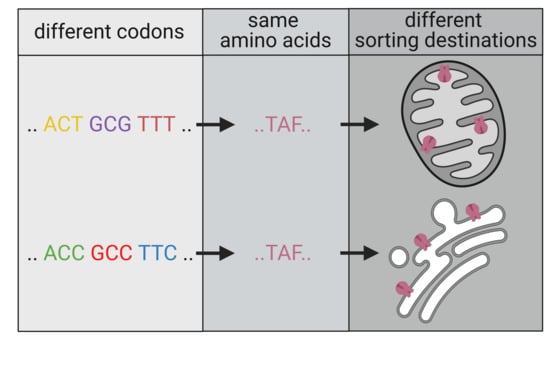
Codon Bias Can Determine Sorting of a Potassium Channel Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Codon-Modified DNA Variants of Channels
2.2. Mutagenesis
2.3. Heterologous Expression
2.4. Cell Culture Media
2.5. Confocal Laser Scanning Microscopy (CLSM)
2.6. Cell Cycle Analysis
2.7. Software Analysis
3. Results
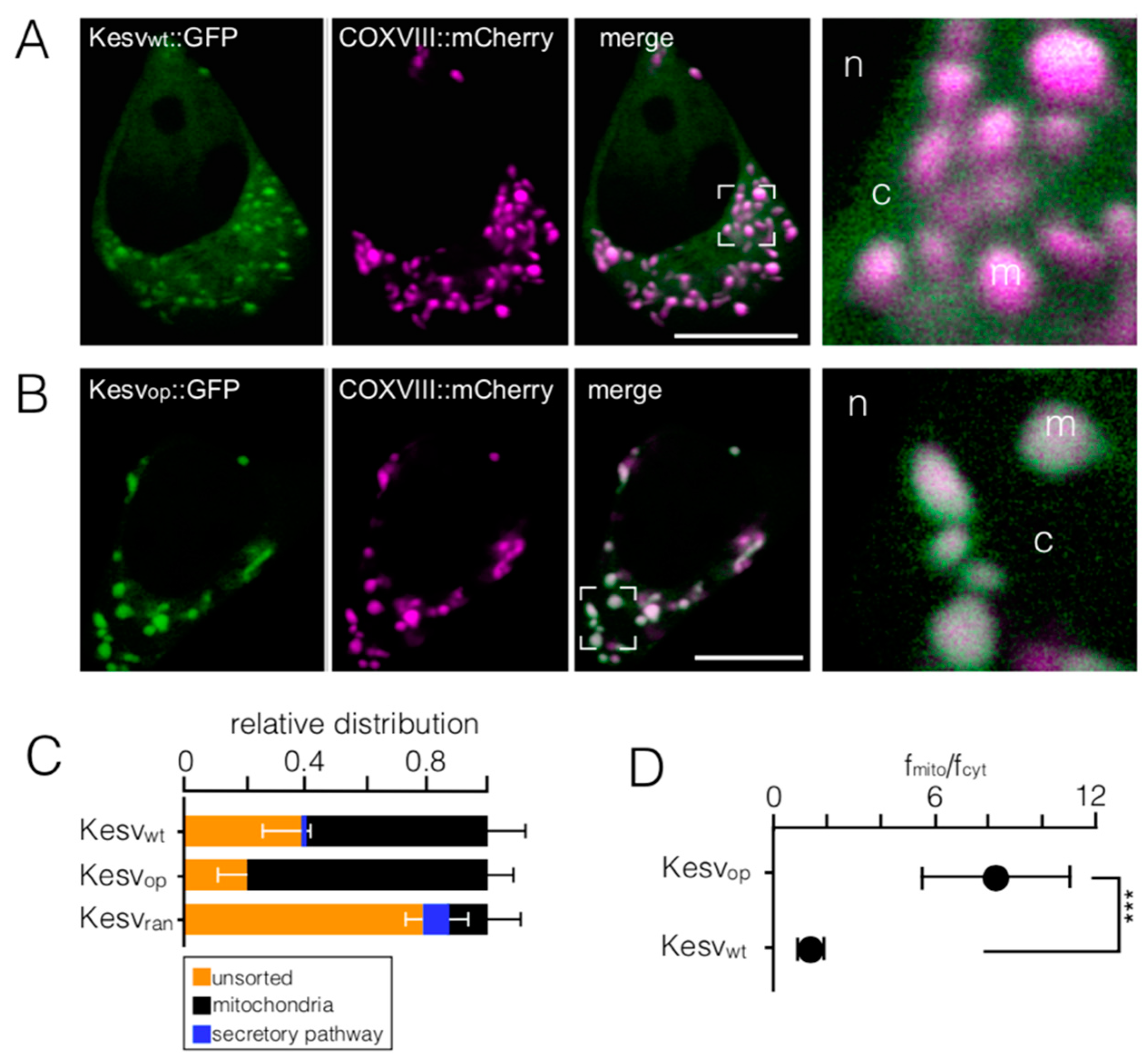
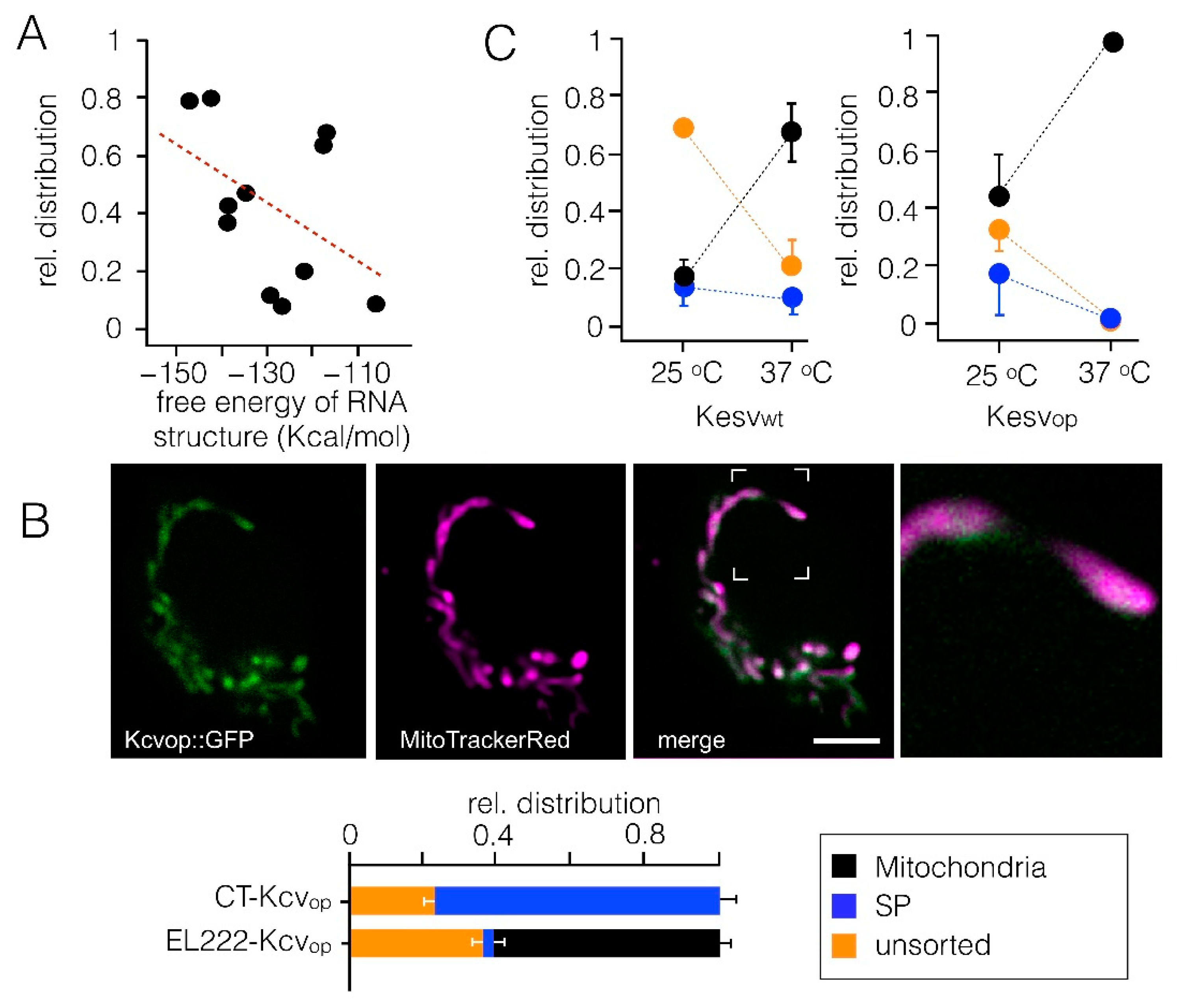
3.1. Mitochondrial Sorting of Channel Protein Is Modulated by Codon Choice
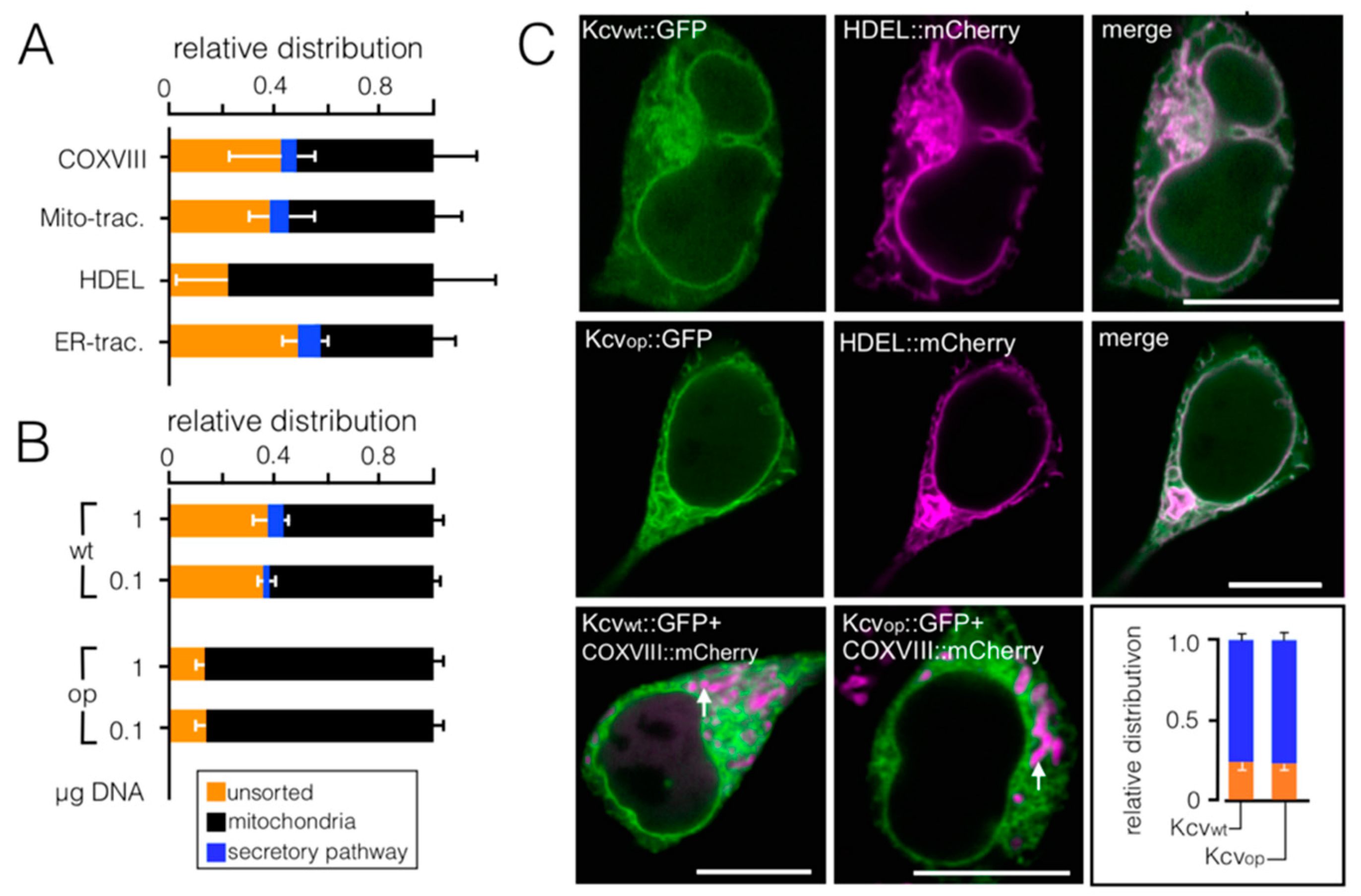
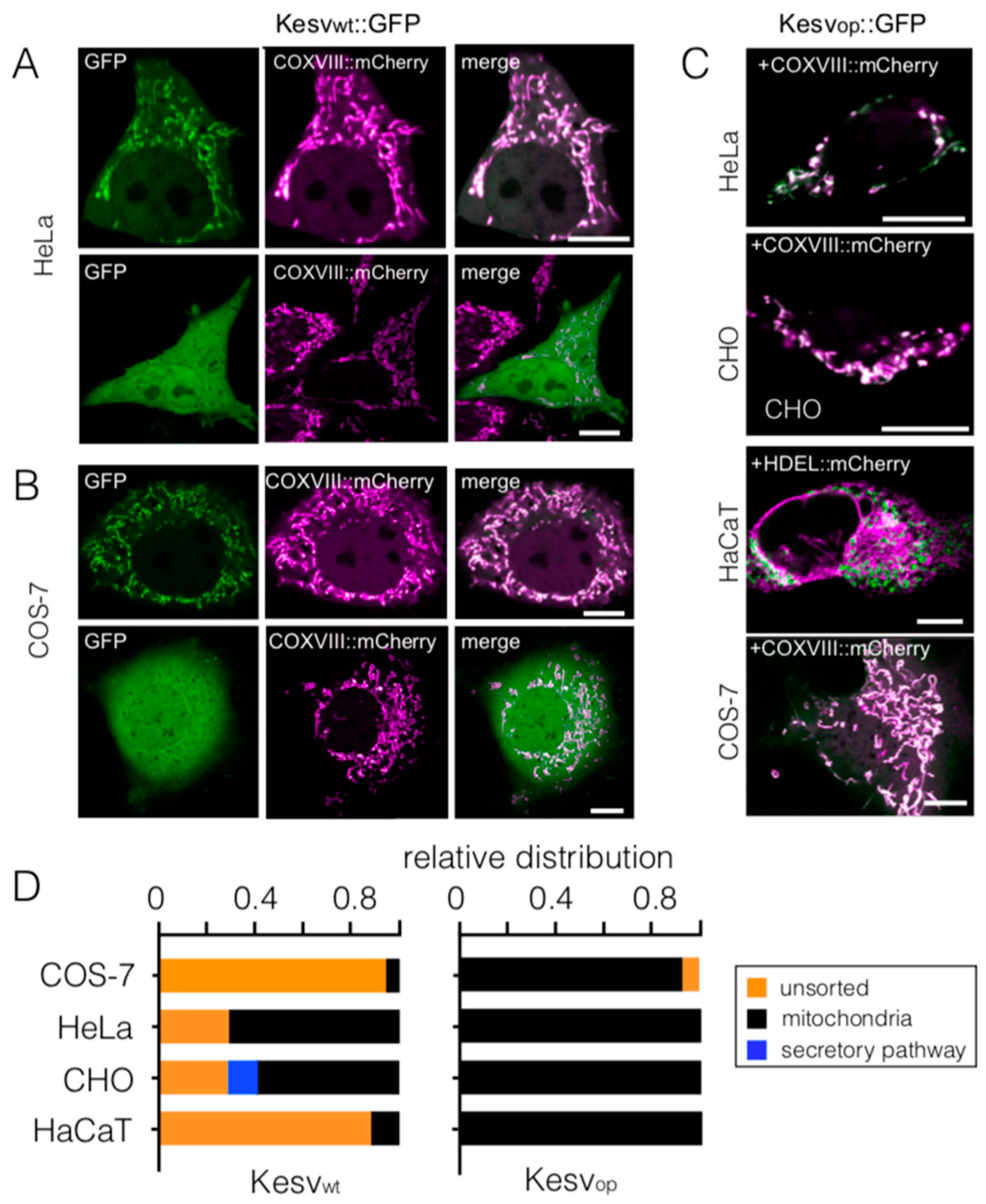
3.2. Codon-Biased Sorting Is a General Phenomenon of Mammalian Cells
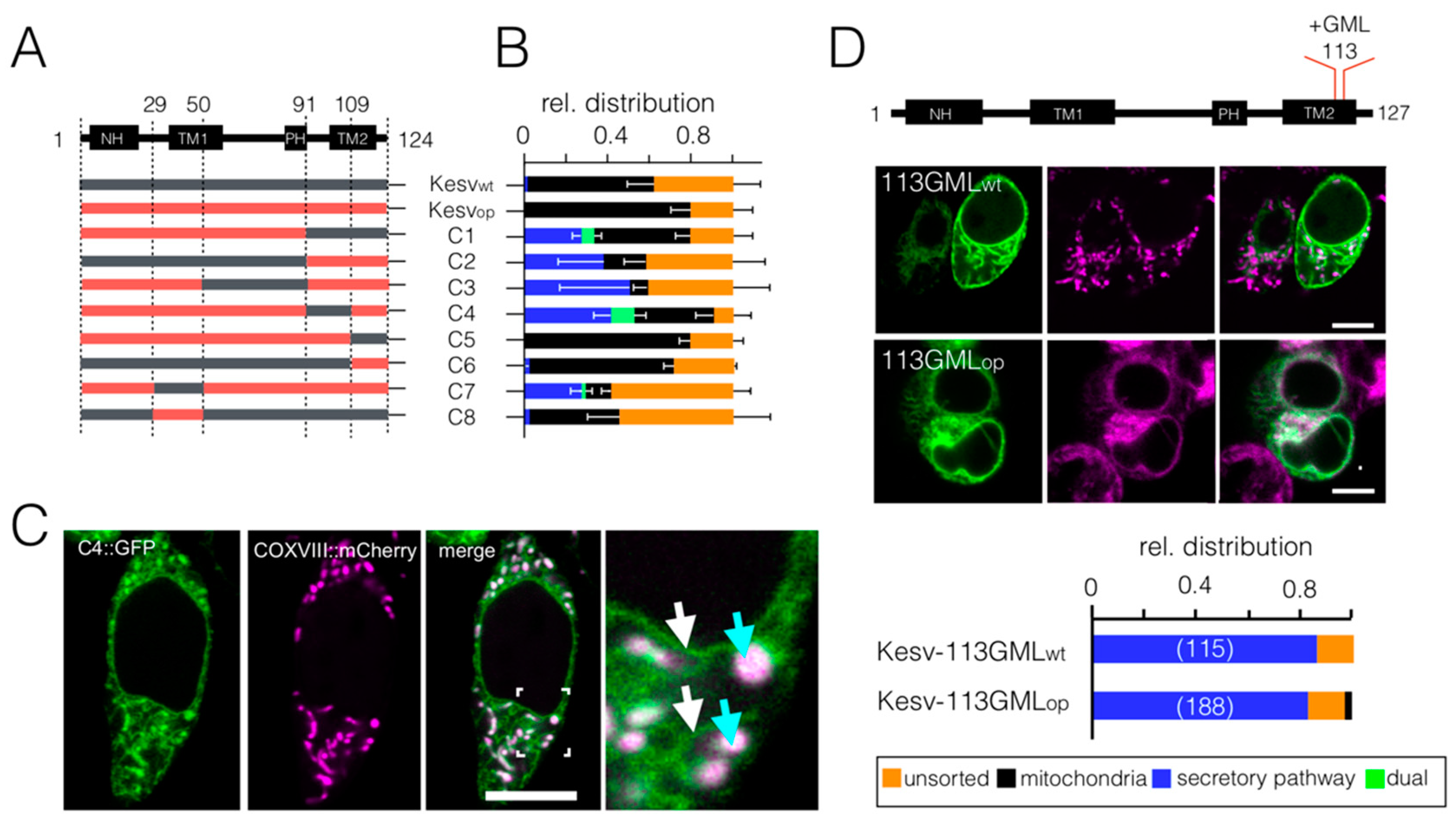
3.3. Chimeras of Genes with Optimized/Non-Optimized Codons Cause Complex Sorting Patterns
3.4. Impact of Codon Usage on Sorting
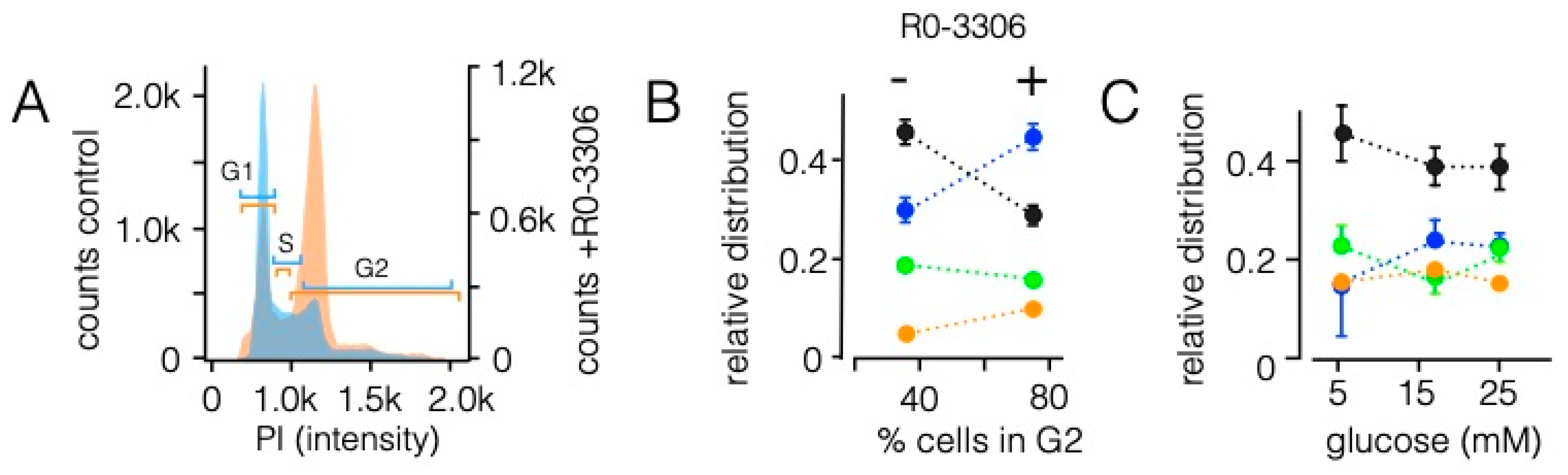
3.5. Sorting Is Affected by State of the Cell Cycle However, Not by the Energy Status of Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Athey, J.; Alexaki, A.; Osipova, E.; Rostovtsev, A.; Santa-Quintero, L.V.; Katneni, U.; Simonyan, V.; Kimchi-Sarfaty, C. A new and updated resource for codon usage tables. BMC Bioinform. 2017, 18, 391. [Google Scholar] [CrossRef]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Morgunov, A.S.; Babu, M. Optimizing membrane-protein biogenesis through nonoptimal-codon usage. Nat. Struct. Mol. Biol. 2014, 21, 1023–1025. [Google Scholar] [CrossRef]
