The virions are covered by spikes, which protrude outside of the lipid bilayer forming the envelope of coronaviruses [,,]. Spikes are built up from the homotrimeric association of the S protein), an acidic class I viral fusion glycoprotein of ~130 kDa which consists of two S1 and S2 subunits [,,,]. The S protein is cleaved between the S1 and S2 subunits (S1/S2 cleavage site) and at an additional site S2’ by host cell protease. The S1 subunit contains a receptor-binding domain (RBD), which allows the virus to specifically recognize host cell receptors via a receptor-binding motif (RBM). The dipeptidyl peptidase 4 (DPP4), also known as CD26, was identified as the host receptor for MERS-CoV [], whereas the angiotensin-converting enzyme 2 (ACE2) serves as the host receptor for SARS-CoV and SARS-CoV-2 []. The S2 subunit is subsequently involved in the fusion of both coronavirus and host cell membranes through the fusion peptide (FP) and the first heptad repeat HR1 [], allowing the entry of the viral RNA genome into the host cell, which will further trigger the replication of the coronavirus at the detriment of the host cell (Figure 1).
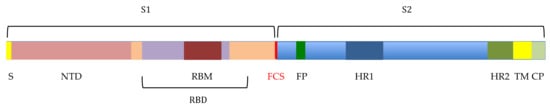
Figure 1.
Molecular organization of the SARS-CoV-2 S-glycoprotein, made up of two linked S1 and S2 subunits that are further separated at the furin cleavage site (FCS). The S1 subunit consists of a signal peptide (S), followed by the N-terminal domain (NTD) containing the receptor-binding domain (RBD) interacting with the ACE2 receptor via the RBM motif (RBM). The S2 subunit comprises the fusion peptide (FP), two heptad repeats HR1 and HR2, the transmembrane spanning domain (TM) that anchors the S protein into the virion membrane, and the cytoplasmic domain CP.
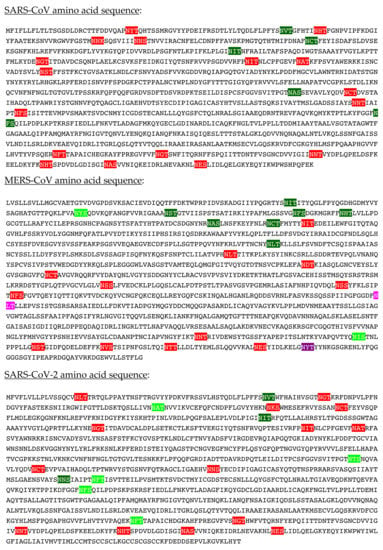
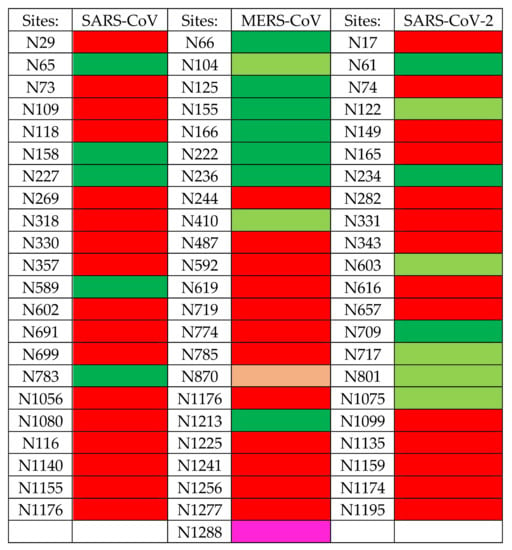
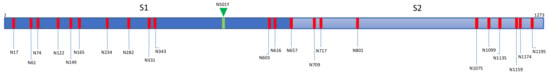
The S proteins forming the coronavirus spikes, consist of heavily N-glycosylated proteins containing a high number of potential N-glycosylation sites NXT/S, actually occupied by complex-type glycans, high-mannose type glycans, and a few hybrid type glycans: 22 sites for SARS-CoV and SARS-CoV-2, and 23 sites for MERS-CoV, respectively [,]. A very limited number of potential O-glycosylation sites occupied by GalNAc and T/Tn-antigens have been identified, especially in the SARS-CoV-2 S-glycoprotein []. According to their heavily N-glycosylated character, S-glycoproteins of coronaviruses represent particularly relevant targets for mannose-specific (Man-specific) lectins, a heterogeneous group of proteins involved in the recognition of Man, mannosides, and complex Man-containing glycans, already known for their potential anti-viral and anti-cancer properties [,,,,,].
The Man-specific lectins are widely distributed in all the living organisms since they have been isolated and characterized from viruses, bacteria, fungi, algae, plants, animals, and humans []. Even though they constitute a disparate group of proteins poorly related by their phylogenetic relationship and structural features, they share a very common property to specifically recognize Man and their derivatives, including mannosides, oligomannosides, and high-mannose glycans []. This specific recognition also concerns other Man-containing glycans like the complex N-glycans which are built around a trimannosyl Manα1,3Manα1,6Man core, readily recognized by Man-specific legume lectins [,]. As shown for plant lectins, the differences observed in the specific recognition of Man-containing glycans, essentially depend on both the axial position of the hydroxyl group at C2 in Man which is more or less strictly recognized by the carbohydrate-binding sites (CBSs) of lectins, and the type of glycosidic bonds α1,2, α1,3 or α1,6, and their internal or external position in the glycan chain, recognized by lectins []. According to these discrepancies and with respect to the diversity of N-glycans occurring at the surface of the coronavirus spikes, many distinct Man-specific lectins from plants, algae, fungi, and bacteria, could serve as relevant glycan probes for SARS-CoV, MERS-CoV, and SARS-CoV-2 viruses.
In addition to their Man-binding capacity, Man-specific lectins from plants, algae, fungi, and bacteria, have been largely studied with respect to their anti-viral properties against different types of enveloped viruses, including HIV-1 [], papilloma virus [], herpes virus, hepatitis C virus [], and Ebola virus []. In this respect, the algal lectin griffithsin [], the cyanobacteria lectins cyanovirin [], actinohivin [], and microvirin [], and various GNA-related lectins like NPA from daffodil (Narcissus pseudonarcissus) and ASA from garlic (Allium sativum) [,,], have been particularly well documented. Most of these Man-specific lectins prevent the virus replication, at least under in vitro conditions, by interfering with the Man-containing N-glycans present on the cell surface of the virion envelope []. Depending on the virus, different Man-containing glycans serve as targets for the Man-specific lectins, e.g., gp120 for HIV-1 [] or hemagglutinin for influenza virus []. Owing to their smaller size, compared to other plant Man-specific lectins, Man-specific lectins from cyanobacteria such as CVN-N (cyanovirin), MVN (microvirin), and AH (actinohivin) from actinobacteria, have been deeply investigated for their anti-viral properties. In addition, only very few publications report the ability of Man-specific lectins from higher plants to interact with glycans of the coronavirus envelope [,,,].
The present review aims at presenting a detailed analysis of the Man-specific lectins from plants, algae, fungi, and bacteria that could serve as relevant probes for targeting the N-glycans coating the spikes of SARS-CoV, MERS-CoV, and SARS-CoV-2 coronaviruses, with a focus on their potential biomedical applications in controlling coronavirus infections, notably the COVID-19 pandemic associated with SARS-CoV-2.
2. Man-Specific Lectins from Higher Plants, Fungi, Algae, and Cyanobacteria
Mannose-specific lectins consist of a group of lectins that are widely distributed in all living organisms and, especially, in higher plants (monocot and dicot plants), in fungi (Ascomycota and Basidiomycota), in the different groups of algae (essentially red and green algae), and in bacteria, mainly the cyanobacteria (formerly known as blue algae). Although they belong to quite different structural scaffolds [], all of these lectins contain CBSs that display a more or less strict specificity for mannose (Man) and Man-containing glycoproteins.
2.1. Man-Specific Lectins from Higher Plants
Seeds and tubers of higher plants often contain Man-specific lectins which have been associated with different structural scaffolds (Figure 2):
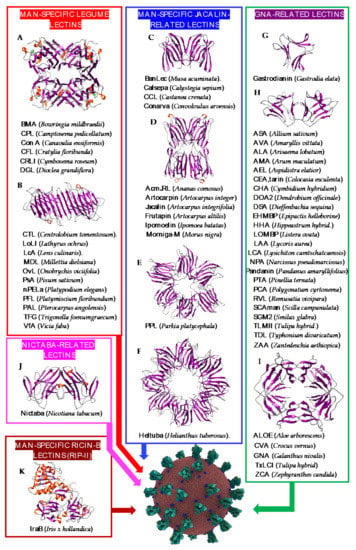
Figure 2.
Overview of lectins from representative genera/species belonging to higher plant families, with their associated structural scaffolds, able to recognize the spike proteins from coronaviruses (Coronavirus Credit: Maria Voigt/RCSB PDB). A-K correspond to lectins used as structural scaffolds ((A): Con A, (B): LcA, (C): BanLec, (D): Morniga-M, (E): PPL, (F): Heltuba, (G): Gastrodianin, (H): ASA, (I): GNA, (J): Nictaba, (K): IraB. References [,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,] correspond to the lectins presented in the figure.
- -
- The β-sandwich structure (jelly-roll structural scaffold), present in legume lectins. They are classically divided into two groups of single- and two-chain lectins, according to the cleavage (two-chain) or the absence of cleavage (single-chain) of their protomers. Single-chain, tetravalent lectins consist of the non-covalent association of four identical protomers (α4), each containing a CBS that specifically recognizes Man and its derivatives. In two-chain lectins, a proteolytic cleavage of the protomeric chain results in the separation of a light (α-chain) and a heavy (β-chain), which remain associated with non-covalent bonds. Two-chain, bivalent lectins (α2β2)) result from the non-covalent association of two identical two-chain protomers, each possessing a CBS specific for Man and its derivatives.
Both Con A (concanavalin A) from jack bean seeds (Canavalia ensiformis) (Figure 2A) and LcA (lentil lectin) from Lens culinaris (Figure 2B) are classical examples of single-chain and two-chain lectins, respectively.
- -
- The β-prism I structure (β-barrel structural scaffold), present in the jacalin-related lectin group. Their protomers are organized in three bundles of four antiparallel β-strands arranged into a β-prism structure along a longitudinal axis. Depending on the number of identical protomers associated by non-covalent bonds, Man-specific jacalin-related lectins consist of bivalent (two CBSs) lectins, e.g., Calsepa from hedge bindweed Calystegia sepium [] (Figure 2C); tetravalent (four CBSs) lectins, e.g., Artocarpin from black mulberry Artocarpus integer [] (Figure 2D); hexavalent (six CBSs) lectins, e.g., PPL from the African locust bean Parkia platycephala [] (Figure 2E); or octavalent (eight CBSs) lectins, e.g., Heltuba from the Jerusalem artichoke Helianthus tuberosus [] (Figure 2F).
- -
- The β-prism II structure (β-trefoil structural scaffold), present in the GNA-related lectin group. The protomer consists of three bundles of four antiparallel β-strands arranged into a flattened β-trefoil structure around a central pseudoaxis. A CBS occurs in each of the bundles of β-strands. Except for gastrodianin, which is composed of a single protomer (Figure 2G), lectins belonging to the GNA-related group result from the non-covalent association of two protomers, e.g., the hexavalent NPA from daffodil (Narcissus pseudonarcissus) bulbs [] (Figure 2H) or four protomers, e.g., the dodecavalent GNA from snowdrop (Galanthus nivalis) bulbs [] (Figure 2I).
- -
- Two covalently linked β-trefoil-domains form the B-chain of the lectin from Iris hollandica, a type II ribosome-inactivating protein (RIP-II) (Figure 2J). Each domain contains a CBS which can accommodate Man and Gal/GalNAc. This Man-binding property confers an unusual Man-specificity to the Iris RIP-II, which readily differs from the Gal/GalNAc-specificity of other RIP-IIs [].
- -
- Nictaba, the jasmonic acid-induced tobacco (Nicotiana tabacum) lectin displays specificity for chitin and GlcNAc oligomers but it can also bind to high mannose glycans with a lower affinity, as shown from glycan array experiments. The molecular structure of Nictaba is still unknown but modeling experiments suggest a β-sandwich structure for this tobacco lectin [] (Figure 2K).
All of these lectins readily interact with Man and oligomannosides but, more interestingly, they display a higher affinity for more complex Man-containing glycan chains that are usually associated with cell surfaces. In this respect, different Man-specific lectins have been co-crystallized in the presence of Man (Figure 3A–C), di-mannoside (Figure 4A), tri-mannoside core Manα1,3Manα1,6Man (Figure 4B,C), and tetra-mannoside (Figure 4D) occurring in N-glycans of the complex and high-mannose type.
