C4d Deposition after Allogeneic Renal Transplantation in Rats Is Involved in Initial Apoptotic Cell Clearance
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model
2.2. General Functional Data
2.3. Histology
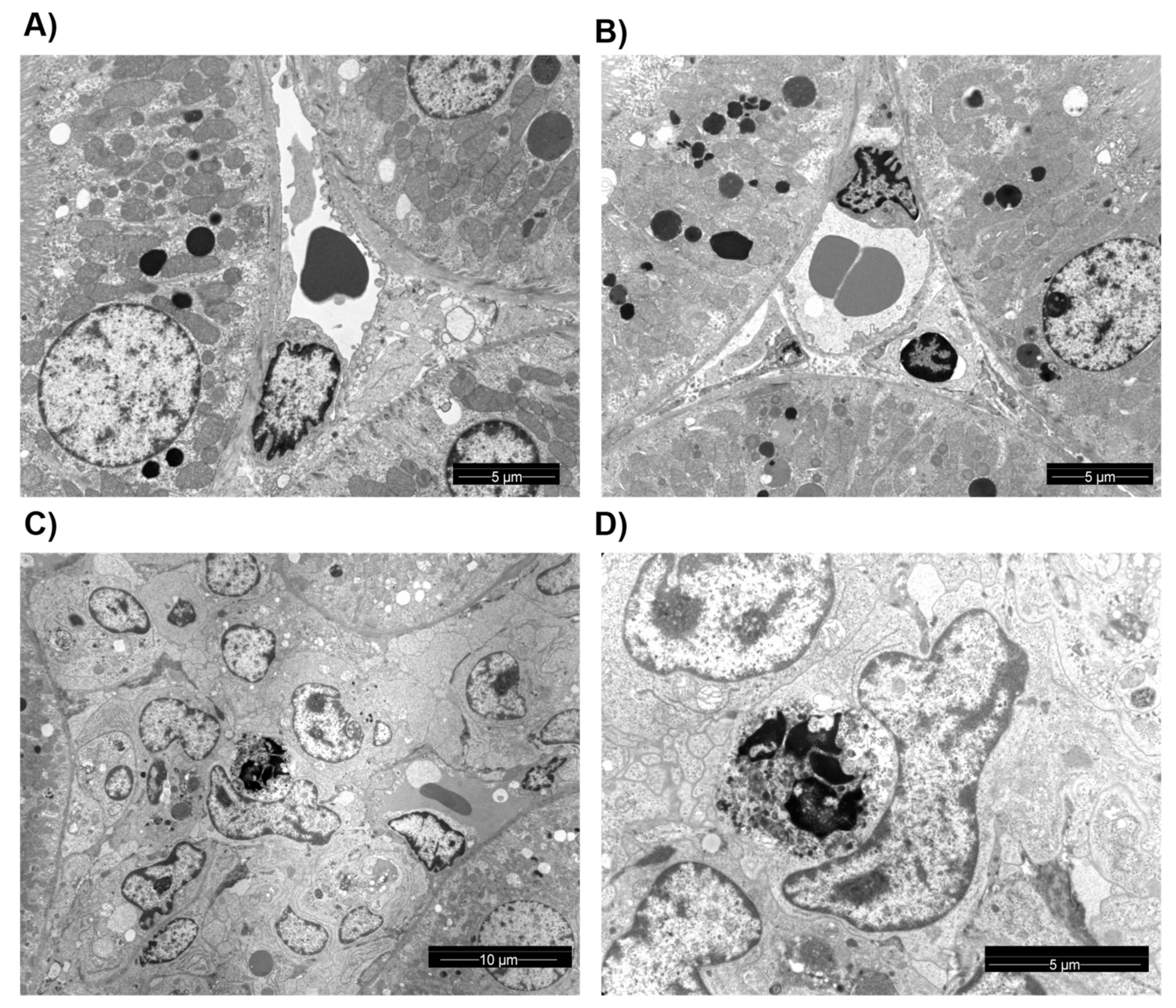
2.4. Transmission Electron Microscopy
2.5. Microarray Analysis
2.6. Real-Time PCR
2.7. Human Biopsy Transcriptome Analysis
2.8. C4d and C3d Deposition
2.9. FACS Analysis
2.10. Statistical Analyses
3. Results
3.1. Human Transcriptome Data
3.2. Animal Model
3.2.1. General Functional Data and Histology
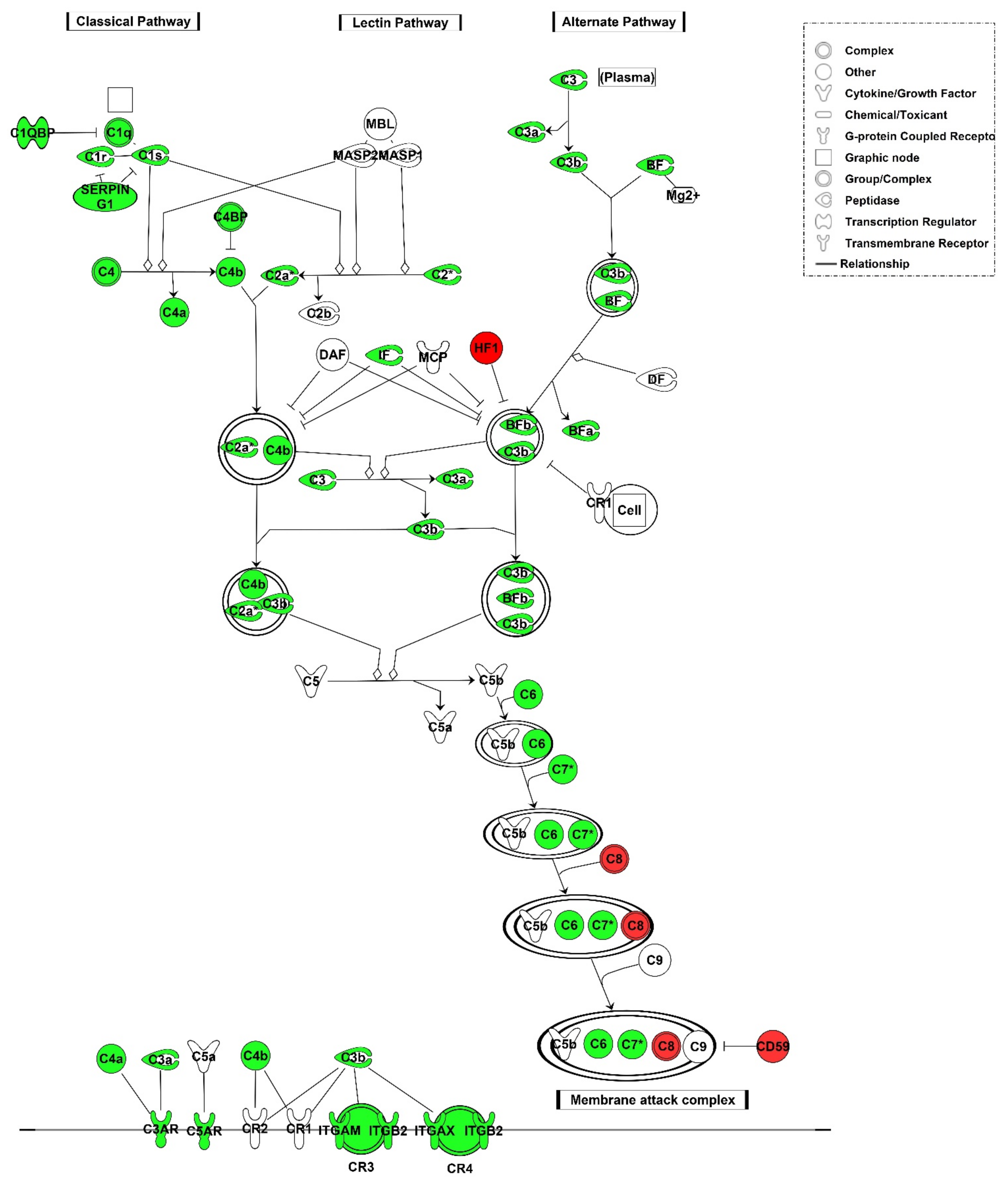
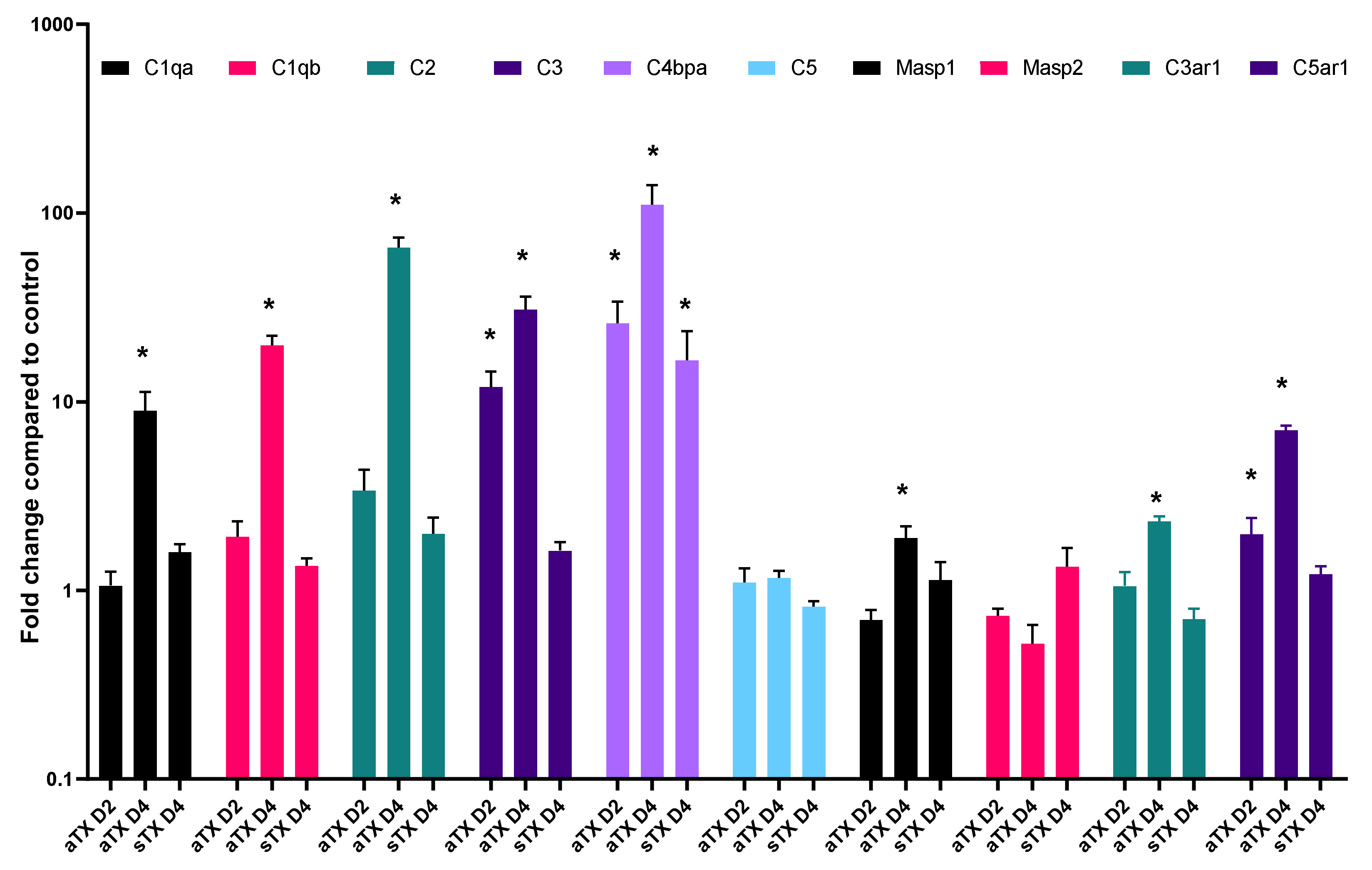
3.2.2. Local Gene Expression of Complement Factors
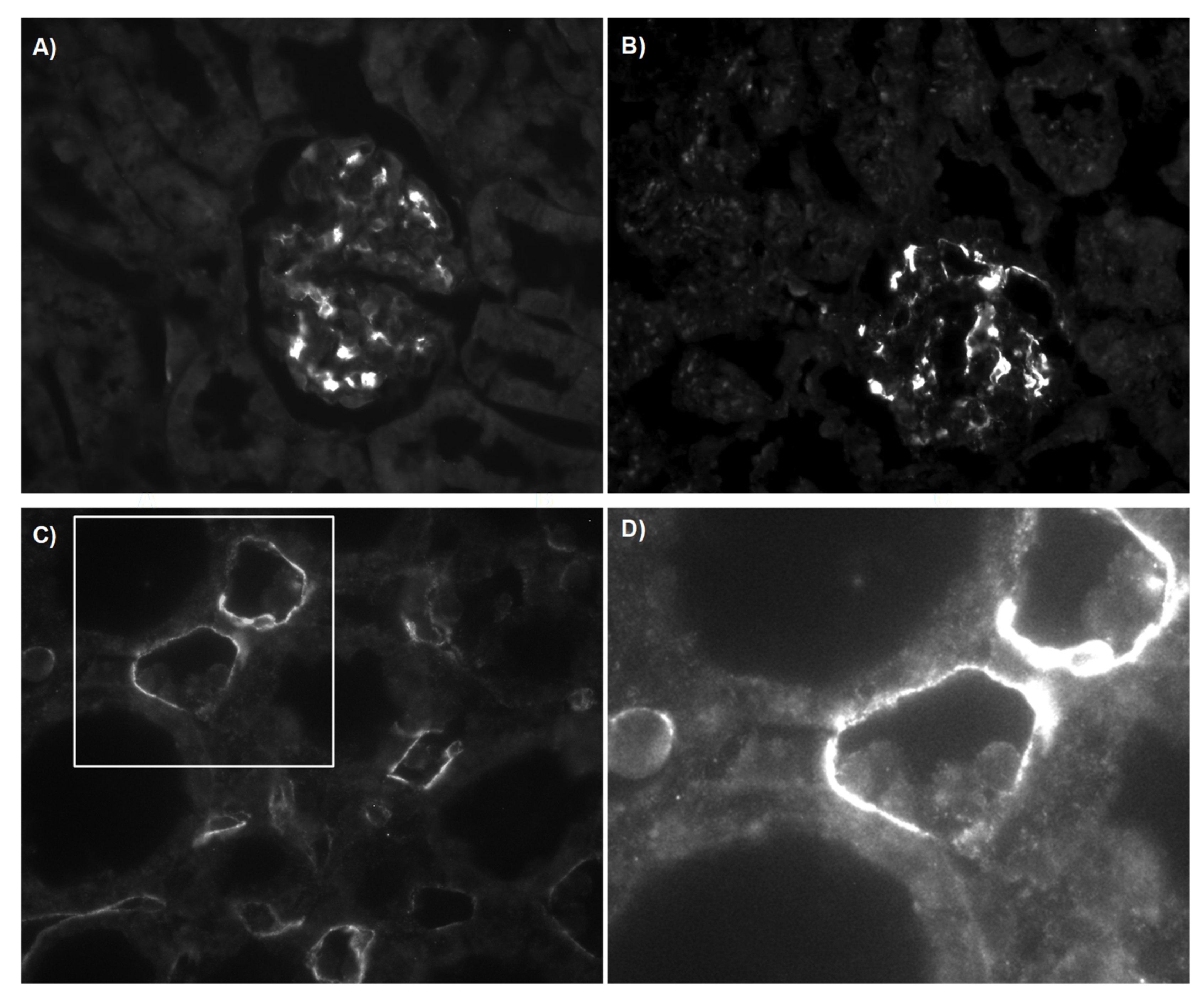
3.2.3. C4d Deposition Indicates Activation of the Complement System
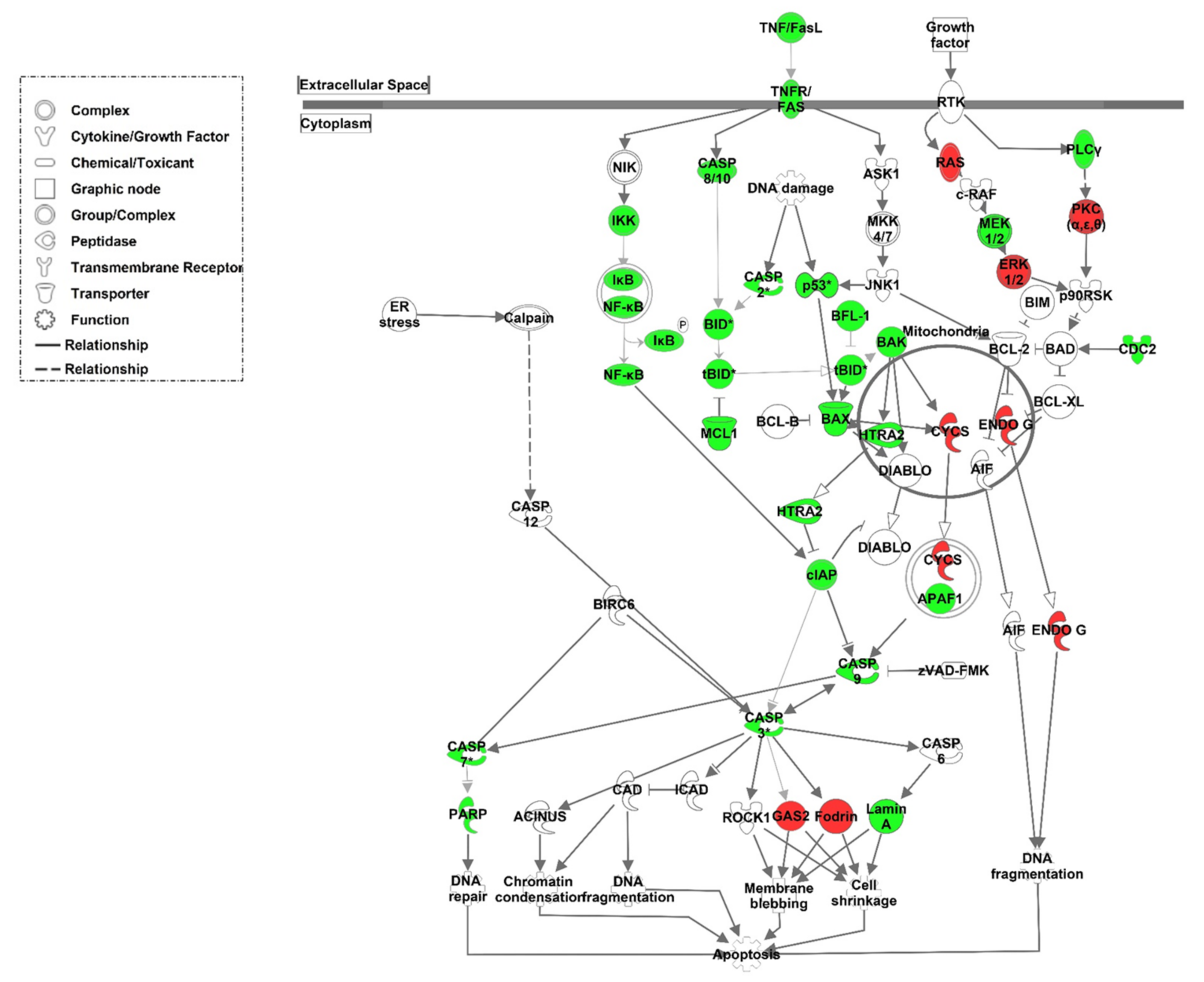
3.3. Structural Analysis and Apoptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rekers, N.V.; de Fijter, J.W.; Claas, F.H.; Eikmans, M. Mechanisms and risk assessment of steroid resistance in acute kidney transplant rejection. Transpl. Immunol. 2016, 38, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J. Diagnosis of renal transplant rejection: Banff classification and beyond. Kidney Res. Clin. Pract. 2020, 39, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Biglarnia, A.R.; Huber-Lang, M.; Mohlin, C.; Ekdahl, K.N.; Nilsson, B. The multifaceted role of complement in kidney transplantation. Nat. Rev. Nephrol. 2018, 14, 767–781. [Google Scholar] [CrossRef]
- Poppelaars, F.; Thurman, J.M. Complement-mediated kidney diseases. Mol. Immunol. 2020, 128, 175–187. [Google Scholar] [CrossRef]