- Mauro, V.; Chappell, S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2015, 20, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lu, G.; Tan, S.; Xia, J.; Xiong, H.; Yu, X.; Qi, Q.; Yu, X.; Li, L.; Yu, H.; et al. A COVID-19 mRNA vaccine encoding SARS-CoV-2 virus-like particles induces a strong antiviral-like immune response in mice. Cell Res. 2020, 30, 936–939. [Google Scholar] [CrossRef]
- Presnyak, V.; Alhusaini, N.; Chen, Y.H.; Martin, S.; Morris, N.; Kline, N.; Olson, S.; Weinberg, D.; Baker, K.E.; Graveley, B.R.; et al. Codon optimality is a major determinant of mRNA stability. Cell 2015, 160, 1111–1124. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Komar, A.A. The Yin and Yang of codon usage. Hum. Mol. Genet. 2016, 25, R77–R85. [Google Scholar] [CrossRef] [PubMed]
- Buhr, F.; Jha, S.; Thommen, S.; Mittelstaet, M.; Kutz, J.; Schwalbe, H.; Rodnina, M.; Komar, A.A. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell 2016, 61, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.M.; de Pouplana, L.R. Speeding with control: Codon usage, tRNAs, and ribosomes. Trends Genet. 2012, 28, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 2015, 59, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.; Siller, E.; Anderson, J.F.; Barral, J.M. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J. Mol. Biol. 2012, 422, 328–335. [Google Scholar] [CrossRef]
- Zhou, M.; Guo, J.; Cha, J.; Chae, M.; Chen, S.; Barral, J.M.; Sachs, M.S.; Liu, Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 2013, 496, 111–116. [Google Scholar] [CrossRef]