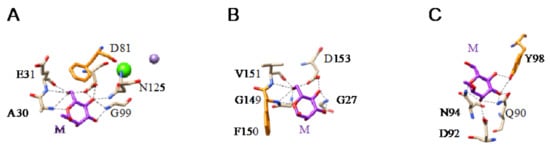
Figure 3.
(A). Network of hydrogen bonds (black dashed lines) anchoring Man (M) to the amino acid residues A30, E31, D81, G99, and N135, forming the CBS of LoLI from Lathyrus ochrus (PDB code 1LOB). Aromatic residue F123 participating in stacking interaction with the pyranose ring of Man is colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring Man (M) to the amino acid residues G27, G149, V151, and D153, forming the CBS of Morniga-M from Morus nigra (PDB code 1XXR). Aromatic residue F150 involved in stacking interaction with the pyranose ring of Man is colored orange. (C). Network of hydrogen bonds (black dashed lines) anchoring Man (M) to the amino acid residues Q90, D92, and N94, forming the CBS of PCL from Polygonatum cyrtonema (PDB code 3A0D). Aromatic residue Y98 participating in stacking interaction with the pyranose ring of Man is colored orange.
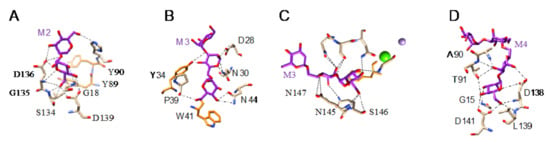
Figure 4.
(A). Network of hydrogen bonds (black dashed lines) anchoring Manα1,2Man (M2) to Heltuba from Helianthus tuberosus (PDB code 1C3N). Aromatic residues Y89 and Y90 participating in stacking interactions with the pyranose rings of mannobiose are colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring Manα1,6Manα1,3Man (M3) to GNA from Galanthus nivalis (PDB code 1JPC). Aromatic residues Y89 and Y90 participating in stacking interactions with the pyranose rings of mannobiose are colored orange. (C). Network of hydrogen bonds (black dashed lines) anchoring Manα1,3Manα1,6Man (M3) to PAL from Pterocarpus angolensis (PDB code 1Q8V). Aromatic residues Y89 and Y90 participating in stacking interactions with the pyranose rings of mannobiose are colored orange. (D). Network of hydrogen bonds (black dashed lines) anchoring mannotetraose (M4) to artocarpin from Artocarpus integer (PDB code 1VBP).
In addition, results from glycan arrays and binding assays, and X-ray crystallography of lectins co-crystallized in the presence of complex glycan chains revealed that Man-binding lectins from higher plants also accommodate more complex N-glycan chains, through a complex network of hydrogen bonds and stacking interactions between the pyranose ring of sugars and aromatic residues (F, W, Y) located in the vicinity of the CBS of lectins (Figure 5A–C).
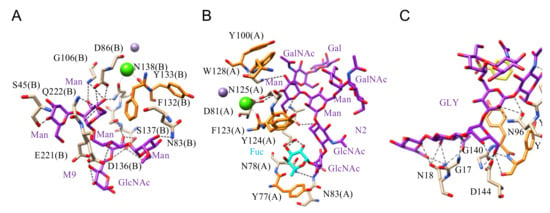
Figure 5.
(A). Network of hydrogen bonds (black dashed lines) anchoring Man9 (M9) to PAL, the Pterocarpus angolensis lectin (PDB code 2PHN). Aromatic residues F132 and Y133 participating in stacking interactions with the pyranose rings of the oligomannoside are colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring N2 oligosaccharide to LoLII, isolectin II from Lathyrus ochrus (PDB code 1LGB). Aromatic residues Y77, Y100, F123, Y124, and W128 involved in stacking interactions with the pyranose rings of the oligosaccharide, are colored orange. The α1,6-linked fucose (Fuc) participates in the H-bond network. (C). Network of hydrogen bonds (black dashed lines) anchoring a biantennary glycan (GLY) to Calsepa from Calystegia sepium (PDB code 5XFI). Aromatic residue Y141 and Y142 participating in stacking interaction with the pyranose rings of the glycan are colored orange.
N-glycans are classified into three types, including oligomannose or high-mannose glycans, complex glycans, and hybrid glycans, according to the extensions of their common core sequence Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAcβ1-Asn (Figure 6A–C):
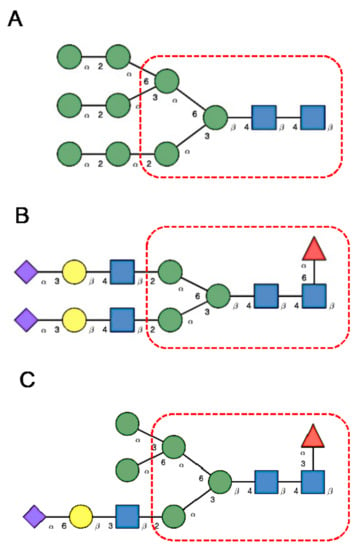
Figure 6.
(A). High-mannose N-glycan, in which only Man (green circles) residues extend the core. (B). Complex N-glycans with two antennae initiated by a GlcNAc residue (blue square) extending the core. (C). Hybrid N-glycan with Man extending the Man1,6 arm of the core and GlcNAc extending the Man1,3 arm of the core. The core is delineated by a dashed red line.
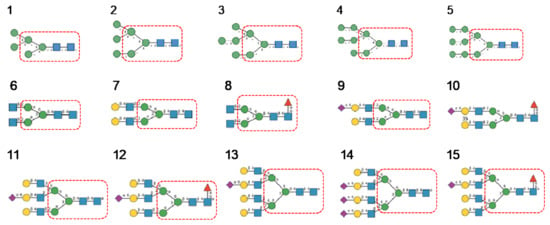
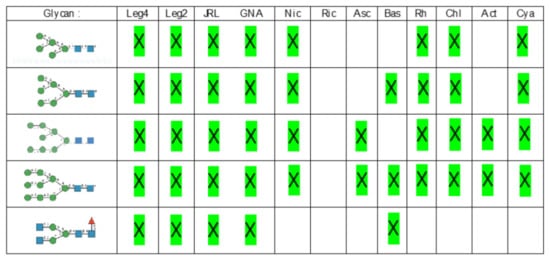
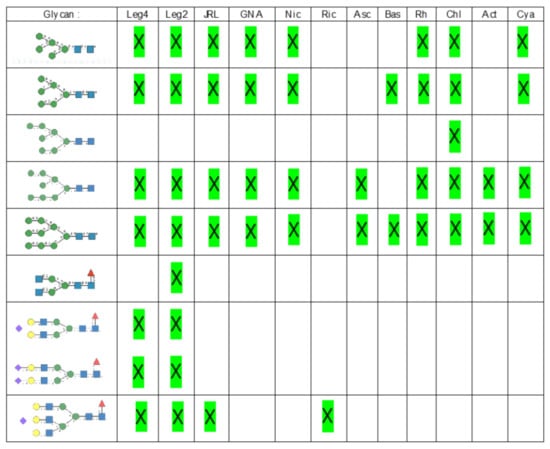
Glycan array analyses were performed by the Consortium for Functional Glycomics (http://www.functionalglycomics.org, accessed on 14 March 2021) for Con A, a typical member of the Man-specific single-chain lectins from the Fabaceae. Con A yielded the best interaction with the following glycans arranged in decreasing order of affinity (Figure 7):
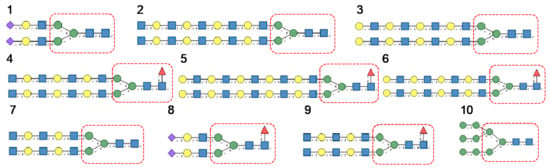
Figure 7.
Glycans recognized by Con A in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
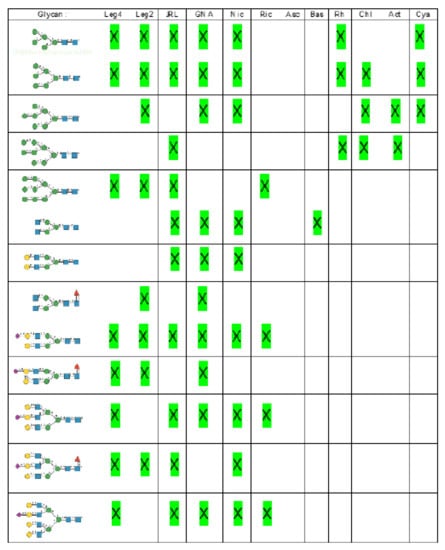
Glycan array analyses performed for PsA, a typical member of the Man-specific two-chain lectins from the Vicieae (a tribe of Leguminosae/Fabaceae), yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 8):
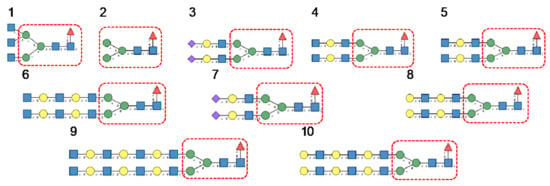
Figure 8.
Glycans recognized by PsA in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Glycan array results obtained for GNA, the Man-specific lectin from snowdrop, the prototype of monocot Man-binding lectins, are in decreasing order of affinity (Figure 9):
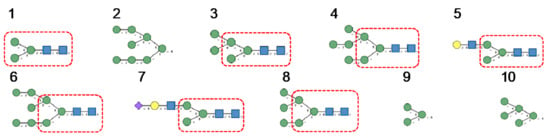
Figure 9.
Glycans recognized by GNA in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Results obtained with Morniga-M, a typical representative of the jacalin-related Man-binding lectins, reveal some reactivity towards bisected hybrid glycans (Figure 10):
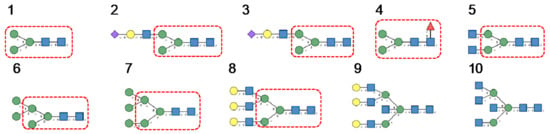
Figure 10.
Glycans recognized by Morniga-M in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
From the glycan array results and X-ray crystallographic analysis of lectins in complex with Man-containing glycans, it clearly appears that beyond a similar affinity for the trimannosyl Manα1,6Manα1,3Man core occurring in both N-glycans of the complex, high-mannose and hybrid types, Man-specific lectins from higher plants are not equivalent and readily differ in their capacity to specifically recognize subtle chemical arrangements in the complex glycan chains. In this respect, Man-specific legume lectins essentially recognize complex glycans α1,6 fucosylated at the GlcNAc residue linked to asparagine, whereas Man-specific lectins belonging to the GNA-related lectin group, essentially recognize more or less branched high-mannose N-glycans. The Man-specific lectins of the jacalin-related group differ from the other lectins by their strong specificity for high-mannose glycans. Depending on the differences observed in the fine specificity towards complex glycans, high-mannose glycans, and hybrid glycans, Man-specific lectins from diverse origins can be used as glycan probes for targeting diverse types of N-glycans covering both host cell and viral surfaces.
2.2. Man-Specific Lectins from Lower Plants and Fungi
Although they are less documented compared to the lectins from higher plants, a few Man-specific lectins have also been identified in lower plants including Bryophyta, Pteridophyta, and Gymnospermae. Man-specific lectins also occur in both groups of fungi, Ascomycota and Basidiomycota. They usually consist of structural scaffolds very similar to those found in higher plant lectins but few of them also exhibit quite unusual structural scaffolds like the β-propeller fold found in fungi or the α + β fold found in the Gymnospermae (Figure 11A–L):
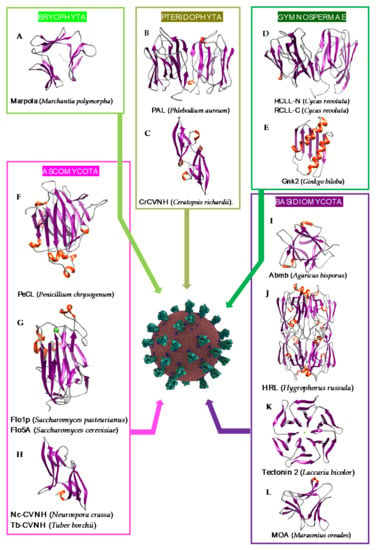
Figure 11.
Overview of lectins from the major representative genera/species of lower plants and fungi, with their associated structural scaffolds, able to recognize the spike proteins from coronaviruses (Coronavirus Credit: Maria Voigt/RCSB PDB). A-L correspond to lectins used as structural scaffolds (A): Marpola, (B): PAL, (C): CrCVNH, (D): RCLL-N, (E): Gnk2, (F): PeCL, (G): Flo1p, (H): TbCVNH, (I): Abmb, (J): HRL, (K): Tectonin 2, (L): MOA. References [,,,,,,,,,,,,] correspond to the lectins presented in the figure.
