Structures, Biosynthesis, and Physiological Functions of (1,3;1,4)-β-d-Glucans
Abstract
1. Introduction
2. Variation of (1,3;1,4)-β-d-Glucans in Different Taxa
2.1. Viridiplantae
2.1.1. Biosynthesis of (1,3;1,4)-β-d-Glucan in Viridiplantae
2.1.2. Physiological Function of (1,3;1,4)-β-d-Glucan in Viridiplantae
2.2. Fungi and Lichens
2.2.1. Biosynthesis of (1,3;1,4)-β-d-Glucan in Fungi and Lichens
2.2.2. Physiological Function of (1,3;1,4)-β-d-Glucan in Fungi and Lichens
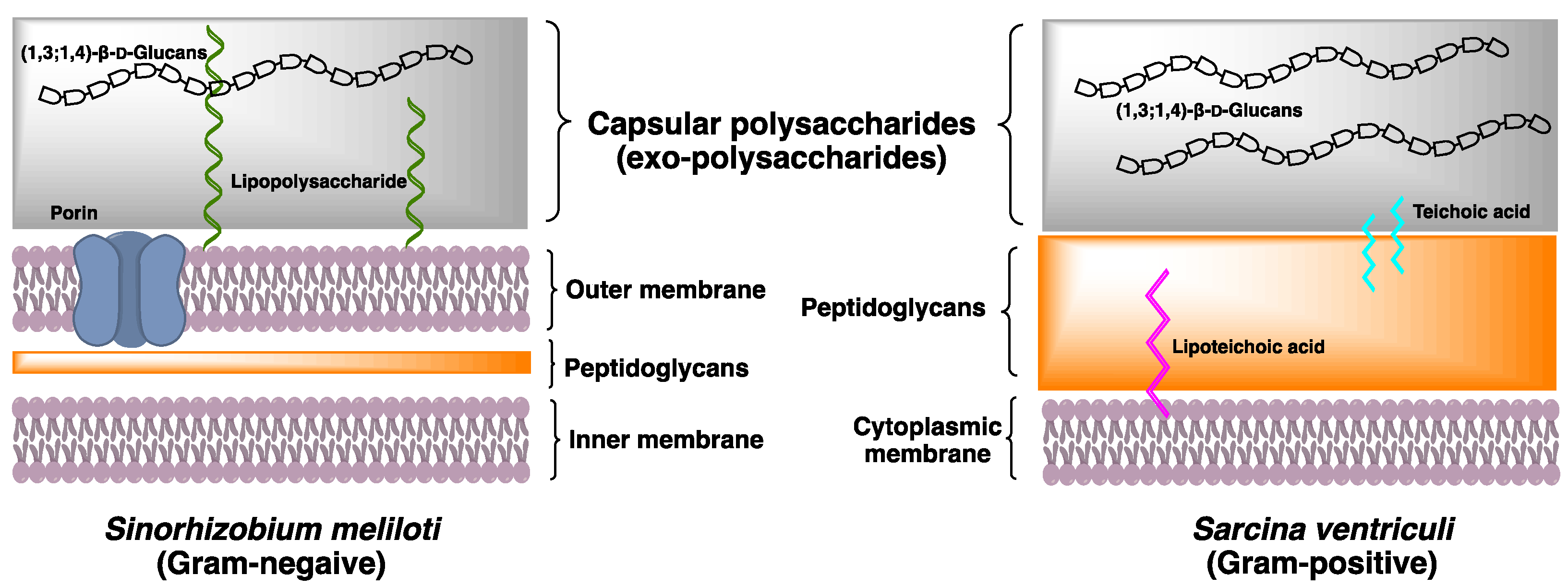
2.3. Bacteria
2.3.1. Biosynthesis of (1,3;1,4)-β-d-Glucan in Bacteria
2.3.2. Physiological Function of (1,3;1,4)-β-d-Glucan in Bacteria
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murphy, E.J.; Rezoagli, E.; Major, I.; Rowan, N.J.; Laffey, J.G. β-Glucan metabolic and immunomodulatory properties and potential for clinical application. J. Fungi 2020, 6, 356. [Google Scholar] [CrossRef]
- Burton, R.A.; Gidley, M.J.; Fincher, G.B. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat. Chem. Biol. 2010, 6, 724–732. [Google Scholar] [CrossRef]
- Sørensen, I.; Pettolino, F.A.; Wilson, S.M.; Doblin, M.S.; Johansen, B.; Bacic, A.; Willats, W.G.T. Mixed-linkage (1→3),(1→4)-β-d-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. Plant J. 2008, 54, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Fincher, G.B. (1,3;1,4)-β-d-glucans in cell walls of the poaceae, lower plants, and fungi: A tale of two linkages. Mol. Plant 2009, 2, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Fincher, G.B. Evolution and development of cell walls in cereal grains. Front. Plant Sci. 2014, 5, 456. [Google Scholar] [CrossRef]
- Lazaridou, A.; Biliaderis, C.G. Molecular aspects of cereal β-glucan functionality: Physical properties, technological applications and physiological effects. J. Cereal Sci. 2007, 46, 101–118. [Google Scholar] [CrossRef]
- Fry, S.C.; Nesselrode, B.H.W.A.; Miller, J.G.; Mewburn, B.R. Mixed-linkage (1→3,1→4)-β-d-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol. 2008, 179. [Google Scholar] [CrossRef]
- Xue, X.; Fry, S.C. Evolution of mixed-linkage (1→3,1→4)-β-d-glucan (MLG) and xyloglucan in Equisetum (horsetails) and other monilophytes. Ann. Bot. 2012, 109, 873–886. [Google Scholar] [CrossRef]