- Zhao, F.; Yu, C.H.; Liu, Y. Codon usage regulates protein structure and function by affecting translation elongation speed in Drosophila cells. Nucleic Acids Res. 2017, 45, 8484–8492. [Google Scholar] [CrossRef] [PubMed]
- Pechmann, S.; Frydman, J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013, 20, 237–243. [Google Scholar] [CrossRef]
- Pechmann, S.; Chartron, J.W.; Frydman, J. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat. Struct. Mol. Biol. 2014, 21, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Baumeister, D.; Schroeder, I.; Kast, S.M.; Van Etten, J.L.; Moroni, A. Minimal art: Or why small viral K+ channels are good tools for understanding basic structure and function relations. Biochim. Biophys. Acta 2011, 1808, 580–588. [Google Scholar] [CrossRef]
- Watson, H.R.; Wunderley, L.; Andreou, T.; Warwicker, J.; High, S. Reorientation of the first signal-anchor sequence during potassium channel biogenesis at the Sec61 complex. Biochem. J. 2013, 456, 297–309. [Google Scholar] [CrossRef]
- Moroni, A.; Viscomi, C.; Sangiorgio, V.; Pagliuca, C.; Meckel, T.; Horvath, F.; Gazzarrini, S.; Valbuzzi, P.; Van Etten, J.L.; DiFrancesco, D.; et al. The short N-terminus is required for functional expression of the virus encoded miniature K+-channel Kcv. FEBS Lett. 2002, 530, 65–69. [Google Scholar] [CrossRef]
- Balss, J.; Mehmel, M.; Baumeister, D.; Hertel, B.; Delaroque, N.; Chatelain, F.C.; Minor, D.J.; Van Etten, J.L.; Moroni, A.; Thiel, G. Transmembrane domain length of viral K+ channels is a signal for mitochondria targeting. Proc. Natl. Acad. Sci. USA 2008, 10, 12313–12318. [Google Scholar] [CrossRef]