- -
- The β-sandwich structure (jelly-roll structural scaffold) typical for legume lectins, also occurs in the Man-specific lectin PeCL from Penicillium chrysogenum, and in floculins Flo1p from the yeast Saccharomyces pasteurianum and Flo5p from Saccharomyces cerevisiae.
- -
- The β-prism II structure (β-trefoil structural scaffold), that builds the GNA-related lectin protomer, also occurs in Marpola, the Man-specific lectin isolated from the liverwort Marchantia polymorpha, and in mushrooms (Abmb from Agaricus bisporus, MOA from Marasmius oreades). Special mention should be made to the apparent β-trefoil structure that was found in the mini-lectin PhosL from the mushroom Pholiota squarrosa. In fact, PhosL results from the trimeric association of three antiparallel β-sheets, each possessing a CBS, that mimics a β-trefoil structure with three identical CBSs. This newly identified lectin fold is involved in the specific recognition of the core α1,6-fucosylation of fucosylated N-glycans [] (Figure 12A,B).
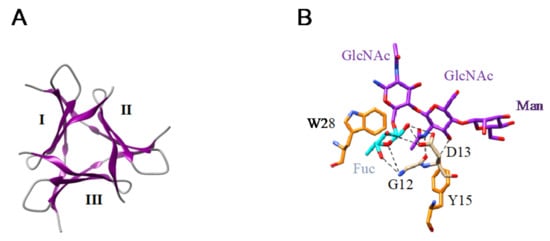 Figure 12. (A). Arrangement of the three antiparallel β-sheets (I, II, III) forming the PhosL homotrimer (PDB code 5XZK) to mimic a pseudo-β-trefoil structure. (B). Network of hydrogen bonds (black dashed lines) anchoring the fucose (Fuc, colored cyan) α1,6-linked for the first GlcNAc from a fucosylated N-glycan to the CBS of PhosL (PDB code 6FX1). Aromatic residues Y89 and Y90 participating in stacking interactions with the pyranose rings of mannobiose are colored orange.
Figure 12. (A). Arrangement of the three antiparallel β-sheets (I, II, III) forming the PhosL homotrimer (PDB code 5XZK) to mimic a pseudo-β-trefoil structure. (B). Network of hydrogen bonds (black dashed lines) anchoring the fucose (Fuc, colored cyan) α1,6-linked for the first GlcNAc from a fucosylated N-glycan to the CBS of PhosL (PDB code 6FX1). Aromatic residues Y89 and Y90 participating in stacking interactions with the pyranose rings of mannobiose are colored orange.
- -
- A rather unusual 6-bladded β-propeller organization was identified in tectonin 2, the Man-specific lectin from the basidiomycete Laccaria bicolor []. As commonly observed in other lectins exhibiting a β-propeller structure, tectonin 2 contains 6 identical Man-specific CBSs located at the top of each of the six blades forming the β-propeller.
- -
- Another quite unusual structural scaffold, which consists of an antiparallel five-stranded β-sheet associated to two α-helices (α + β fold), occurs in the monomeric ginkbilobin 2, a Man-specific lectin from Ginkgo biloba []. Hydrophilic residues occurring on the loops connecting the β-strands associated with charged residues of two β-strands, to form a Man-binding pocket located at the top of the monomer [].
The mannose-binding specificity of lectins from lower plants (Bryophyta, Pteridophyta, Gymnospermae), yeasts, and mushrooms (Ascomycota and Basidiomycota), is directed against both Man, mannosides, and more complex high-mannose glycans, as shown from the X-ray crystallographic studies for various Man-specific lectins complexed to Man and oligomannosides (Figure 13 and Figure 14).
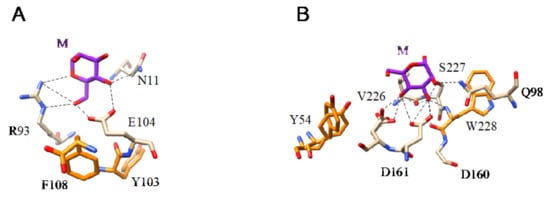
Figure 13.
(A). Network of hydrogen bonds (black dashed lines) anchoring Man (M) to the amino acid residues N11, R93, and E104, forming the CBS of ginkbilobin from Ginkgo biloba (PDB code 4XRE). Aromatic residue Y103 and F108 participating in weak stacking interactions with the pyranose ring of Man are colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring Man (M) to the amino acid residues Q98, D160, D161, V226, and S227, forming the CBS of flocculin Flo5 from Saccharomyces cerevisiae (PDB code 2XJP). Aromatic residues Y54 and W228 involved in stacking interactions with the pyranose ring of Man are colored orange.
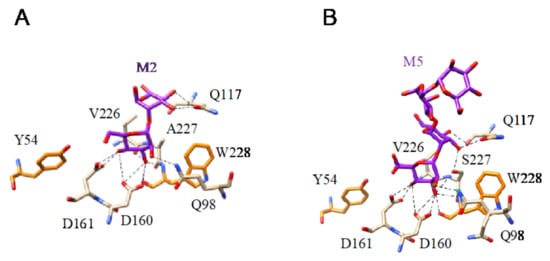
Figure 14.
(A). Network of hydrogen bonds (black dashed lines) anchoring Mannobiose (M2) to the amino acid residues forming the CBS of flocculin Flo5 from Saccharomyces cerevisiae (PDB code 2XJU). Aromatic residue Y54 and FW228 participating in stacking interactions with the pyranose ring of the first Man residue are colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring Mannopentaose (M5) to the amino acid residues forming the CBS of flocculin Flo5 from Saccharomyces cerevisiae (PDB code 2XJT). Aromatic residue Y54 and W228 involved in stacking interactions with the pyranose ring of the first Man of the oligomannoside are colored orange.
In the glycan array analyses performed at Consortium for Functional Glycomics for TbCVN, the Tuber borchii Man-specific lectin yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 15):
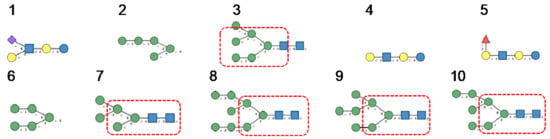
Figure 15.
Glycans recognized by TbCVN in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Glycan array studies performed for ABA, the Agaricus bisporus lectin, yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 16):
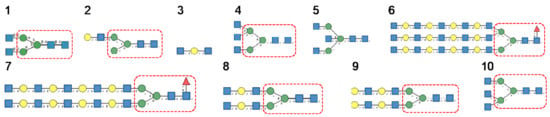
Figure 16.
Glycans recognized by ABA in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Glycan array analyses performed for the lectin MOA from the Marasmius oreades mushroom, yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 17):
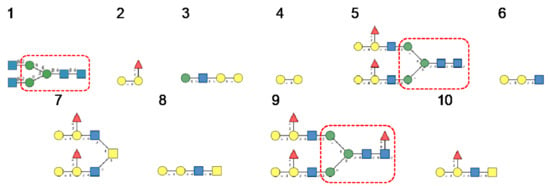
Figure 17.
Glycans recognized by MOA in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Mannose-specific lectins from Ascomycota, such as the TbCVN and NcCVN cyanovirin-related lectins from Tuber borchii (TbCVN) and Neurospora crassa (NcCVN), usually differ from lectins of the Basidiomycota by a more pronounced affinity to high-mannose glycans and a lesser affinity for complex type glycans, such as ABA from Agaricus bisporus and MOA from Marasmius oreades.
2.3. Man-Specific Lectins from Algae and Bacteria
Seaweeds belonging to the different classes of algae, including red algae (Rhodophyta), brown algae (Phaeophyta), green algae (Chlorophyta), and yellow-green algae (Chrysophyta), are well-known sources of Man-specific lectins, together with the lectins from Actinobacteria and Cyanobacteria, formerly classified as blue algae.
Seaweed Man-specific lectins exhibit well-identified structural scaffolds but some of them also exhibit unrelated structural scaffolds that still remain to be characterized.
- -
- The β-prism I or β-barrel scaffold occurs predominantly in red algae (Rhodophyta) but is less common in other classes of algae (Figure 18A–M). Algae of the genera Agardhiella, Eucheuma, Kappaphycus, Meristiella, Meristotheca, and Solieria exhibit this type of molecular organization, in which the CBS is located at the top of the barrel structure.
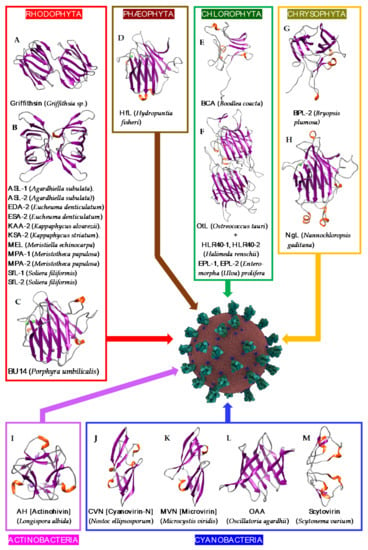 Figure 18. Overview of lectins from representative genera/species of algae and cyanobacteria, with their associated structural scaffolds, able to recognize the spike proteins from coronaviruses (Coronavirus Credit: Maria Voigt/RCSB PDB). A-M correspond to lectins used as structural scaffolds ((A): Griffithsin, (B): ASL-1, (C): BU14, (D): HfL, (E): BCA, (F): OtL, (G): BPL-2, (H): NgL, (I): Actinohivin, (J): CVN, (K): MVN, (L): OAA, (M): Scytovirin. References [,,,,,,,,,,,,,,,,,,,,] correspond to the lectins presented in the figure.
Figure 18. Overview of lectins from representative genera/species of algae and cyanobacteria, with their associated structural scaffolds, able to recognize the spike proteins from coronaviruses (Coronavirus Credit: Maria Voigt/RCSB PDB). A-M correspond to lectins used as structural scaffolds ((A): Griffithsin, (B): ASL-1, (C): BU14, (D): HfL, (E): BCA, (F): OtL, (G): BPL-2, (H): NgL, (I): Actinohivin, (J): CVN, (K): MVN, (L): OAA, (M): Scytovirin. References [,,,,,,,,,,,,,,,,,,,,] correspond to the lectins presented in the figure. - -
- A less common β−prism II organization occurs in red algae (Grateloupia chiangii) and green algae (Boodlea coacta), in which CBSs are located in the grooves delineated by the bundles of β-strands. The MFP2-like structural scaffold, related to the β-prism II organization, occurs in the green alga Bryopsis plumosa.
- -
- A rather uncommon β-sandwich organization was found in the red alga Hydropuntia (Gracilaria) fisheri, and in the green alga Ostreococcus tauri.
- -
- It is interesting to note that the structural scaffolds of many other Man-specific lectins, identified in red algae (Carpopeltis flabellata, Gracilaria bursa-pastoris) and green algae (Enteromopha prolifera, Halimeda renschii), still remain unknown and are not apparently related to any of the known structural scaffolds reported in algae.
Man-specific lectins from the actinobacteria (actinohivin) and cyanobacteria/ex-blue algae group (cyanovirin CVN, microvirin MVN, scytovirin SVN, and the Oscillatoria agardhii agglutinin OAA), exhibit a β-prism II (β-trefoil) structure and a β-prism I (β-barrel) structure, respectively (Figure 18).
The three-dimensional structures for different bacterial lectins in complex with Man, mannosides, and more complex oligomannoside chains are available at the PDB (Figure 19 and Figure 20).
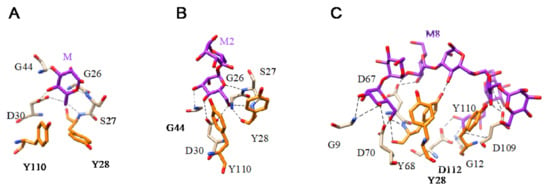
Figure 19.
(A). Network of hydrogen bonds (black dashed lines) anchoring Man (M) to the amino acid residues forming the CBS of griffithsin (PDB code 2RDK). Aromatic residue Y28 and Y110 participating in stacking interactions with the pyranose ring of Man are colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring 6α-mannobiose (M2) to the amino acid residues of the CBS of griffithsin (PDB code 2HYQ). Aromatic residue Y28 and Y110 involved in stacking interactions with the pyranose ring of the first Man residue are colored orange. (C). Network of hydrogen bonds (black dashed lines) anchoring a high-mannose branched glycan (M8) to the amino acid residues of the CBS of griffithsin (PDB code 3LL2). Aromatic residue Y28, Y68, and Y110, participating in stacking interactions with the pyranose rings of Man residues, are colored orange.