- Salmeán, A.A.; Duffieux, D.; Harholt, J.; Qin, F.; Michel, G.; Czjzek, M.; Willats, W.G.T.; Hervé, C. Insoluble (1→3),(1→4)-β-d-glucan is a component of cell walls in brown algae (Phaeophyceae) and is masked by alginates in tissues. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Pérez-Mendoza, D.; Rodríguez-Carvajal, M.Á.; Romero-Jiménez, L.; De Araujo Farias, G.; Lloret, J.; Gallegos, M.T.; Sanjuán, J. Novel mixed-linkage β-glucan activated by c-di-GMP in Sinorhizobium Meliloti. Proc. Natl. Acad. Sci. USA 2015, 112, E757–E765. [Google Scholar] [CrossRef]
- Wood, P.J.; Weisz, J.; Blackwell, B.A. Molecular characterization of cereal beta-d-glucans. Structural analysis of oat beta-d-glucan and rapid structural evaluation of beta-d-glucans from different sources by high-performance liquid chromatography of oligosaccharides released by lichenase. Cereal Chem. 1991, 68, 31–39. [Google Scholar]
- Wood, P.J.; Braaten, J.T.; Scott, F.W.; Riedel, K.D.; Wolynetz, M.S.; Collins, M.W. Effect of dose and modification of viscous properties of oat gum on plasma glucose and insulin following an oral glucose load. Br. J. Nutr. 1994, 72, 731–743. [Google Scholar] [CrossRef]
- Lee, J.; Hollingsworth, R.I. Oligosaccharide β-glucans with unusual linkages from Sarcina ventriculi. Carbohydr. Res. 1997, 304, 133–141. [Google Scholar] [CrossRef]
- Cui, W.; Wood, P.J. Relationships between structural features, molecular weight and rheological properties of cereal β-d-glucans. Hydrocolloids 2000. [Google Scholar] [CrossRef]
- Bohm, N.; Kulicke, W.M. Rheological studies of barley (1→3)(1→4)-β-glucan in concentrated solution: Mechanistic and kinetic investigation of the gel formation. Carbohydr. Res. 1999, 315, 302–331. [Google Scholar] [CrossRef]
- Lazaridou, A.; Biliaderis, C.G.; Izydorczyk, M.S. Molecular size effects on rheological properties of oat β-glucans in solution and gels. Food Hydrocoll. 2003, 17, 693–712. [Google Scholar] [CrossRef]
- Wood, P.J. Oat Beta-Glucan: Structure, Location, and Properties; American Association of Cereal Chemists, Inc.: St. Paul, MN, USA, 1986; pp. 121–152. [Google Scholar]
- Inglett, G.E. Hypocholesterolemic β-glucan-amylodextrins from oats as dietary fat-replacements. In ACS Book of Abstracts from 199th National Meeting; ACS publications: Washington, DC, USA, 1990. [Google Scholar]