- Engel, A.J.; Winterstein, L.M.; Kithil, M.; Langhans, M.; Moroni, A.; Thiel, G. Light-regulated transcription of mitochondrial targeted K+ channel. Cells 2020, 9, 2507. [Google Scholar] [CrossRef]
- Zhang, Y.; Schäffer, T.; Wölfle, T.; Fitzke, E.; Thiel, G.; Rospert, S. Cotranslational intersection between the SRP and GET targeting pathways to the endoplasmic reticulum of Saccharomyces cerevisiae. Mol. Cell. Biol. 2016, 36, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- von Chappuis, C.; Meckel, T.; Moroni, A.; Thiel, G. The sorting of a small potassium channel in mammalian cells can be shifted between mitochondria and plasma membrane. Cell Calcium 2014, 58, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Palmer, E.; Freeman, T. Investigation into the use of C- and N-terminal GFP fusion proteins for subcellular localization studies using reverse transfection microarrays. Comp. Funct. Genom. 2004, 5, 342–353. [Google Scholar] [CrossRef]
- Motta-Mena, L.B.; Reade, A.; Mallory, M.J.; Giantz, S.; Weiner, O.D.; Lynch, K.W.; Gardner, K.H. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat. Chem. Biol. 2014, 10, 196–202. [Google Scholar] [CrossRef]