- Budge, K.; Dellepiane, S.; Yu, S.M.; Cravedi, P. Complement, a Therapeutic Target in Diabetic Kidney Disease. Front. Med. 2020, 7, 599236. [Google Scholar] [CrossRef] [PubMed]
- Halloran, P.F.; Wadgymar, A.; Ritchie, S.; Falk, J.; Solez, K.; Srinivasa, N.S. The significance of the anti-class I antibody response. I. Clinical and pathologic features of anti-class I-mediated rejection. Transplantation 1990, 49, 85–91. [Google Scholar] [CrossRef]
- Feucht, H.E.; Schneeberger, H.; Hillebrand, G.; Burkhardt, K.; Weiss, M.; Riethmuller, G.; Land, W.; Albert, E. Capillary deposition of C4d complement fragment and early renal graft loss. Kidney Int. 1993, 43, 1333–1338. [Google Scholar] [CrossRef] [PubMed]
- Feucht, H.E.; Felber, E.; Gokel, M.J.; Hillebrand, G.; Nattermann, U.; Brockmeyer, C.; Held, E.; Riethmuller, G.; Land, W.; Albert, E. Vascular deposition of complement-split products in kidney allografts with cell-mediated rejection. Clin. Exp. Immunol. 1991, 86, 464–470. [Google Scholar] [CrossRef]
- Murata, K.; Iwata, T.; Nakashima, S.; Fox-Talbot, K.; Qian, Z.; Wilkes, D.S.; Baldwin, W.M. C4d deposition and cellular infiltrates as markers of acute rejection in rat models of orthotopic lung transplantation. Transplantation 2008, 86, 123–129. [Google Scholar] [CrossRef]
- Nickeleit, V.; Mihatsch, M.J. Kidney transplants, antibodies and rejection: Is C4d a magic marker? Nephrol. Dial. Transplant. 2003, 18, 2232–2239. [Google Scholar] [CrossRef]
- Trpkov, K.; Campbell, P.; Pazderka, F.; Cockfield, S.; Solez, K.; Halloran, P.F. Pathologic features of acute renal allograft rejection associated with donor-specific antibody, Analysis using the Banff grading schema. Transplantation 1996, 61, 1586–1592. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Patel, K.; Tu, Z.; Yang, X.; Kulik, L.; Alawieh, A.; Allen, P.; Cheng, Q.; Wallace, C.; Kilkenny, J.; et al. A novel injury site-natural antibody targeted complement inhibitor protects against lung transplant injury. Am. J. Transplant. 2020, 21, 2067–2078. [Google Scholar] [CrossRef]
- Atkinson, C.; Qiao, F.; Yang, X.; Zhu, P.; Reaves, N.; Kulik, L.; Goddard, M.; Holers, V.M.; Tomlinson, S. Targeting pathogenic postischemic self-recognition by natural IgM to protect against posttransplantation cardiac reperfusion injury. Circulation 2015, 131, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Pushpakumar, S.B.; Perez-Abadia, G.; Soni, C.; Wan, R.; Todnem, N.; Patibandla, P.K.; Fensterer, T.; Zhang, Q.; Barker, J.H.; Maldonado, C. Enhancing complement control on endothelial barrier reduces renal post-ischemia dysfunction. J. Surg. Res. 2011, 170, e263–e270. [Google Scholar] [CrossRef][Green Version]
- Kotimaa, J.P.; van Werkhoven, M.B.; O’Flynn, J.; Klar-Mohamad, N.; van Groningen, J.; Schilders, G.; Rutjes, H.; Daha, M.R.; Seelen, M.A.; van Kooten, C. Functional assessment of mouse complement pathway activities and quantification of C3b/C3c/iC3b in an experimental model of mouse renal ischaemia/reperfusion injury. J. Immunol. Methods 2015, 419, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zang, G.; Jiang, J.; He, W.; Johnston, N.J.; Ling, H.; Chen, R.; Zhang, X.; Liu, Y.; Haig, A.; et al. Attenuating Ischemia-Reperfusion Injury in Kidney Transplantation by Perfusing Donor Organs With siRNA Cocktail Solution. Transplantation 2016, 100, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Edemir, B.; Kurian, S.M.; Eisenacher, M.; Lang, D.; Muller-Tidow, C.; Gabriels, G.; Salomon, D.R.; Schlatter, E. Activation of counter-regulatory mechanisms in a rat renal acute rejection model. BMC. Genom. 