- Morgan, K.R. Cereal β-glucans. In Handbook of Hydrocolloids; Philips, G., Williams, P., Eds.; Woodhead Publishing & CRC Press: Cambridge, UK, 2000. [Google Scholar]
- Brennan, C.S.; Cleary, L.J. The potential use of cereal (1→3,1→4)-β-d-glucans as functional food ingredients. J. Cereal Sci. 2005, 42, 1–13. [Google Scholar] [CrossRef]
- Harris, P.J.; Smith, B.G. Plant cell walls and cell-wall polysaccharides: Structures, properties and uses in food products. Int. J. Food Sci. Technol. 2006, 41, 129–143. [Google Scholar] [CrossRef]
- Othman, R.A.; Moghadasian, M.H.; Jones, P.J.H. Cholesterol-lowering effects of oat β-glucan. Nutr. Rev. 2011, 69, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.J.; Fincher, G.B. Distribution, fine structure and function of (1,3;1,4)-β-Glucans in the grasses and other taxa. In Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides; Elsevier Science Publishing Co. Inc.: San Diego, CA, USA, 2009. [Google Scholar] [CrossRef]
- Tamura, K.; Hemsworth, G.R.; Déjean, G.; Rogers, T.E.; Pudlo, N.A.; Urs, K.; Jain, N.; Davies, G.J.; Martens, E.C.; Brumer, H. Molecular mechanism by which prominent human gut bacteroidetes utilize mixed-linkage beta-glucans, major health-promoting cereal polysaccharides. Cell Rep. 2017, 21, 417–430. [Google Scholar] [CrossRef]
- Meier, H.; Reid, J.S.G. Reserve polysaccharides other than starch in higher plants. In Plant Carbohydrates I. Intracellular Carbohydrates; Springer Science & Business Media: Berlin, Germany, 1982. [Google Scholar]
- Kim, J.B.; Olek, A.T.; Carpita, N.C. Cell wall and membrane-associated exo-β-d-glucanases from developing maize seedlings. Plant Physiol. 2000, 123, 471–485. [Google Scholar] [CrossRef]
- Fincher, G.B.; Stone, B.A. CEREALS|Chemistry of Nonstarch Polysaccharides. In Encyclopedia of Grain Science; Wrigley, C., Ed.; Elsevier: Amsterdam, The Netherlands, 2004; pp. 206–223. [Google Scholar] [CrossRef]
- Li, W.; Cui, S.W.; Kakuda, Y. Extraction, fractionation, structural and physical characterization of wheat β-d-glucans. Carbohydr. Polym. 2006, 63, 408–416. [Google Scholar] [CrossRef]
- Popper, Z.A.; Fry, S.C. Primary cell wall composition of bryophytes and charophytes. Ann. Bot. 2003, 91, 1–12. [Google Scholar] [CrossRef]