- Heckman, K.L.; Pease, L.R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Prot. 2007, 2, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carrereas, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Clarke, T.F.; Clark, P.L. Rare codons cluster. PLoS ONE 2008, 3, e3412. [Google Scholar] [CrossRef] [PubMed]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple rules for passive diffusion through the nuclear pore complex. J. Cell Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef] [PubMed]
- Plugge, B.; Gazzarini, S.; Cerana, R.; Van Etten, J.; Nelson, M.; DiFrancesco, D.; Moroni, A.; Thiel, G. A potassium ion channel protein encoded by chlorella virus PBCV-1. Science 2000, 287, 1641–1644. [Google Scholar] [CrossRef]
- Stein, K.C.; Frydman, J. The stop-and-go traffic regulating protein biogenesis: How translation kinetics controls proteostasis. J. Biol. Chem. 2019, 294, 2076–2084. [Google Scholar] [CrossRef] [PubMed]
- Geiger, T.; Wehner, A.; Schaab, C.; Cox, J.; Mann, M. Comparative proteomoic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteom. 2012, 11, M111-014050. [Google Scholar] [CrossRef]
- Murillo, I.; Henderson, L.M. Expression of gp91phox/Nox2 in COS-7 cells: Cellular localization of the protein and the detection of outward proton currents. Biochem. J. 2005, 385, 649–657. [Google Scholar] [CrossRef]
- Dittmar, K.A.; Goodenbour, J.M.; Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006, 2, e221. [Google Scholar] [CrossRef] [PubMed]
- Szabo, I.; Bock, J.; Jekle, A.; Soddemann, M.; Adams, C.; Lang, F.; Zoratti, M.; Gubins, E. A novel potassium channel in lymphocyte mitochondria. J. Biol. Chem. 2005, 280, 12790–12798. [Google Scholar] [CrossRef]