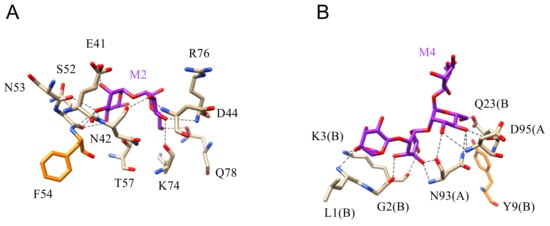
Figure 20.
(A). Network of hydrogen bonds (black dashed lines) anchoring mannobiose (M2) to the amino acid residues forming the CBS of cyanovirin CVN (PDB code 2PYS). Aromatic residue F54 participating in a very weak stacking interaction with the pyranose ring of Man is colored orange. (B). Network of hydrogen bonds (black dashed lines) anchoring mannotetraose (M4) to cyanovirin (PDB code 3GXZ). Amino acid residues of A and B protomers forming the swapped CVN dimer, participate in the interaction with the oligomannoside.
High-mannose glycan-binding activity measured by Hori et al. [] for ESA-2, the Eucheuma serra lectin, yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 21):
Figure 21.
Glycans recognized by ESA-2 in glycan-binding activity measurements, arranged from 1 to 10 in decreasing order of affinity.
Glycan array analyses performed at Consortium for Functional Glycomics for cyanovirin-N CVN-N, a typical member of the cyanobacterial Man-specific lectins, yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 22):
Figure 22.
Glycans recognized by CVN-N in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Glycan array studies performed for actinohivin, a typical member of the actinobacteria Man-specific lectins, yielded the best results with the following glycans arranged in decreasing order of affinity (Figure 23):
Figure 23.
Glycans recognized by actinohivin in glycan array experiments, arranged from 1 to 10 in decreasing order of affinity.
Man-specific lectins from algae, actinobacteria, and cyanobacteria, almost exclusively interact with high-mannose glycans and, to a very little extent, with complex N-glycans. Most of the high-mannose glycans recognized by algal and bacterial lectins readily participate in the glycan equipment of the S proteins forming the spikes that decorate the surface of SARS-CoV, MERS-CoV, and SARS-CoV-2 virions.
6. Biomedical Perspectives
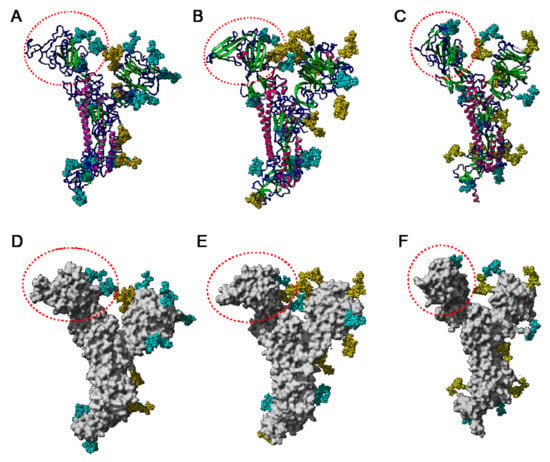
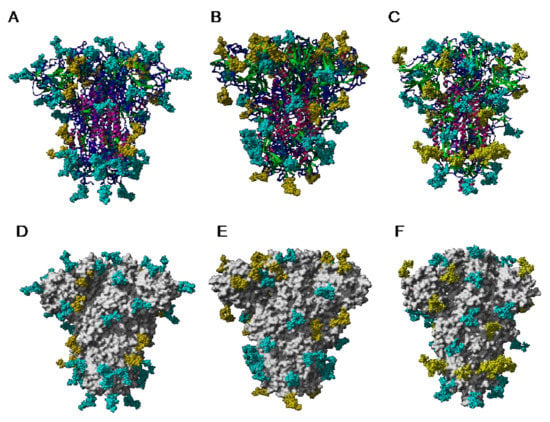
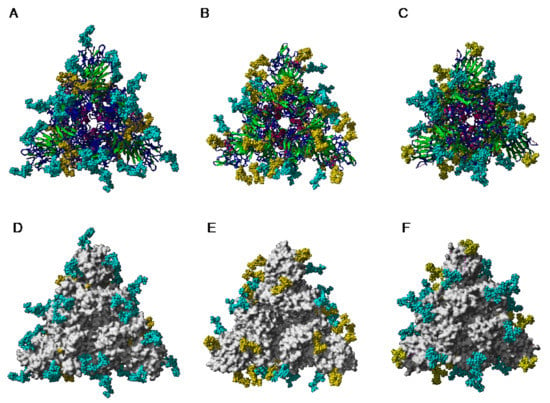
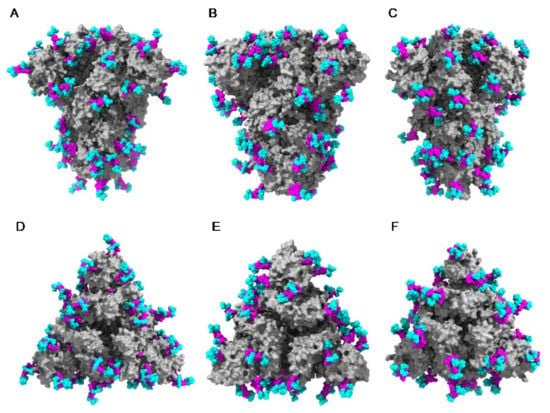
The extensive glycosylation at the surface of the S protein offers a shielding effect to SARS-CoV, MERS-CoV, and SARS-CoV-2. In addition to this protective role, a possible stabilization function has been postulated for some of the N-glycans covering the SARS-CoV-2 S protein, that could be exploited in the future for targeting the glycan coat by therapeutic drugs []. However, the distribution of the glycan shield at the surface of the spikes and especially, at the top of the spikes, does not consistently support a protective function susceptible to reduce the vulnerability of the coronaviruses towards antibody epitopes and/or exogenous proteases. In this respect, the exposed area of the RBDs at the top of the spikes is substantially free of glycans and thus, remains available for specific recognition by the opened spike conformation of the corresponding ACE2 (SARS-CoV and SARS-CoV-2) and DPP4 (MERS-CoV) host cell receptors (Figure 36A–C).
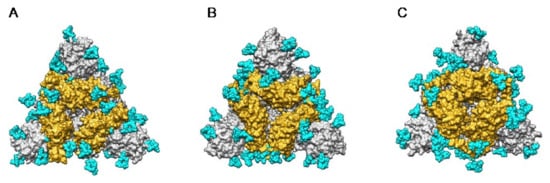
Figure 36.
(A–C). Top face of the ribbon diagram of the homotrimeric spike of SARS-CoV (PDB code 6ACD) (A), MERS-CoV (PDB code 5W9H) (B), and SARS-CoV-2 (PDB code 6VXX) (C). High-mannose glycans and complex glycans are colored cyan. Molecular surfaces corresponding to the RBDs are colored gold and the remaining molecular surfaces are colored grey.
Nevertheless, the steric hindrance created by the interaction of Man-specific lectins such as the dimeric PsA (50 kDa), the tetrameric GNA (50 kDa), and Morniga-M (60 kDa) or the octameric Heltuba (120 kDa), with the high-mannose and complex N-glycans located in the vicinity of the RBD area, should interfere with the receptor recognition process and could disturb or prevent the entry of coronaviruses in host cells. In this respect, the di-, tetra-, hexa- (Parkia platicephala lectin), and octameric Man-specific lectins from higher plants, which exhibit high molecular weights in the range between 50–100 kDa, could serve as blocking agents that can prevent the coronavirus infection. Other Man-specific lectins from algae, fungi, and cyanobacteria seem to be less adapted to act as a blocking agent owing to their smaller size, e.g., 25 kDa for the griffithsin swapped homodimer and 20 kDa for the cyanovirin swapped homodimer, respectively (Figure 37).
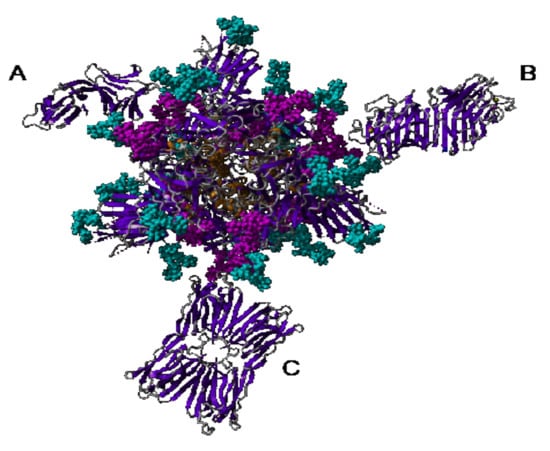
Figure 37.
Cartoon showing the relative size of different Man-binding lectins (dimeric GNA-like lectin (A), bivalent legume lectin β2α2 (B), and Jacalin-related lectin (C)), complexed to the homotrimeric spike (front face) from SARS-CoV-2.
As blocking agents for coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2, Man-specific lectins from higher plants, algae, fungi, and cyanobacteria could be used to entrap and immobilize the virions using air-conditioned filters impregnated with lectins to prevent the coronavirus dissemination by contaminated people. However, the covalent fixation of lectins to air-conditioned filters necessitates large amounts of purified lectins, which may be problematic in view of low yields usually obtained during purification of Man-specific lectins from higher plants. Recent improvements performed in the large-scale purification of the algal lectins from Eucheuma [], and griffithsin from tobacco leaves, could provide the bulk production of Man-specific lectins required for the manufacturing of lectin-impregnated filters []. However, the blocking activity of dried lectins immobilized on air-conditioned filters seems unlikely because these proteins need water to retain both their conformation and carbohydrate-binding activity. At most, one can imagine a filtration device equipped with a wet filter containing immobilized lectins, which would remove the air from the virions before entering the air-conditioned filter.
Another potential biomedical application for Man-specific lectins could consist of their use as glycan probes for the detection of coronaviruses in the environment, e.g., on various domestic surfaces such as doorknobs, home furnishings, handrails, mirrors, computers, etc. Virions deposited on these surfaces could be detected under UV illumination, e.g., using specific antibodies labeled with a fluorochrome.
It is possible to imagine further therapeutic issues for Man-specific lectins with respect to the coronavirus infection. Recently, a risk of contamination by SARS-CoV-2 has been identified via spoiled blood or blood products transfused to healthy people []. Although the risk of a transfusion transmission is essentially theoretical [], a plasmapheresis through a column of immobilized Man-specific lectin would be sufficient to eliminate the eventual blood contaminants. A similar filtration of the suspected blood samples through a column containing an immobilized Man-specific lectin could be performed in blood centers or blood banks and would be a control measure sufficient to purge the blood from coronavirus particles and thus prevent possible contamination for the transfused people. In this respect, up to 80% of MERS-CoV and 70% of MARV (Marburg virus), were successfully eliminated by lectin affinity plasmapheresis using GNA as a blocking agent immobilized in the extra-capillary space of a standard plasma filter [].
From the scarce in vitro experiments conducted to study the effects of Man-specific lectins from higher plants [,,], and griffithsin [,], towards MERS-CoV and SARS-CoV-2, the anti-viral activity of lectins appeared to prevent both attachment and entry of the virus in the host cell but does not interfere with the coronavirus replication process within the cell. Consequently, Man-specific lectins cannot be considered as replication blockers for coronaviruses. A very different situation occurs for other enveloped viruses such as HIV-1 [], influenza virus [], and Herpes virus [], whose replication is apparently blocked in vitro, in the presence of Man-specific lectins.
In addition, some of the biomedical applications presented here could be adapted to veterinary medicine by addressing animal coronavirus concerns [] as part of the “one health” approach.
7. Bioinformatics
The X-ray coordinates available for lectins, lectins in complex with simple sugars, oligosaccharides, and more complex glycans, and S-glycoproteins from SARS-CoV, MERS-CoV, and SARS-CoV-2, were taken from the PDB.
Homology modeling of other lectins was performed with the YASARA Structure program [] using various structurally related proteins as templates from the PDB [], depending on the overall structural scaffold to which they belong. PROCHECK [], ANOLEA [], and the calculated QMEAN scores [,] were used to assess the geometric and thermodynamic qualities of the resulting three-dimensional models.
Docking of simple sugars and oligosaccharides was performed with YASARA and SwissDock [,]. The molecular surface of the lectins and S-glycoproteins from SARS-CoV, MERS-CoV, and SARS-CoV-2, were calculated and displayed with Chimera []. Molecular cartoons were drawn with Chimera and YASARA.