2008, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.R.; Abe, K.; Miyazaki, M.; Zhou, W.; Sacks, S.H. In situ localization of C3 synthesis in experimental acute renal allograft rejection. Am. J. Pathol. 2000, 157, 825–831. [Google Scholar] [CrossRef]
- Serinsöz, E.; Bock, O.; Gwinner, W.; Schwarz, A.; Haller, H.; Kreipe, H.; Mengel, M. Local complement C3 expression is upregulated in humoral and cellular rejection of renal allografts. Am. J. Transpl. 2005, 5, 1490–1494. [Google Scholar] [CrossRef] [PubMed]
- Edemir, B.; Reuter, S.; Borgulya, R.; Schroter, R.; Neugebauer, U.; Gabriels, G.; Schlatter, E. Acute Rejection Modulates Gene Expression in the Collecting Duct. J. Am. Soc. Nephrol. 2008, 19, 538–546. [Google Scholar] [CrossRef]
- Reuter, S.; Velic, A.; Edemir, B.; Schroter, R.; Pavenstadt, H.; Gabriels, G.; Bleich, M.; Schlatter, E. Protective role of NHE-3 inhibition in rat renal transplantation undergoing acute rejection. Pflug. Arch. 2008, 456, 1075–1084. [Google Scholar] [CrossRef]
- Grabner, A.; Kentrup, D.; Schnöckel, U.; Gabriëls, G.; Schröter, R.; Pavenstädt, H.; Schober, O.; Schlatter, E.; Schäfers, M.; Reuter, S. Non-invasive imaging of acute allograft rejection after rat renal transplantation using 18F-FDG PET. J. Vis. Exp. 2013, e4240. [Google Scholar] [CrossRef]
- Huang, G.; Wilson, N.A.; Reese, S.R.; Jacobson, L.M.; Zhong, W.; Djamali, A. Characterization of transfusion-elicited acute antibody-mediated rejection in a rat model of kidney transplantation. Am. J. Transpl. 2014, 14, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Lam Peng, A. BRB Array Tools. Available online: http://linus.nci.nih.gov/BRB-ArrayTools.html (accessed on 1 April 2008).