- Eder, M.; Tenhaken, R.; Driouich, A.; Lütz-Meindl, U. Occurrence and characterization of arabinogalactan-like proteins and hemicelluloses in Micrasterias (Streptophyta). J. Phycol. 2008, 44, 1221–1234. [Google Scholar] [CrossRef]
- Lechat, H.; Amat, M.; Mazoyer, J.; Buléon, A.; Lahaye, M. Structure and distribution of glucomannan and sulfated glucan in the cell walls of the red alga Kappaphycus alvarezii (Gigartinales, Rhodophyta). J. Phycol. 2000, 36, 891–902. [Google Scholar] [CrossRef]
- Ford, C.W.; Percival, E. 551. Polysaccharides synthesised by Monodus subterraneus. Part II. The cell-wall glucan. J. Chem. Soc. 1965. [Google Scholar] [CrossRef]
- Nevo, Z.; Sharon, N. The cell wall of Peridinium westii, a non cellulosic glucan. BBA Biomembr. 1969, 173, 161–175. [Google Scholar] [CrossRef]
- Doblin, M.S.; Kurek, I.; Jacob-Wilk, D.; Delmer, D.P. Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiol. 2002, 43, 1407–1420. [Google Scholar] [CrossRef]
- Burton, R.A.; Wilson, S.M.; Hrmova, M.; Harvey, A.J.; Shirley, N.J.; Medhurst, A.; Stone, B.A.; Newbigin, E.J.; Bacic, A.; Fincher, G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science 2006, 311, 1940–1942. [Google Scholar] [CrossRef]
- Doblin, M.S.; Pettolino, F.A.; Wilson, S.M.; Campbell, R.; Burton, R.A.; Fincher, G.B.; Newbigin, E.; Bacic, A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-d-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 2009. [Google Scholar] [CrossRef]
- Carpita, N.C.; Gibeaut, D.M. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3, 1–30. [Google Scholar] [CrossRef]
- Meikle, P.J.; Hoogenraad, N.J.; Bonig, I.; Clarke, A.E.; Stone, B.A. A (1→3,1→4)-β-glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3,1→4)-β-glucans. Plant J. 1994, 5, 1–9. [Google Scholar] [CrossRef]
- Wilson, S.M.; Burton, R.A.; Doblin, M.S.; Stone, B.A.; Newbigin, E.J.; Fincher, G.B.; Bacic, A. Temporal and spatial appearance of wall polysaccharides during cellularization of barley (Hordeum vulgare) endosperm. Planta 2006, 224, 655–667. [Google Scholar] [CrossRef]
- Tonooka, T.; Aoki, E.; Yoshioka, T.; Taketa, S. A novel mutant gene for (1-3, 1-4)-β-d-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breed. Sci. 2009, 59, 47–54. [Google Scholar] [CrossRef][Green Version]