Assuming that putative N-glycosylation sites Asn-X-Thr/Ser of SARS-CoV, MERS-CoV, and SARS-CoV2, are actually glycosylated, a classic bi-antennary high-mannose glycan chain with a tri-mannoside core (Man)2-(Man)3-(GlcNAc)2 was modeled using the GlyProt server (http://www.glycosciences.de/modeling/glyprot/php/main.php, accessed on 1 March 2021) [], and represented in CPK on the molecular surface of the spike S-glycoproteins. According to the known differential distribution of high-mannose N-glycans and complex N-glycans on the surface of the S-glycoproteins, another run of modeling was performed using standard high-mannose glycans and complex glycans as models.
Cartoons for high-mannose glycans, complex N-glycans, and hybrid glycans, were built and represented with the DrawGlycan SNFG package for Mac []. Classical colored symbols were used for representing Fuc (red triangle), Gal (yellow circle), GalNAc (yellow square), GlcNAc (blue square), Man (green circle), and sialic acid/Neu5Ac (purple losange), respectively.
8. Discussion
Glycans of the S-glycoproteins forming the spikes of SARS-CoV, MERS-CoV and SARS-CoV-2, consist of high-mannose glycans and often sialylated N-glycans that predominantly occupy their N-glycosylation sites [,]. However, depending on the coronaviruses, some discrepancies occur between the distribution of the two types of glycans on the surface of the virion, which introduces some diversity in the glycan shield covering the coronavirus spikes []. Accordingly, Man-specific lectins from plants, algae, fungi, and bacteria, which differ slightly due to their fine sugar-binding specificities, offer a vast panel of glycan probes more or less adapted to the specific recognition of the different coronaviruses. In this respect, GNA-related lectins together with Man-specific lectins from algae and cyanobacteria, appear as glycan probes nicely adapted to the recognition of the high-mannose shield which predominates at the top of the MERS-CoV spike. Otherwise, legume lectins with a higher affinity for N-glycans possessing the trimannoside Manα1,3Manα1,6Man core, seem better adapted to the recognition of the N-glycans distributed predominantly at the top of the glycan shield from SARS-CoV and SARS-CoV-2.
Besides Man-specific lectins, other lectins displaying different carbohydrate-binding specificities have previously been identified as glycan probes for MERS-CoV [,]. In this respect, lectins exhibiting Man/Glc-specificity (CLA from Cladrastris lutea), Gal/GalNAc-specificity (Morniga-G from Morus nigra), GlcNAc-specificity (Nictaba from Nicotiana tabacum), chitin-specificity (UDA from Urtica dioica), display an antiviral activity often higher or similar to those displayed by genuine Man-specific lectins, essentially the GNA-related lectins from monocot plants []. The occurrence of (sialylated)complex glycans and hybrid glycans arrayed on the surface of coronavirus spikes, readily accounts for the differences observed among the carbohydrate-binding specificities towards the coronaviruses. Inhibition of the SARS-CoV replication was observed in a lethal SARS-CoV-infected BALB/c mouse model after treatment with the (GlcNAc)n-binding stinging nettle (Urtica dioica) lectin []. Nictaba from Nicotiana tabacum and UDA, two GlcNAc-binding lectins, also displayed antiviral properties against several families of enveloped viruses including influenza A/B, Dengue virus type 2, herpes simplex virus, and HIV, respectively []. Accordingly, a mixture of lectins with different carbohydrate-binding specificities should provide a glycan probe tool well adapted to all of the coronavirus types and strains.
Author Contributions
A.B., M.S. and H.B., provided and analyzed the bibliographic information for lectins. S.L.P. and B.K. provided and analyzed the bibliographic information dealing with coronaviruses and D.P. analyzed the technical aspects of the bibliographic information. P.R. performed the in silico molecular modelling and docking experiments, and provided the molecular cartoons and the docking pictures. E.J.M.V.D. and P.R. wrote the review. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Belouard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef]
- Lu, G.; Wang, Q.; Gao, G.F. Bat-to-human: Spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015, 23, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Tortorici, M.A.; Bosch, B.J.; Frenz, B.; Rottier, P.J.M.; DiMaio, F.; Rey, F.A.; Veesler, D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 2016, 531, 114–117. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar]
- Glowacka, I.; Bertram, S.; Herzog, P.; Pfefferle, S.; Steffen, I.; Muench, M.O.; Simmons, G.; Hofmann, H.; Kuru, T.; Wever, F.; et al. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010, 84, 1198–1205. [Google Scholar] [CrossRef]
- Li, F. Receptor recognition and cross-species infections of SARS coronavirus. Antivir. Res. 2013, 100, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Kee Wong, S.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.M.; Rottier, P.J.M. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Berndsen, Z.; Raghwani, J.; Seabright, G.; Allen, J.D.; Pybus, O.G.; McLellan, J.S.; Wilson, I.A.; Bowden, T.A.; Ward, A.B.; et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020, 11, 2688. [Google Scholar] [CrossRef] [PubMed]
- Shajahan, A.; Supekar, N.T.; Gleinich, A.S.; Azadi, P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 2020, 30, 981–988. [Google Scholar] [CrossRef]
- Keyaerts, E.; Vijgen, L.; Pannecouque, C.; Van Damme, E.; Peumans, W.; Egberink, H.; Balzarini, J.; Van Ranst, M. Plant lectins are potent inhibitors of coronaviruses by interfering with two targets in the viral replication cycle. Antivir. Res. 2007, 75, 179–187. [Google Scholar] [CrossRef]
- Balzarini, J. Carbohydrate-binding agents: A potential future cornerstone for the chemotherapy of enveloped viruses? Antivir. Chem. Chemother. 2007, 18, 1–11. [Google Scholar] [CrossRef]
- Barre, A.; Van Damme, E.J.M.; Simplicien, M.; Benoist, H.; Rougé, P. Man-specific, GalNAc/T-Tn-specific and Neu5Ac-specific seaweed lectins as glycan probes for the SARS-CoV-2 (COVID19) coronavirus. Mar. Drugs 2019, 18, 543. [Google Scholar]
- Sohrab, S.S.; Suhail, M.; Kamal, M.A.; Ahmad, F.; Azhar, E.I. Emergence of human pathogenic coronaviruses: Lectins as antivirals for SARS-CoV-2. Curr. Pharm. Des. 2020, 26, 5286–5292. [Google Scholar] [CrossRef]
- Kumar Gupta, R.; Apte, G.R.; Bharat Lokhande, K.; Mishra, S.; Pal, J.K. Carbohydrate-binding agents: Potential of repurposing for COVID-19 therapy. Curr. Protein Pept. Sci. 2020, 21, 1085–1096. [Google Scholar] [CrossRef]
- Tsaneva, M.; Van Damme, E.J.M. 130 years of plant lectin research. Glycoconj. J. 2020, 37, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Liener, I.E.; Sharon, N.; Goldstein, I.J. The Lectins, Properties, Functions, and Applications in Biology and Medicine; Academic Press Inc.: Orlando, FL, USA; San Diego, CA, USA; New York, NY, USA; Austin, TX, USA, 1986; pp. 1–600. [Google Scholar]
- Peumans, W.J.; Van Damme, E.J.M.; Barre, A.; Rougé, P. Classification of plant lectins in families of structurally and evolutionary related proteins. Adv. Exp. Med. Biol. 2001, 491, 27–54. [Google Scholar] [PubMed]
- Debray, H.; Decout, D.; Strecker, G.; Spik, G.; Montreuil, J. Specificity of twelve lectins towards oligosaccharides and glycopeptides related to N-glycosylproteins. Eur. J. Biochem. 1981, 117, 41–55. [Google Scholar] [CrossRef]
- Bourne, Y.; Anguille, C.; Fontecilla-Camps, J.C.; Rougé, P.; Cambillau, C. Co-crystallization and preliminary X-ray diffraction studies of Lathyrus ochrus isolectin I with di- and trisaccharides, and a biantennary octasaccharide. J. Mol. Biol. 1990, 213, 211–213. [Google Scholar] [CrossRef]
- Barre, A.; Bourne, Y.; Van Damme, E.J.M.; Peumans, W.J.; Rougé, P. Mannose-binding plant lectins: Different structural scaffolds for a common sugar-recognition process. FEBS Lett. 2001, 83, 645–651. [Google Scholar] [CrossRef]
- Akkouh, O.; Ng, T.B.; Singh, S.S.; Yin, C.; Dan, X.; Chan, Y.S.; Pan, W.; Cheung, R.C.F. Lectins with anti-HIV activity: A review. Molecules 2015, 20, 648–668. [Google Scholar] [CrossRef] [PubMed]
- Levendosky, K.; Mizenina, O.; Martinelli, E.; Jean-Pierre, N.; Kizima, L.; Rodriguez, A.; Kleinbeck, K.; Bonnaire, T.; Robbiani, M.; Zydowsky, T.M.; et al. Griffithsin and carrageenan combination to target Herpes simplex virus 2 and human papilloma virus. Antimicrob. Agents Chemother. 2015, 59, 7290–7298. [Google Scholar] [CrossRef] [PubMed]
- Meuleman, P.; Albecka, A.; Belouzard, S.; Vercauteren, K.; Verhoye, L.; Wychowski, C.; Leroux-Roels, G.; Palmer, K.E.; Dubuisson, G. Griffithsin has antiviral activity against hepatitis C virus. Antimicrob. Agents Chemother. 2011, 55, 5159–5167. [Google Scholar] [CrossRef]
- Barton, C.; Kouokam, J.C.; Lasnik, A.B.; Foreman, O.; Cambon, A.; Brock, G.; Montefiori, D.C.; Vojdani, F.; McCormick, A.A.; O’Keefe, B.R.; et al. Activity of and effect of subcutaneous treatment with the broad-spectrum antiviral lectin griffithsin in two laboratory rodent models. Antimicrob. Agents Chemother. 2014, 58, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; O’Keefe, B.R.; Sowder, R.C., 2nd; Bringans, S.; Gardella, R.; Berg, S.; Cochran, P.; Turpin, J.A.; Buckheit, R.W., Jr.; McMahon, J.B.; et al. Isolation and characterization of griffithsin, a novel HIV-inactivating protein, from the red alga Griffithsia sp. J. Biol. Chem. 2005, 280, 9345–9353. [Google Scholar] [CrossRef]
- Boyd, M.R.; Gustafson, K.R.; McMahon, J.B.; Shoemaker, R.H.; O’Keefe, B.R.; Mori, T.; Gulakowski, R.J.; Wu, L.; Rivera, M.I.; Laurencot, C.M.; et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: Potential applications to microbicide development. Antimicrob. Agents Chemother. 1997, 41, 1521–1530. [Google Scholar] [CrossRef]
- Chiba, H.; Inokoshi, J.; Okamoto, M.; Asanuma, S.; Matsuzaki, K.I.; Iwama, M.; Mizumoto, K.; Tanaka, H.; Oheda, M.; Fujita, K.; et al. Actinohivin, a novel anti-HIV protein from actinomycete that inhibits syncytium formation: Isolation, characterization, and biological activities. Biochem. Biophys. Res. Commun. 2001, 282, 595–601. [Google Scholar] [CrossRef]
- Huskens, D.; Férir, G.; Vermeire, K.; Kehr, J.-C.; Balzarini, J.; Dittmann, E.; Schols, D. Microvirin, a novel α(1,2)-mannose-specific lectin isolated from Microcystis aeruginosa, has anti-HIV-1 activity comparable with that of cyanovirin-N but a much higher safety profile. J. Biol. Chem. 2010, 285, 24845–24854. [Google Scholar] [CrossRef]
- Balzarini, J.; Schols, D.; Neyts, J.; Van Damme, E.; Peumans, W.; De Clercq, E. α-(1-3)- and α-(1–6)-D-mannose-specific plant lectins are markedly inhibitory to human immunodeficiency virus and cytomegalovirus infections in vitro. Antimicrob. Agents Chemother. 1991, 35, 410–416. [Google Scholar] [CrossRef]