- Weis, B.L.; Schleiff, E.; Zerges, W. Protein targeting to subcellular organelles via mRNA localization. Biochim. Biophys. Acta 2013, 1833, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692. [Google Scholar] [CrossRef]
- Costa, F.; Castella, P.; Colombo, S.F.; Borgese, N. Discrimination between the endoplasmatic reticulum and mitochondria by spontaneous inserting tail-anchored proteins. Traffic 2018, 19, 182–197. [Google Scholar] [CrossRef] [PubMed]
- Eliscovich, C.; Singer, R. RNP transport in cell biology: The long and winding road. Curr. Opin. Cell Biol. 2017, 45, 38–46. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Spiridonov, N.A.; Kashina, A. Sounds of sillence: Synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013, 41, 2073–2094. [Google Scholar] [CrossRef]
- Pop, C.; Rouskin, S.; Ingolia, N.T.; Han, L.; Phizicky, E.M.; Weissman, J.S.; Koller, D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014, 10, 770. [Google Scholar] [CrossRef]
- Hamilton, R.S.; Davis, I. Identifying and searching for conserved RNA localization signals. Methods Mol. Biol. 2011, 714, 447–466. [Google Scholar]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef]
- Rullan, M.; Benzinger, D.; Schmidt, G.W.; Milias-Argeitis, A.; Khammash, M. An optogenetic platform for real time singel-cell interrogation of stochastic transcriptional regulation. Mol. Cell 2018, 70, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Körner, C.; Friedel, P. Quantitative analysis of transcription and translation in gene amplified Chinese hamster ovary cells on the basis of a kinetic model. Cytotechnology 1990, 29, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef]