- Nemeth, C.; Freeman, J.; Jones, H.D.; Sparks, C.; Pellny, T.K.; Wilkinson, M.D.; Dunwell, J.; Andersson, A.A.M.; Åman, P.; Guillon, F.; et al. Down-regulation of the CSLF6 gene results in decreased (1,3;1,4)-β-d-glucan in endosperm of wheat. Plant Physiol. 2010, 152, 1209–1218. [Google Scholar] [CrossRef]
- Hu, G.; Burton, C.; Hong, Z.; Jackson, E. A mutation of the cellulose-synthase-like (CslF6) gene in barley (Hordeum vulgare L.) partially affects the β-glucan content in grains. J. Cereal Sci. 2014, 59, 189–195. [Google Scholar] [CrossRef]
- Taketa, S.; Yuo, T.; Tonooka, T.; Tsumuraya, Y.; Inagaki, Y.; Haruyama, N.; Larroque, O.; Jobling, S.A. Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-β-d-glucan biosynthesis. J. Exp. Bot. 2012, 63, 381–392. [Google Scholar] [CrossRef]
- Vega-Sánchez, M.E.; Verhertbruggen, Y.; Christensen, U.; Chen, X.; Sharma, V.; Varanasi, P.; Jobling, S.A.; Talbot, M.; White, R.G.; Joo, M.; et al. Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012, 159, 56–69. [Google Scholar] [CrossRef]
- Kim, S.J.; Zemelis, S.; Keegstra, K.; Brandizzi, F. The cytoplasmic localization of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan. Plant J. 2015, 81, 537–547. [Google Scholar] [CrossRef]
- Kim, S.J.; Zemelis-Durfee, S.; Jensen, J.K.; Wilkerson, C.G.; Keegstra, K.; Brandizzi, F. In the grass species Brachypodium distachyon, the production of mixed-linkage (1,3;1,4)-β-glucan (MLG) occurs in the Golgi apparatus. Plant J. 2018, 93, 1062–1075. [Google Scholar] [CrossRef]
- Dimitroff, G.; Little, A.; Lahnstein, J.; Schwerdt, J.G.; Srivastava, V.; Bulone, V.; Burton, R.A.; Fincher, G.B. (1,3;1,4)-β-glucan biosynthesis by the CSLF6 enzyme: Position and flexibility of catalytic residues influence product fine structure. Biochemistry 2016, 55, 2054–2061. [Google Scholar] [CrossRef] [PubMed]
- Lazaridou, A.; Biliaderis, C.G.; Micha-Screttas, M.; Steele, B.R. A comparative study on structure-function relations of mixed-linkage (1→3), (1→4) linear β-d-glucans. Food Hydrocoll. 2004, 18, 837–855. [Google Scholar] [CrossRef]
- Buckeridge, M.S.; Rayon, C.; Urbanowicz, B.; Tiné, M.A.S.; Carpita, N.C. Mixed linkage (1→3),(1→4)-β-d-glucans of grasses. Cereal Chem. 2004, 81, 115–127. [Google Scholar] [CrossRef]
- Carpita, N.C.; Defernez, M.; Findlay, K.; Wells, B.; Shoue, D.A.; Catchpole, G.; Wilson, R.H.; McCann, M.C. Cell wall architecture of the elongating maize coleoptile. Plant Physiol. 2001, 127, 551–565. [Google Scholar] [CrossRef]