- Balzarini, J.; Neyts, J.; Schols, D.; Hosoya, M.; Van Damme, E.; Peumans, W.; De Clercq, E. The mannose-specific plant lectins from Cymbidium hybrid and Epipactis helleborine and the (N-acetylglucosamine)n-specific plant lectin from Urtica dioica are potent and selective inhibitors of human immunodeficiency virus and cytomegalovirus replication in vitro. Antivir. Res. 1992, 18, 191–207. [Google Scholar]
- Balzarini, J.; Hatse, S.; Vermeire, K.; Princen, K.; Aquaro, S.; Perno, C.F.; De Clercq, E.; Egberink, H.; Vanden Mooter, G.; Peumans, W.; et al. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrob. Agents Chemother. 2004, 48, 3858–3870. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Simplicien, M.; Benoist, H.; Van Damme, E.J.M.; Rougé, P. Mannose-specific lectins from marine algae: Diverse structural scaffolds associated to common virucidal and anti-cancer properties. Mar. Drugs 2019, 17, 440. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, K.B.; Gray, E.S.; Lambson, B.E.; nMoore, P.L.; Choge, I.A.; Mlisana, K.; Karim, S.S.; McMahon, J.; O’Keefe, B.; Chikwamba, R.; et al. Mannose-rich glycosylation patterns on HIV-1 subtype C gp120 and sensitivity to lectins, griffithsin, cyanovirin-N and scytovirin. Virology 2010, 402, 187–196. [Google Scholar] [CrossRef]
- Sato, Y.; Morimoto, K.; Kubo, T.; Sakaguchi, T.; Nishizono, A.; Hirayama, M.; Hori, K. Entry inhibition of influenza viruses with high mannose binding lectin ESA-2 from the red alga Eucheuma serra through the recognition of viral hemagglutinin. Mar. Drugs 2015, 13, 3454–3465. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tang, G.; Tang, J.; Wang, L.-X. Targeting N-glycan cryptic sugar moieties for broad-spectrum virus neutralization: Progress in identifying conserved molecular targets in viruses of distinct phylogenetic origins. Molecules 2015, 20, 4610–4622. [Google Scholar] [CrossRef]
- Mitchell, C.A.; Ramessar, K.; O’Keefe, B.R. Antiviral lectins: Selective inhibitors of viral entry. Antivir. Res. 2017, 142, 37–54. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, G.S. Status of mannose-binding lectin (MBL) and complement system in COVID-19 patients and therapeutic applications of antiviral plant MBLs. Mol. Cell Biochem. 2021, 21, 1–26. [Google Scholar]
- Animashaun, T.; Hughes, R.C. Bowringia milbraedii agglutinin. Specificity of binding to early processing intermediates of asparagine-linked oligosaccharide and use as a marker of endoplasmic reticulum glycoproteins. J. Biol. Chem. 1989, 264, 4657–4663. [Google Scholar] [CrossRef]
- Souza Teixeira, C.; da Silva, H.C.; de Moura, T.R.; Pereira-Júnior, F.N.; do Nascimento, K.S.; Nagano, C.S.; Sampaio, A.H.; Delatorre, P.; Rocha, B.A.; Cavada, B.S. Crystal structure of the lectin of Camptosema pedicellatum: Implication of a conservative substitution at the hydrophobic subsite. J. Biochem. 2012, 152, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, B.B.; Goldstein, I.J. Physical and chemical characterization of concanavalin A, the hemagglutinin from jack bean (Canavalia ensiformis). Biochim. Biophys. Acta 1967, 133, 376–379. [Google Scholar] [CrossRef]
- Del Sol, F.G.; Cavada, B.S.; Calvete, J.J. Crystal structures of Cratylia floribunda seed lectin at acidic and basic pHs. Insights into the structural basis of the pH-dependent dimer-tetramer transition. J. Struct. Biol. 2007, 158, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rocha, B.A.; Delatorre, P.; Oliveira, T.M.; Benevides, R.G.; Pires, A.F.; Sousa, A.A.; Souza, L.A.; Assreuy, A.M.; Debray, H.; de Azevedo, X.W.F.; et al. Structural basis for both pro- and anti-inflammatory response induced by mannose-specific legume lectin from Cymbosema roseum. Biochimie 2011, 93, 806–816. [Google Scholar] [CrossRef]
- Rozwarski, D.A.; Swami, B.M.; Brewer, C.F.; Sacchetini, J.C. Crystal structure of the lectin from Dioclea grandiflora complexed with core trimannoside of asparagine-linked carbohydrates. J. Biol. Chem. 1998, 273, 32818–32825. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.C.; Osterne, V.J.; Santiago, M.Q.; Pinto-Junior, V.R.; Silva-Filho, J.C.; Lossio, C.F.; Nascimento, F.L.; Almeida, R.P.; Teixeira, C.S.; Leal, R.B.; et al. Structural analysis of Centrolobium tomentosum seed lectin with inflammatory activity. Arch. Biochem. Biophys. 2016, 596, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Bourne, Y.; Abergel, C.; Cambillau, C.; Frey, M.; Rougé, P.; Fontecilla-Camps, J.C. X-ray crystal structure determination and refinement at 1.9 Å resolution of isolectin I from the seeds of Lathyrus ochrus. J. Mol. Biol. 1990, 214, 571–584. [Google Scholar] [CrossRef]
- Foriers, A.; Van Driessche, E.; De Neve, R.; Kanarek, L.; Strosberg, A.D. The subunit structure and N-terminal sequences of the α- and β-subunits of the lentil lectin (Lens culinaris). FEBS Lett. 1977, 75, 237–240. [Google Scholar] [CrossRef]
- Gao, S.; An, J.; Wu, C.F.; Gu, Y.; Chen, F.; Yu, Y.; Wu, Q.Q.; Bao, J.K. Effect of amino acid residue and oligosaccharide chain chemical modifications on spectral and hemagglutinating activity of Millettia dielsiana Harms. ex Diels. lectin. Acta Biochim. Biophys. Sin. 2005, 37, 47–54. [Google Scholar] [CrossRef]
- Young, N.M.; Williams, R.E.; Roy, C.; Yaguchi, M. Structural comparison of the lectin from sainfoin (Onobrychis viciifolia) with concanavalin A and other D-mannose specific lectins. Can. J. Biochem. 1982, 60, 933–941. [Google Scholar] [CrossRef]
- Einspahr, H.; Pareks, E.H.; Suguna, K.; Subramanian, E.; Suddath, F.L. The crystal structure of pea lectin at 3.0-Å resolution. J. Biol. Chem. 1986, 261, 16518–16527. [Google Scholar] [CrossRef]
- Cavada, B.S.; Araripe, D.A.; Silva, I.B.; Pinto-Junior, V.R.; Osterne, V.J.S.; Neco, A.H.B.; Laranjeira, E.P.P.; Lossio, C.F.; Correia, J.L.A.; Pires, A.F.; et al. Structural studies and nociceptive activity of a native lectin from Platypodium elegans seeds (nPELa). Int. J. Biol. Macromol. 2018, 107, 236–246. [Google Scholar] [CrossRef]
- Pereira-Junior, F.N.; Silva, H.C.; Freitas, B.T.; Rocha, B.A.; Nascimento, K.S.; Nagano, C.S.; Leal, R.B.; Sampaio, A.H.; Cavada, B.S. Purification and characterization of a mannose/N-acetyl-D-glucosamine-specific lectin from the seeds of Platymiscium floribundum Vogel. J. Mol. Recognit. 2012, 25, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Loris, R.; Imberty, A.; Beeckmans, S.; Van Driessche, E.; Read, J.S.; Bouckaert, J.; De Greve, H.; Buts, L.; Wyns, L. Crystal structure of Pterocarpus angolensis lectin in complex with glucose, sucrose, and turanose. J. Biol. Chem. 2003, 278, 16297–16303. [Google Scholar] [CrossRef]
- Naeem, A.; Ahmad, E.; Ashraf, M.T.; Khan, R.H. Purification and characterization of mannose/glucose-specific lectin from seeds of Trigonella foenumgraecum. Biochemistry 2007, 72, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Reeke, G.N., Jr.; Becker, J.W. Three-dimensional structure of favin: Saccharide binding-cyclic permutation in leguminous lectins. Science 1986, 234, 1108–1111. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Peumans, W.J.; Hause, B.; Bras, J.; Kumar, M.; Proost, P.; Barre, A.; Rougé, P.; Van Damme, E.J.M. Jasmonic acid methyl ester induces the synthesis of a cytoplasmic/nuclear chito-oligosaccharide binding lectin in tobacco leaves. FASEB J. 2002, 16, 905–907. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Briké, F.; Winter, H.C.; Van Leuven, F.; Goldstein, I.J.; Peumans, W.J. Molecular cloning of two different mannose-binding lectins from tulip bulbs. Eur. J. Biochem. 1996, 236, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Meagher, J.L.; Winter, H.C.; Ezell, P.; Goldstein, I.J.; Stuckey, J.A. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology 2005, 15, 1033–1042. [Google Scholar] [CrossRef]
- Bourne, Y.; Roig-Zamboni, V.; Barre, A.; Peumans, W.J.; Astoul, C.H.; Van Damme, E.J.; Rougé, P. The crystal structure of the Calystegia sepium agglutinin reveals a novel quaternary arrangement of lectin subunits with a β-prism fold. J. Biol. Chem. 2004, 279, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Nomura, K.; Ashida, H.; Uemura, N.; Kushibe, S.; Ozaki, T.; Yoshida, M. Purification and characterization of a mannose/glucose-specific lectin from Castanea crenata. Phytochemistry 1998, 49, 667–673. [Google Scholar] [CrossRef]
- Nakamura-Tsuruta, S.; Uchiyama, N.; Peumans, W.J.; Van Damme, E.J.M.; Totani, K.; Ito, Y.; Hirabayashi, J. Analysis of the sugar-binding specificity of mannose-binding-type jacalin-related lectins by frontal affinity chromatography—An approach to functional classification. FEBS J. 2008, 275, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Azarkan, M.; Feller, G.; Vandenameele, J.; Herman, R.; El Mahyaoui, R.; Sauvage, E.; Vanden Broeck, A.; Matagne, A.; Charlier, P.; Kerff, F. Biochemical and structural characterization of a mannose-binding jacalin-related lectin with two-sugar binding sites from pineapple (Anana comosus) stem. Sci. Rep. 2018, 8, 11508. [Google Scholar] [CrossRef]
- Misquith, S.; Rani, P.G.; Surolia, A. Carbohydrate binding specificity of the B-cell maturation mitogen from Artocarpus integrifolia seeds. J. Biol. Chem. 1994, 269, 30393–30401. [Google Scholar] [CrossRef]
- Barre, A.; Peumans, W.J.; Rossignol, M.; Borderies, M.; Culerrier, R.; Van Damme, E.J.M.; Rougé, P. Artocarpin is a polyspecific jacalin-related lectin with a monosaccharide preference for mannose. Biochimie 2004, 86, 685–691. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, F.D.; Bezerra da Silva, B.; Pessoa Furtado, G.; de Sa Carneiro, I.; Lobo, M.D.P.; Guan, Y.; Guo, J.; Coker, A.R.; Lourenzoni, M.R.; Florindo Guedes, M.I.; et al. Frutapin, a lectin from Artocarpus incisa (breadfruit): Cloning, expression and molecular analysis. Biosci. Rep. 2017, 37, BSR20170969. [Google Scholar] [CrossRef]
- Chang, W.C.; Liu, K.L.; Hsu, F.C.; Jeng, S.T.; Cheng, Y.S. Ipomoelin, a jacalin-related lectin with a compact tetrameric association and versatile carbohydrate binding properties regulated by its N terminus. PLoS ONE 2012, 7, e40618. [Google Scholar] [CrossRef]
- Rabijns, A.; Barre, A.; Van Damme, E.J.M.; Peumans, W.J.; De Ranter, C.J.; Rougé, P. Structural analysis of the jacalin-related lectin Morniga-M from the black mulberry (Morus nigra) in complex with mannose. FEBS J. 2005, 272, 3725–3732. [Google Scholar] [CrossRef]
- Mann, K.; Farias, C.M.; Del Sol, F.G.; Santos, C.F.; Grangeiro, T.B.; Nagano, C.S.; Cavada, B.S.; Calvete, J.J. The amino acid sequence of the glucose/mannose-specific lectin isolated from Parkia platycephala seeds reveals three tandemly arranged jacalin-related domains. Eur. J. Biochem. 2001, 268, 4414–4422. [Google Scholar] [CrossRef]