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Reeve, J.; Sellarés, J.; Mengel, M.; Sis, B.; Skene, A.; Hidalgo, L.; de Freitas, D.G.; Famulski, K.S.; Halloran, P.F. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am. J. Transpl. 2013, 13, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Murata, K.; Lee, C.Y.; Fox-Talbot, K.; Wasowska, B.A.; Pescovitz, M.D.; Baldwin, W.M., III. C4d deposition and clearance in cardiac transplants correlates with alloantibody levels and rejection in rats. Am. J. Transplant. 2006, 6, 923–932. [Google Scholar] [CrossRef]
- Reuter, S.; Schnockel, U.; Schroter, R.; Schober, O.; Pavenstadt, H.; Schafers, M.; Gabriels, G.; Schlatter, E. Non-invasive imaging of acute renal allograft rejection in rats using small animal F-FDG-PET. PLoS ONE 2009, 4, e5296. [Google Scholar] [CrossRef]
- Nagata, S.; Hanayama, R.; Kawane, K. Autoimmunity and the clearance of dead cells. Cell 2010, 140, 619–630. [Google Scholar] [CrossRef]
- Nonaka, M.; Kimura, A. Genomic view of the evolution of the complement system. Immunogenetics 2006, 58, 701–713. [Google Scholar] [CrossRef]
- Banda, N.K.; Takahashi, K. Analysis of the Complement Activation in Mice. In The Complement System: Methods and Protocols; Gadjeva, M., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 365–371. [Google Scholar]
- Collins, A.B.; Schneeberger, E.E.; Pascual, M.A.; Saidman, S.L.; Williams, W.W.; Tolkoff-Rubin, N.; Cosimi, A.B.; Colvin, R.B. Complement activation in acute humoral renal allograft rejection: Diagnostic significance of C4d deposits in peritubular capillaries. J. Am. Soc. Nephrol. 1999, 10, 2208–2214. [Google Scholar] [CrossRef] [PubMed]