- Richards, H.A.; Halfhill, M.D.; Millwood, R.J.; Stewart, C.N. Quantitative GFP fluorescence as an indicator of recombinant protein synthesis in transgenic plants. Plant Cell Rep. 2003, 22, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Little, M.H.; Wilkinson, L.; Brown, D.L.; Piper, M.; Yamada, T.; Stow, J.L. Dual trafficking of Slit3 to mitochondria and cell surface demonstrates novel localization for Slit protein. Am. J. Physiol.-Cell Physiol. 2001, 281, C486–C495. [Google Scholar] [CrossRef]
- Frenkel-Morgenstern, M.; Danon, T.; Christian, T.; Igarashi, T.; Cohen, L.; Hou, Y.M.; Jensen, L.J. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012, 8, 572. [Google Scholar] [CrossRef]
- Vassilev, L.T. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle 2006, 5, 2555–2556. [Google Scholar] [CrossRef]
- Sakowicz-Burkiewicz, M.; Grden, M.; Maciejewska, I.; Szutowicz, A.; Pawelczyk, T. High glucose impairs ATP formation on the surface of human peripheral blood B lymphocytes. Intern. J. Biochem. Cell Biol. 2013, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Graham, N.A.; Tahmasian, M.; Kohli, B.; Komisopoulou, E.; Zhu, M.; Vivanco, I.; Teitell, M.A.; Wu, H.; Ribas, A.; Lo, R.S.; et al. Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Mol. Syst. Biol. 2012, 8, 589. [Google Scholar] [CrossRef]