- Bourne, Y.; Zamboni, V.; Barre, A.; Peumans, W.J.; Van Damme, E.J.; Rougé, P. Helianthus tuberosus lectin reveals a widespread scaffold for mannose-binding lectins. Structure 1999, 7, 1473–1482. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Smeets, K.; Torrekens, S.; van Leuven, F.; Goldstein, I.J.; Peumans, W.J. The closely related homomeric and heterodimeric mannose-binding lectins from garlic are encoded by one-domain and two-domain lectin genes, respectively. Eur. J. Biochem. 1992, 206, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-F.; An, J.; He, X.-J.; Deng, J.; Hong, Z.-X.; Liu, C.; Hong-Zhou, L.; Li, Y.-J.; Wang, C.-J.; Chen, F.; et al. Molecular cloning of a novel mannose-binding lectin gene from bulbs of Amaryllis vittata (Amaryllidaceae). Acta Bot. Sin. 2004, 46, 1301–1306. [Google Scholar]
- Lin, J.; Zhou, X.; Pang, Y.; Gao, H.; Fei, J.; Shen, G.A.; Wang, J.; Li, X.; Sun, X.; Tang, K. Cloning and characterization of an agglutinin gene from Arisaema lobatum. Biosci. Rep. 2005, 25, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Goossens, K.; Smeets, K.; Van Leuven, F.; Verhaert, P.; Peumans, W.J. The major tuber storage protein of Araceae species is a lectin. Characterization and molecular cloning of the lectin from Arum maculatum L. Plant Physiol. 1995, 107, 1147–1158. [Google Scholar] [CrossRef]
- Xu, X.; Wu, C.; Liu, C.; Luo, C.; Luo, Y.; Li, J.; Zhao, X.; Van Damme, E.; Bao, J. Purification and characterization of a mannose-binding lectin from the rhizomes of Aspidistra elatior Blume with antiproliferative activity. Acta Biochim. Biophys. Sin. 2007, 39, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.R.; Winter, H.C.; Vericimo, M.A.; Meagher, J.L.; Stckey, J.A.; Goldstein, I.J.; Paschoalin, V.M.; Silva, J.T. Structural analysis and binding properties of isoforms of tarin, the GNA-related lectin from Colocasia esculenta. Biochim. Biophys. Acta 2015, 1854, 20–30. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Smeets, K.; Torrekens, S.; Van Leuven, F.; Peumans, W.J. Characterization and molecular cloning of mannose-binding lectins from the Orchidaceae species Listera ovata, Epipactis helleborine and Cymbidium hybrid. Eur. J. Biochem. 1994, 221, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, X.; Tang, K. Cloning and expression of a novel cDNA encoding a mannose-binding lectin from Dendrobium officinale. Toxicon 2005, 45, 535–540. [Google Scholar] [CrossRef]
- Kaku, H.; Van Damme, E.J.; Peumans, W.J.; Goldstein, I.J. Carbohydrate-binding specificity of the daffodil (Narcissus pseudonarcissus) and amaryllis (Hippeastrum hybr.) bulb lectins. Arch. Biochem. Biophys. 1990, 279, 298–304. [Google Scholar] [CrossRef]
- Liu, J.; Xu, X.; Liu, J.; Balzarini, J.; Luo, Y.; Kong, Y.; Li, J.; Chen, F.; Van Damme, E.J.; Bao, J. A novel tetrameric lectin from Lycoris aurea with four mannose binding sites per monomer. Acta Biochim. Pol. 2007, 54, 159–166. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Sakamoto, H.; Tateno, H.; Hirabayashi, J.; Oguri, S. Purification, characterization, and molecular cloning of lectin from winter buds of Lysichiton camtschatcensis (L.) Schott. Biosci. Biotechnol. Biochem. 2012, 76, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.S.; Sun, S.S.; Ooi, V.E. Purification and characterization of a new antiviral protein from the leaves of Pandanus amaryllifolius (Pandanaceae). Int. J. Biochem. Cell Biol. 2004, 36, 1440–1446. [Google Scholar] [CrossRef]
- Yao, J.H.; Zhao, X.Y.; Liao, Z.H.; Lin, J.; Chen, Z.H.; Chen, F.; Song, J.; Sun, X.F.; Tang, K.X. Cloning and molecular characterization of a novel lectin gene from Pinellia ternata. Cell Res. 2003, 13, 301–308. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Barre, A.; Rougé, P.; Van Leuven, F.; Balzarini, J.; Peumans, W.J. Molecular cloning of the lectin and a lectin-related protein from common Solomon’s seal (Polygonatum multiflorum). Plant Mol. Biol. 1996, 31, 657–672. [Google Scholar] [CrossRef]
- Ding, J.; Bao, J.; Zhu, D.; Zhang, Y.; Wang, D.C. Crystal structure of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure. J. Struct. Biol. 2010, 171, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Shetty, K.N.; Bhat, G.G.; Inamdar, S.R.; Swamy, B.M.; Suguna, K. Crystal structure of a β-prism II lectin from Remusatia vivipara. Glycobiology 2012, 22, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.M.; Wood, S.D.; Reynolds, C.D.; Rizkallah, P.J.; Peumans, W.J.; Van Damme, E.J.; Allen, A.K. Purification, crystallization and preliminary X-ray analysis of a mannose-binding lectin from bluebell (Scilla campanulata) bulbs. Acta Crystallogr. D Biol. Crystallogr. 1996, 52, 1021–1023. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.S.; Sun, S.S.; Wang, H.; Ooi, V.E. New mannose-binding lectin isolated from the rhizome of Sarsaparilla Smilax glabra Roxb. (Liliaceae). J. Agric. Food Chem. 2004, 52, 6091–6095. [Google Scholar] [CrossRef]
- Luo, Y.; Xu, X.; Liu, J.; Li, J.; Sun, Y.; Liu, Z.; Liu, J.; Van Damme, E.; Balzarini, J.; Bao, J. A novel mannose-binding tuber lectin from Typhonium divaricatum (L.) Decne (family Araceae) with antiviral activity against HSV-II and anti-proliferative effect on human cancer cell lines. J. Biochem. Mol. Biol. 2007, 40, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Pang, Y.; Liu, X.; Wang, X.; Deng, Z.; Sun, X.; Tang, K. Molecular cloning and characterization of a novel mannose-binding lectin cDNA from Zantedeschia aethiopica. Biocell 2005, 29, 187–193. [Google Scholar] [CrossRef]
- Koike, T.; Titani, K.; Suzuki, M.; Beppu, H.; Kuzuya, H.; Maruta, K.; Shimpo, K.; Fujita, K. The complete amino acid sequence of a mannose-binding lectin from “Kidachi Aloe” (Aloe arborescens Miller var. natalensis Berger). Biochem. Biophys. Res. Commun. 1995, 214, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Astoul, C.H.; Barre, A.; Rougé, P.; Peumans, W.J. Cloning and characterization of a monocot mannose-binding lectin from Crocus vernus (family Iridaceae). Eur. J. Biochem. 2000, 267, 5067–5077. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Kaku, H.; Perini, F.; Goldstein, I.J.; Peeters, B.; Yagi, F.; Decock, B.; Peumans, W.J. Biosynthesis, primary structure and molecular cloning of snowdrop (Galanthus nivalis L.) lectin. Eur. J. Biochem. 1991, 202, 23–30. [Google Scholar] [CrossRef]
- Wu, C.-F.; Li, J.; An, J.; Chang, L.-Q.; Che, F.; Bao, J.-K. Purification, biological activities, and molecular cloning of a novel mannose-binding lectin from bulbs of Zephyranthes candida herb (Amaryllidaceae). J. Integr. Plant Biol. 2006, 48, 223–231. [Google Scholar] [CrossRef]
- Lannoo, N.; Peumans, W.J.; Van Pamel, E.V.; Alvarez, R.; Xiong, T.C.; Hause, G.; Mazars, C.; Van Damme, E.J. Localization and in vitro binding studies suggest that the cytoplasmic/nuclear tobacco lectin can interact with high-mannose and complex N-glycans. FEBS Lett. 2006, 580, 6329–6337. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Van Damme, E.J.; Hause, B.; Barre, A.; Chen, Y.; Rougé, P.; Peumans, W.J. Iris bulbs express type 1 and type 2 ribosome-inactivating proteins with unusual properties. Plant Physiol. 2001, 125, 866–876. [Google Scholar] [CrossRef]
- Schouppe, D.; Rougé, P.; Lasanajak, Y.; Barre, A.; Smith, D.F.; Proost, P.; Van Damme, E.J.M. Mutational analysis of the carbohydrate binding activity of the tobacco lectin. Glycoconj. J. 2010, 27, 613–623. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peumans, W.J.; Barre, A.; Bras, J.; Rougé, P.; Proost, P.; Van Damme, E.J. The liverwort contains a lectin that is structurally and evolutionary related to the monocot mannose-binding lectins. Plant Physiol. 2002, 129, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Yamasaki, T.; Tateno, H. The trimeric solution structure and fucose-binding mechanism of the core fucosylation-specific lectin PhoSL. Sci. Rep. 2018, 8, 7740. [Google Scholar] [CrossRef] [PubMed]
- Tateno, H.; Winter, H.C.; Petryniak, J.; Goldstein, I.J. Purification, characterization, molecular cloning, and expression of novel members of jacalin-related lectins from rhizomes of the true fern Phlebodium aureum (L) J. Smith (Polypodiaceae). J. Biol. Chem. 2003, 278, 10891–10899. [Google Scholar] [CrossRef]
- Yagi, F.; Iwaya, T.; Haraguchi, T.; Goldstein, I.J. The lectin from leaves of Japanese cycad, Cycas revoluta Thunb. (gymnosperm) is a member of the jacalin-related family. Eur. J. Biochem. 2002, 269, 4335–4341. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Hatano, K.; Miyauchi, Y.; Suwa, Y.; Sawano, Y.; Tanokura, M. A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol. 2014, 166, 766–778. [Google Scholar] [CrossRef]
- Francis, F.; Jaber, K.; Colinet, F.; Portetelle, D.; Haubruge, E. Purification of a new mannose-specific lectin from Penicillium chrysogenum and its aphicidal properties. Fungal Biol. 2011, 115, 1093–1099. [Google Scholar] [CrossRef]
- Goossens, K.V.Y.; Ielasi, F.S.; Nookaew, I.; Stals, I.; Alonso-Sarduy, L.; Daenen, L.; Van Mulders, S.E.; Stassen, C.; van Eijsden, R.G.E.; Siewers, V.; et al. Molecular mechanism of flocculation self-recognition in yeast and its role in mating and survival. mBio 2015, 6, e00427-15. [Google Scholar] [CrossRef] [PubMed]
- Veelders, M.; Brückner, S.; Ott, D.; Unverzagt, C.; Mösch, H.U.; Essen, L.O. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 2010, 107, 22511–22516. [Google Scholar] [CrossRef] [PubMed]
- Rachmawati, H.; Sundari, S.; Nabila, N.; Tandrasasmita, O.M.; Amalia, R.; Siahaan, T.J.; Tjandrawinata, R.T.; Ismaya, W. Orf239342 from the mushroom Agaricus bisporus is a mannose binding protein. Biochem. Biophys. Res. Commun. 2019, 515, 99–103. [Google Scholar] [CrossRef]
- Suzuki, T.; Sugiyama, K.; Hirai, H.; Ito, H.; Morita, T.; Dohra, H.; Murata, T.; Usui, T.; Tateno, H.; Hirabayashi, J.; et al. Mannose-specific lectin from the mushroom Hygrophorus russula. Glycobiology 2012, 22, 616–629. [Google Scholar] [CrossRef]
- Wohlschlager, T.; Titz, A.; Künzler, M.; Varrot, A. Expression, purification, and functional characterization of tectonin 2 from Laccaria bicolor: A six-bladed b-propeller lectin specific for O-methylated glycans. Methods Mol. Biol. 2020, 2132, 669–682. [Google Scholar] [PubMed]
- Sommer, R.; Makshakova, O.N.; Wohlschlager, T.; Hutin, S.; Marsh, M.; Titz, A.; Künzler, M.; Varrot, A. Crystal structure of fungal tectonin in complex with O-methylated glycans suggest key role in innate immune defense. Structure 2018, 26, 391–402. [Google Scholar] [CrossRef]
- Shimokawa, M.; Fukudome, A.; Yamashita, R.; Minami, Y.; Yagi, F.; Tateno, H.; Hirabayashi, J. Characterization and cloning of GNA-like lectin from the mushroom Marasmius oreades. Glycoconj. J. 2012, 29, 457–465. [Google Scholar] [CrossRef]
- Koharudin, L.M.; Viscomi, A.R.; Jee, J.G.; Ottonello, S.; Gronenborn, A.M. The evolutionary conserved family of cyanovirin-N homologs: Structures and carbohydrate specificity. Structure 2008, 16, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Chaves, R.P.; da Silva, S.R.; Nascimento Neto, L.G.; Carneiro, R.F.; Coelho da Silva, A.L.; Sampaio, A.H.; Lopes de Sousa, B.; Cabral, M.G.; Videira, P.A.; Teixeira, E.H.; et al. Structural characterization of two isolectins from the marine red alga Solieria filiformis (Kützing) P.W. Gabrielson and their anticancer effect on MCF-7 breast cancer cells. Int. J. Biol. Macromol. 2018, 107, 1320–1329. [Google Scholar] [CrossRef] [PubMed]