- Fleming, S.D.; Shea-Donohue, T.; Guthridge, J.M.; Kulik, L.; Waldschmidt, T.J.; Gipson, M.G.; Tsokos, G.C.; Holers, V.M. Mice Deficient in Complement Receptors 1 and 2 Lack a Tissue Injury-Inducing Subset of the Natural Antibody Repertoire. J. Immunol. 2002, 169, 2126–2133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Farrar, C.A.; Abe, K.; Pratt, J.R.; Marsh, J.E.; Wang, Y.; Stahl, G.L.; Sacks, S.H. Predominant role for C5b-9 in renal ischemia/reperfusion injury. J. Clin. Investig. 2000, 105, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wang, X.; Liang, T.; Bao, D.; Wang, Y.; Du, Y.; Li, H.; Du, J.; Chen, A.; Fu, Z.; et al. Effect of Hyperbaric Oxygen on Tissue Damage and Expression of Adhesion Molecules and C3 in a Rat Model of Renal Ischemia-Reperfusion Injury After Kidney Transplantation. Ann. Transpl. 2020, 25, e919385. [Google Scholar] [CrossRef]
- Farrar, C.A.; Zhou, W.; Lin, T.; Sacks, S.H. Local extravascular pool of C3 is a determinant of postischemic acute renal failure. FASEB J. 2006, 20, 217–226. [Google Scholar] [CrossRef]
- Pratt, J.R.; Basheer, S.A.; Sacks, S.H. Local synthesis of complement component C3 regulates acute renal transplant rejection. Nat. Med. 2002, 8, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Sacks, S.; Zhou, W. New Boundaries for Complement in Renal Disease. J. Am. Soc. Nephrol. 2008, 19, 1865–1869. [Google Scholar] [CrossRef]
- Baldwin, W.M.; Ota, H.; Rodriguez, E.R. Complement in transplant rejection: Diagnostic and mechanistic considerations. Springer Semin. Immunopathol. 2003, 25, 181–197. [Google Scholar] [CrossRef]