- Gibeaut, D.M.; Pauly, M.; Bacic, A.; Fincher, G.B. Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 2005, 221, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Kiemle, S.N.; Zhang, X.; Esker, A.R.; Toriz, G.; Gatenholm, P.; Cosgrove, D.J. Role of (1,3)(1,4)-β-glucan in cell walls: Interaction with cellulose. Biomacromolecules 2014, 15, 1727–1736. [Google Scholar] [CrossRef]
- Olafsdottir, E.S.; Ingólfsdottir, K. Polysaccharides from lichens: Structural characteristics and biological activity. Planta Med. 2001, 67, 199–208. [Google Scholar] [CrossRef]
- Honegger, R.; Haisch, A. Immunocytochemical location of the (1→3) (1→4)-β-glucan lichenin in the lichen-forming ascomycete Cetraria islandica (Icelandic moss). New Phytol. 2001, 150, 739–746. [Google Scholar] [CrossRef]
- Fontaine, T.; Simenel, C.; Dubreucq, G.; Adam, O.; Delepierre, M.; Lemoine, J.; Vorgias, C.E.; Diaquin, M.; Latgé, J.P. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 2000, 275, 27594–27607. [Google Scholar] [CrossRef]
- Ao, J.; Free, S.J. Genetic and biochemical characterization of the GH72 family of cell wall transglycosylases in Neurospora Crassa. Fungal Genet. Biol. 2017, 101, 46–54. [Google Scholar] [CrossRef]
- Maddi, A.; Free, S.J. α-1,6-mannosylation of N-linked oligosaccharide present on cell wall proteins is required for their incorporation into the cell wall in the filamentous fungus Neurospora crassa. Eukaryot. Cell 2010, 9, 1766–1775. [Google Scholar] [CrossRef]
- Murphy, E.J.; Masterson, C.; Rezoagli, E.; O’Toole, D.; Major, I.; Stack, G.D.; Lynch, M.; Laffey, J.G.; Rowan, N.J. β-Glucan extracts from the same edible shiitake mushroom Lentinus edodes produce differential in-vitro immunomodulatory and pulmonary cytoprotective effects—Implications for coronavirus disease (COVID-19) immunotherapies. Sci. Total Environ. 2020, 732, 139330. [Google Scholar] [CrossRef] [PubMed]