- Kawakubo, A.; Makino, H.; Ohnishi, J.; Hirohara, H.; Kanji, H. Occurrence of highly yielded lectins homologous within genus Eucheuma. J. Appl. Phycol. 1999, 11, 149–156. [Google Scholar] [CrossRef]
- Sato, Y.; Morimoto, K.; Hirayama, M.; Hori, K. High mannose-specific lectin (KAA-2) from the red alga Kappaphycus alvarezii potently inhibits influenza virus infection in a strain-independent manner. Biochem. Biophys. Res. Commun. 2011, 405, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Hung, L.D.; Sato, Y.; Hori, K. High-mannose N-glycan-specific lectin from the red alga Kappaphycus striatum (Carrageenophyte). Phytochemistry 2011, 72, 855–861. [Google Scholar] [CrossRef]
- Chaves, R.P.; da Silva, S.R.; da Silva, J.P.F.A.; Carneiro, R.F.; de Sousa, B.L.; Abreu, J.O.; de Carvalho, F.C.T.; Rocha, C.R.C.; Farias, W.R.L.; de Sousa, O.V.; et al. Meristiella echinocarpa lectin (MEL): A new member of the OAAH-lectin family. J. Appl. Phycol. 2018, 30, 2629–2638. [Google Scholar] [CrossRef]
- Van Holle, S.; Van Damme, E.J.M. Messages from the past: New insights in plant lectin evolution. Front. Plant Sci. 2019, 10, 36. [Google Scholar] [CrossRef]
- Suttisrisung, S.; Senapin, S.; Withyachumnarnkul, B.; Wongprasert, K. Identification and characterization of a novel legume-like lectin cDNA sequence from the red marine algae Gracilaria fisheri. J. Biosci. 2011, 36, 833–843. [Google Scholar] [CrossRef]
- Hori, K.; Miyazawa, K.; Ito, K. Isolation and characterization of glycoconjugate-specific isoagglutinins from a marine green alga Boodlea coacta (Dickie) Murray et De Toni. Bot. Mar. 1986, XXIX, 323–328. [Google Scholar] [CrossRef]
- Sato, Y.; Hirayama, M.; Morimoto, K.; Yamamoto, N.; Okuyama, S.; Hori, K. High-mannose-binding lectin with preference for the cluster of α1-2-mannose from the green alga Boodlea coacta is a potent entry inhibitor of HIV-1 and influenza viruses. J. Biol. Chem. 2011, 286, 19446–19458. [Google Scholar] [CrossRef]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouze, P.; Worden, A.Z.; Robbens, S.; Partensky, F.; Degroeve, S.; Echeynie, S.; Cooke, R.; et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Hirayama, M.; Sato, Y.; Morimoto, K.; Hori, K. A novel high-mannose specific lectin from the green alga Halimeda renschii exhibits a potent anti-influenza virus activity through high-affinity binding to the viral hemagglutinin. Mar. Drugs 2017, 15, 255. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, A.L.; Sanz, L.; Sánchez, E.L.; Wolfenstein-Todel, C.; Calvete, J.J. Isolation of two novel mannan- and L-fucose-binding lectins from the green alga Enteromorpha prolifera: Biochemical characterization of EPL-2. Arch. Biochem. Biophys. 2003, 415, 245–250. [Google Scholar] [CrossRef]
- Hwang, H.J.; Han, J.W.; Jeon, H.; Han, J.W. Induction of recombinant lectin expression by an artificially constructed tandem repeat structure: A case study using Bryopsis plumosa mannose-binding lectin. Biomolecules 2018, 8, 146. [Google Scholar] [CrossRef]
- Corteggiani Carpinelli, E.; Telatin, A.; Vitulo, N.; Forcato, C.; D’Angelo, M.; Schiavon, R.; Vezzi, A.; Giacometti, G.M.; Morosinotto, T.; Valle, G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 2014, 7, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A.; Cai, M.; Ray, S.; Ghirlando, R.; Yamaguchi, M.; Muramoto, K. New carbohydrate specificity and HIV-1 fusion blocking activity of the cyanobacterial protein MVL: NMR, ITC and sedimentation equilibrium studies. J. Mol. Biol. 2004, 339, 901–914. [Google Scholar] [CrossRef]
- Sato, Y.; Okuyama, S.; Hori, K. Primary structure and carbohydrate binding specificity of a potent anti-HIV lectin isolated from the filamentous cyanobacterium, Oscillatoria agardhii. J. Biol. Chem. 2007, 282, 11021–11029. [Google Scholar] [CrossRef]
- Bokesch, H.R.; O’Keefe, B.R.; McKee, T.C.; Pannell, L.K.; Patterson, G.M.; Gardella, R.S.; Sowder, R.C.; Turmin, J.; Watson, K.; Buckheit, R.W., Jr.; et al. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry 2003, 42, 2578–2584. [Google Scholar] [CrossRef]
- Hori, K.; Sato, Y.; Ito, K.; Fujiwara, Y.; Iwamoto, Y.; Makino, H.; Kawakubo, A. Strict specificity for high-mannose type N-glycans and primary structure of a red alga Eucheuma serrata lectin. Glycobiology 2007, 17, 479–491. [Google Scholar] [CrossRef]
- Wang, P.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; Graham, B.S.; et al. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. Cell Host Microbe 2021, 29, 1–5. [Google Scholar] [CrossRef]
- van der Meer, F.J.U.M.; de Haan, C.A.M.; Schuurman, N.M.P.; Haijema, B.J.; Peumans, W.J.; Van Damme, E.J.M.; Delputte, P.L.; Balzarini, J.; Egberink, H.F. Antiviral activity of carbohydrate-binding agents against Nidovirales in cell cultures. Antivir. Res. 2007, 76, 21–29. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Shahed-Al-Mahmud, M.; Chen, X.; Chen, T.-H.; Liao, K.-S.; Lo, J.M.; Wu, Y.-M.; Ho, M.-C.; Wu, C.-Y.; Wong, C.-H.; et al. A carbohydrate-binding protein from the edible lablab beans effectively blocks the infections of influenza viruses and SARS-CoV-2. Cell Rep. 2020, 32, 108016. [Google Scholar] [CrossRef] [PubMed]
- Debray, H.; Rougé, P.; Debray, H.; Rougé, P. The fine sugar specificity of the Lathyrus ochrus seed lectin and isolectins. FEBS Lett. 1994, 176, 120–124. [Google Scholar] [CrossRef]
- Bourne, Y.; Mazurier, J.; Legrand, D.; Rougé, P.; Montreuil, J.; Spik, G.; Cambillau, C. Structure of a legume lectin complexed with the human lactotransferrin N2 fragment, and with an isolated biantennary glycopeptide: Role of the fucose moiety. Structure 1994, 2, 209–219. [Google Scholar] [CrossRef]
- Zlółkowska, N.E.; O’Keefe, B.R.; Mori, T.; Zhu, C.; Giomarelli, B.; Vojdani, F.; Palmer, K.E.; McMahon, J.B.; Wlodawer, A. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure 2006, 14, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, B.R.; Giomarelli, B.; Barnard, D.L.; Shenoy, S.R.; Chan, P.K.; McMahon, J.B.; Palmer, K.E.; Barnett, B.W.; Meyerholz, D.K.; Vohlford-Lenane, C.L.; et al. Broad-spectrum in vitro activity and in vivo efficacy of the antiviral protein griffithsin against emerging viruses of the family Coronaviridae. J. Virol. 2010, 84, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Millet, J.K.; Séron, K.; Labitt, R.N.; Danneels, A.; Palmer, K.E.; Whittaker, G.R.; Dubuisson, J.; Belouzard, S. Middle East respiratory syndrome coronavirus infection is inhibited by griffithsin. Antivir. Res. 2016, 133, 1–8. [Google Scholar] [CrossRef]
- Rahimi, N. C-type lectin CD209L/L-SIGN and CD209/DC-SIGN; Cell adhesion molecules turned to pathogen recognition receptors. Biology 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Roehrich, H.; Yuan, C.; Hou, J.H. Immunohistochemical study of SARS-CoV-2 viral entry factors in the cornea and ocular surface. Cornea 2020, 39, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Lenza, M.P.; Oyenarte, I.; Diercks, T.; Quintana, J.I.; Gimeno, A.; Coelho, H.; Diniz, A.; Peccati, F.; Delgado, S.; Bosch, A.; et al. Structural characterization of the N-linked glycans in the receptor binding domain of the SARS-CoV-2 spike protein and their interactions with human lectins using NMR spectroscopy. Angew. Chem. Int. Ed. 2020, 59, 23763–23771. [Google Scholar] [CrossRef] [PubMed]
- Caniglia, J.L.; Guda, M.R.; Asuthkar, S.; Tsung, A.J.; Velpula, K.K. A potential role for galectin-3 inhibitors in the treatment of COVID-19. PeerJ 2020, 8, e9392. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, F.J.U.M.; de Haan, C.A.M.; Schuurman, N.M.P.; Haijema, B.J.; Verheije, M.H.; Bosch, B.J.; Balzarini, J.; Egberink, H.F. The carbohydrate-binding plant lectins and the non-peptidic antibiotic pradimicin a target the glycans of the coronavirus envelope glycoproteins. J. Antimicrob. Chemother. 2007, 60, 741–749. [Google Scholar] [CrossRef][Green Version]
- Casalino, L.; Gaieb, Z.; Goldsmith, J.A.; Hjorth, C.K.; Dommer, A.C.; Harbison, A.M.; Fogarty, C.A.; Barros, E.P.; Taylor, B.C.; McLellan, J.S.; et al. Beyond shielding: The roles of glycans in SARS-CoV-2 spike protein. ACS Cent. Sci. 2020, 6, 1722–1734. [Google Scholar] [CrossRef]
- O’Keefe, B.R.; Vojdani, F.; Buffa, V.; Shattock, R.J.; Montefiori, D.C.; Bakke, J.; Mirsalis, J.; d’Andrea, A.L.; Hume, S.D.; Bratcher, B.; et al. Scaleable manufacture of HIV-1 entry inhibitor griffithsin and validation of its safety and efficacy as a topical micrbicide component. Proc. Natl. Acad. Sci. USA 2009, 106, 6099–6104. [Google Scholar] [CrossRef]
- Leblanc, J.-F.; Germain, M.; Delage, G.; O’Brien, S.; Drews, S.J.; Lewin, A. Risk of transmission of severe acute respiratory syndrome coronavirus 2 by transfusion: A literature review. Transfusion 2020, 60, 3046–3054. [Google Scholar] [CrossRef] [PubMed]
- Gaussen, A.; Hornby, L.; Rockl, G.; O’Brien, S.; Delage, G.; Sapir-Pichhadze, R.; Drews, S.J.; Weiss, M.J.; Lewin, A. Evidence of SARS-CoV-2 infection in cells, tissues and organs and the risk of transmission through transplantation. Transplantation 2021, 105, 1405–1422. [Google Scholar] [CrossRef]
- Koch, B.; Schult-Dietrich, P.; Büttner, S.; Dilmaghani, B.; Lohmann, D.; Baer, P.C.; Dietrich, U.; Geiger, H. Lectin affinity plasmapheresis for middle east respiratory syndrome-coronavirus and Marburg virus glycoprotein elimination. Blood Purif. 2018, 46, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 244, 108693. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parametrizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry of protein structures. J. Appl. Crystallogr. 1993, 126, 283–291. [Google Scholar] [CrossRef]
- Melo, F.; Feytmans, E. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 1998, 277, 1141–1152. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Biasiin, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Böhm, M.; Bohne-Lang, A.; Frank, M.; Loss, A.; Rojas-Macias, M.A.; Lütteke, T. Glycosciences.DB: An annotated data collection linking glycomics and proteomics data (2018 update). Nucleic Acids Res. 2019, 47, D1195–D1201. [Google Scholar] [CrossRef]
- Cheng, K.; Zhou, Y.; Neelamegham, S. DrawGlycan-SNFG: A robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology 2017, 27, 200–205. [Google Scholar] [CrossRef]
- Kumaki, Y.; Wandersee, M.K.; Smith, A.J.; Zhou, Y.; Simmons, G.; Nelson, N.M.; Bailey, K.W.; Vest, Z.G.; Li, J.K.-K.; Kay-Sheung Chan, P.; et al. Inhibition of severe acute respiratory syndrome coronavirus replication in a lethal SARS-CoV BALB/c mouse model by stinging nettle lectin, Urtica dioica agglutinin. Antivir. Res. 2011, 90, 22–32. [Google Scholar] [CrossRef]
- Gordts, S.C.; Renders, M.; Férir, G.; Huskens, D.; Van Damme, E.J.M.; Peumans, W.J.; Balzarini, J.; Schols, D. NICTABA and UDA, two GlcNAc-binding lectins with unique antiviral activity profiles. J. Antimicrob. Chemother. 2015, 70, 1674–1685. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).