- Arora, M.; Munoz, E.; Tenner, A.J. Identification of a site on mannan-binding lectin critical for enhancement of phagocytosis. J Biol. Chem. 2001, 276, 43087–43094. [Google Scholar] [CrossRef]
- Csencsits, K.; Burrell, B.E.; Lu, G.; Eichwald, E.J.; Stahl, G.L.; Bishop, D.K. The classical complement pathway in transplantation: Unanticipated protective effects of C1q and role in inductive antibody therapy. Am. J. Transplant. 2008, 8, 1622–1630. [Google Scholar] [CrossRef]
- Botto, M.; Dell’Agnola, C.; Bygrave, A.E.; Thompson, E.M.; Cook, H.T.; Petry, F.; Loos, M.; Pandolfi, P.P.; Walport, M.J. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 1998, 19, 56–59. [Google Scholar] [CrossRef]
- Korb, L.C.; Ahearn, J.M. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: Complement deficiency and systemic lupus erythematosus revisited. J. Immunol. 1997, 158, 4525–4528. [Google Scholar] [PubMed]
- Taylor, P.R.; Carugati, A.; Fadok, V.A.; Cook, H.T.; Andrews, M.; Carroll, M.C.; Savill, J.S.; Henson, P.M.; Botto, M.; Walport, M.J. A hierarchical role for classical pathway complement proteins in the clearance of apoptotic cells in vivo. J. Exp. Med. 2000, 192, 359–366. [Google Scholar] [CrossRef]
- Ling, G.S.; Crawford, G. C1q restrains autoimmunity and viral infection by regulating CD8(+) T cell metabolism. Science 2018, 360, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Liptak, P.; Kemeny, E.; Morvay, Z.; Szederkenyi, E.; Szenohradszky, P.; Marofka, F.; Toldi, J.; Exner, M.; Ivanyi, B. Peritubular capillary damage in acute humoral rejection: An ultrastructural study on human renal allografts. Am. J. Transplant. 2005, 5, 2870–2876. [Google Scholar] [CrossRef]