- Mohammadi-Farani, A.; Ghazi-Khansari, M.; Sahebgharani, M. Glucose concentration in culture medium affects mRNA expression of TRPV1 and CB1 receptors and changes capsaicin toxicity in PC12 cells. Iran. J. Basic Med. Sci. 2014, 17, 673. [Google Scholar]
- Kramer, E.B.; Vallabhaneni, H.; Mayer, L.M.; Farabaugh, P.J. A comprehensive analysis of translational missense errors in the yeast Saccharomyces cerevisiae. RNA 2010, 16, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Komar, A.A. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci. 2009, 34, 16–24. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engel, A.J.; Kithil, M.; Langhans, M.; Rauh, O.; Cartolano, M.; Van Etten, J.L.; Moroni, A.; Thiel, G. Codon Bias Can Determine Sorting of a Potassium Channel Protein. Cells 2021, 10, 1128. https://doi.org/10.3390/cells10051128
Engel AJ, Kithil M, Langhans M, Rauh O, Cartolano M, Van Etten JL, Moroni A, Thiel G. Codon Bias Can Determine Sorting of a Potassium Channel Protein. Cells. 2021; 10(5):1128. https://doi.org/10.3390/cells10051128
Chicago/Turabian StyleEngel, Anja J., Marina Kithil, Markus Langhans, Oliver Rauh, Matea Cartolano, James L. Van Etten, Anna Moroni, and Gerhard Thiel. 2021. "Codon Bias Can Determine Sorting of a Potassium Channel Protein" Cells 10, no. 5: 1128. https://doi.org/10.3390/cells10051128
APA StyleEngel, A. J., Kithil, M., Langhans, M., Rauh, O., Cartolano, M., Van Etten, J. L., Moroni, A., & Thiel, G. (2021). Codon Bias Can Determine Sorting of a Potassium Channel Protein. Cells, 10(5), 1128. https://doi.org/10.3390/cells10051128