- Samar, D.; Kieler, J.B.; Klutts, J.S. Identification and deletion of Tft1, α predicted glycosyltransferase necessary for cell wall β-1,3;1,4-glucan synthesis in Aspergillus fumigatus. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Guerriero, G.; Silvestrini, L.; Legay, S.; Maixner, F.; Sulyok, M.; Hausman, J.F.; Strauss, J. Deletion of the celA gene in Aspergillus nidulans triggers overexpression of secondary metabolite biosynthetic genes. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Liu, H.; Li, Y.; Chen, D.; Qi, Z.; Wang, Q.; Wang, J.; Jiang, C.; Xu, J.R. A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2017, 114, E7756–E7765. [Google Scholar] [CrossRef]
- Backes, A.; Hausman, J.F.; Renaut, J.; Barka, E.A.; Jacquard, C.; Guerriero, G. Expression analysis of cell wall-related genes in the plant pathogenic fungus Drechslera teres. Genes 2020, 11, 300. [Google Scholar] [CrossRef]
- Canale-Parola, E. Biology of the sugar-fermenting Sarcinae. Bacteriol. Rev. 1970, 34, 82–97. [Google Scholar] [CrossRef]
- Whitney, J.C.; Howell, P.L. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013, 21, 63–72. [Google Scholar] [CrossRef]
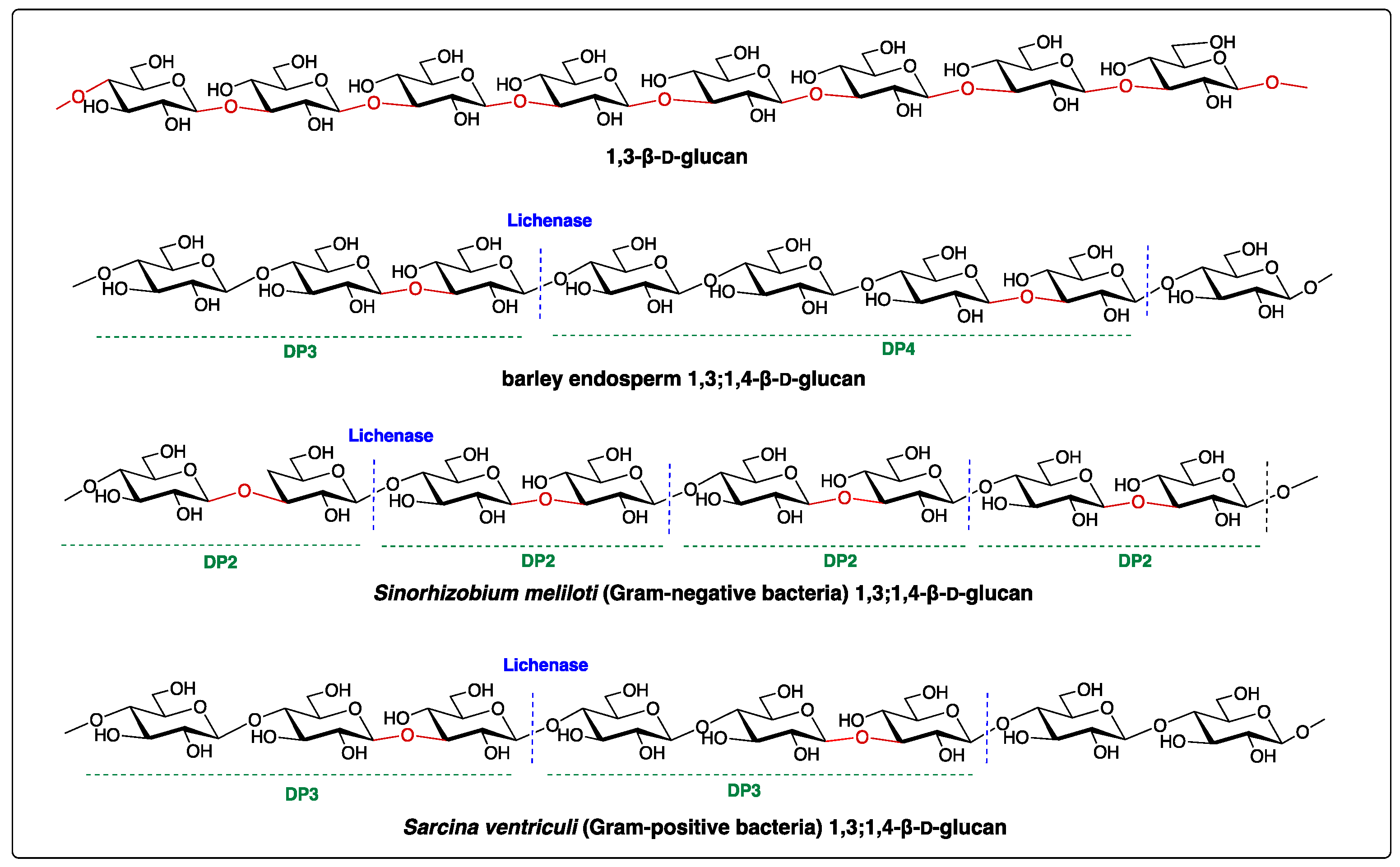
| Species | DP2 | DP3 | DP4 | References |
|---|---|---|---|---|
| Hordeum vulgare | 0 | 1.8–3.5 | 1 | [6] |
| Triticum aestivum | 0 | 3.0–4.5 | 1 | [11,12] |
| Avena sativa | 0 | 1.5–2.3 | 1 | [6] |
| Secale cereale | 0 | 1.9–3 | 1 | [6] |
| Equisetum arvense | 0 | 0.05–0.1 | 1 | [3] |
| Equisetum fluviatile | 0 | 0.1 | 1 | [7] |
| Cetraria islandica | 0 | 20.2–24.6 | 1 | [6] |
| Sinorhizobium meliloti | 1 | 0 | 0 | [10] |
| Sarcina ventriculi | 0 | 1 | 0 | [13] |
| Ectocarpus sp. | 0 | 1 | 0 | [9] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, S.-C.; Saldivar, R.K.; Liang, P.-H.; Hsieh, Y.S.Y. Structures, Biosynthesis, and Physiological Functions of (1,3;1,4)-β-d-Glucans. Cells 2021, 10, 510. https://doi.org/10.3390/cells10030510
Chang S-C, Saldivar RK, Liang P-H, Hsieh YSY. Structures, Biosynthesis, and Physiological Functions of (1,3;1,4)-β-d-Glucans. Cells. 2021; 10(3):510. https://doi.org/10.3390/cells10030510
Chicago/Turabian StyleChang, Shu-Chieh, Rebecka Karmakar Saldivar, Pi-Hui Liang, and Yves S. Y. Hsieh. 2021. "Structures, Biosynthesis, and Physiological Functions of (1,3;1,4)-β-d-Glucans" Cells 10, no. 3: 510. https://doi.org/10.3390/cells10030510
APA StyleChang, S.-C., Saldivar, R. K., Liang, P.-H., & Hsieh, Y. S. Y. (2021). Structures, Biosynthesis, and Physiological Functions of (1,3;1,4)-β-d-Glucans. Cells, 10(3), 510. https://doi.org/10.3390/cells10030510





_Rebecka_Karmakar_Saldivar.png)