- Racusen, L.C.; Colvin, R.B.; Solez, K.; Mihatsch, M.J.; Halloran, P.F.; Campbell, P.M.; Cecka, M.J.; Cosyns, J.P.; Demetris, A.J.; Fishbein, M.C.; et al. Antibody-mediated rejection criteria—An addition to the Banff 97 classification of renal allograft rejection. Am. J. Transplant. 2003, 3, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Blom, A.M.; Villoutreix, B.O.; Dahlbäck, B. Complement inhibitor C4b-binding protein—Friend or foe in the innate immune system? Mol. Immunol. 2004, 40, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.H.; Blom, A.M.; Dahlbäck, B. Vitamin K-dependent protein S localizing complement regulator C4b-binding protein to the surface of apoptotic cells. J. Immunol. 2002, 169, 2580–2586. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reuter, S.; Kentrup, D.; Grabner, A.; Köhler, G.; Buscher, K.; Edemir, B. C4d Deposition after Allogeneic Renal Transplantation in Rats Is Involved in Initial Apoptotic Cell Clearance. Cells 2021, 10, 3499. https://doi.org/10.3390/cells10123499
Reuter S, Kentrup D, Grabner A, Köhler G, Buscher K, Edemir B. C4d Deposition after Allogeneic Renal Transplantation in Rats Is Involved in Initial Apoptotic Cell Clearance. Cells. 2021; 10(12):3499. https://doi.org/10.3390/cells10123499
Chicago/Turabian StyleReuter, Stefan, Dominik Kentrup, Alexander Grabner, Gabriele Köhler, Konrad Buscher, and Bayram Edemir. 2021. "C4d Deposition after Allogeneic Renal Transplantation in Rats Is Involved in Initial Apoptotic Cell Clearance" Cells 10, no. 12: 3499. https://doi.org/10.3390/cells10123499
APA StyleReuter, S., Kentrup, D., Grabner, A., Köhler, G., Buscher, K., & Edemir, B. (2021). C4d Deposition after Allogeneic Renal Transplantation in Rats Is Involved in Initial Apoptotic Cell Clearance. Cells, 10(12), 3499. https://doi.org/10.3390/cells10123499





