Central Suppression of the GH/IGF Axis and Abrogation of Exercise-Related mTORC1/2 Activation in the Muscle of Phenotype-Selected Male Marathon Mice (DUhTP)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, Study Design, and Sample Preparation
2.2. Next-Generation Sequencing (NGS), Differential Gene Expression Analysis, and Data Processing
2.3. IGF1 Assay
2.4. Protein Isolation and SDS-PAGE
2.5. Analysis of Signal Transduction by Western Immunoblotting (WIB)
2.6. Analysis of Signal Transduction by Capillary Immuno-Electrophoresis (WESTM)
2.7. Statistical Analysis
3. Results
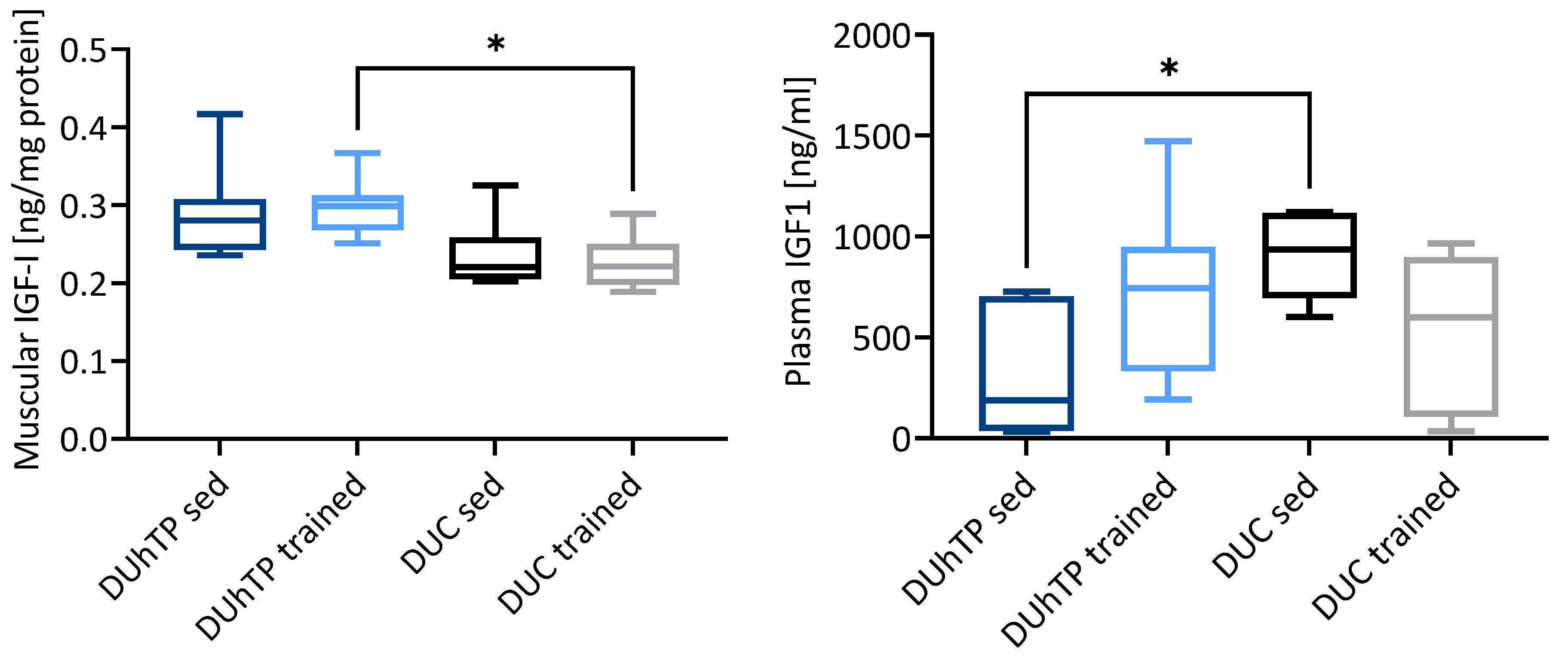
3.1. Effects of Phenotype Selection and Running on the GH/IGF Axis in the Pituitary Gland and Muscle Tissue
3.2. Effects of Phenotype Selection and Endurance Exercise on mRNA Expression of Intracellular Signaling Compounds in the Pituitary Gland and Muscle
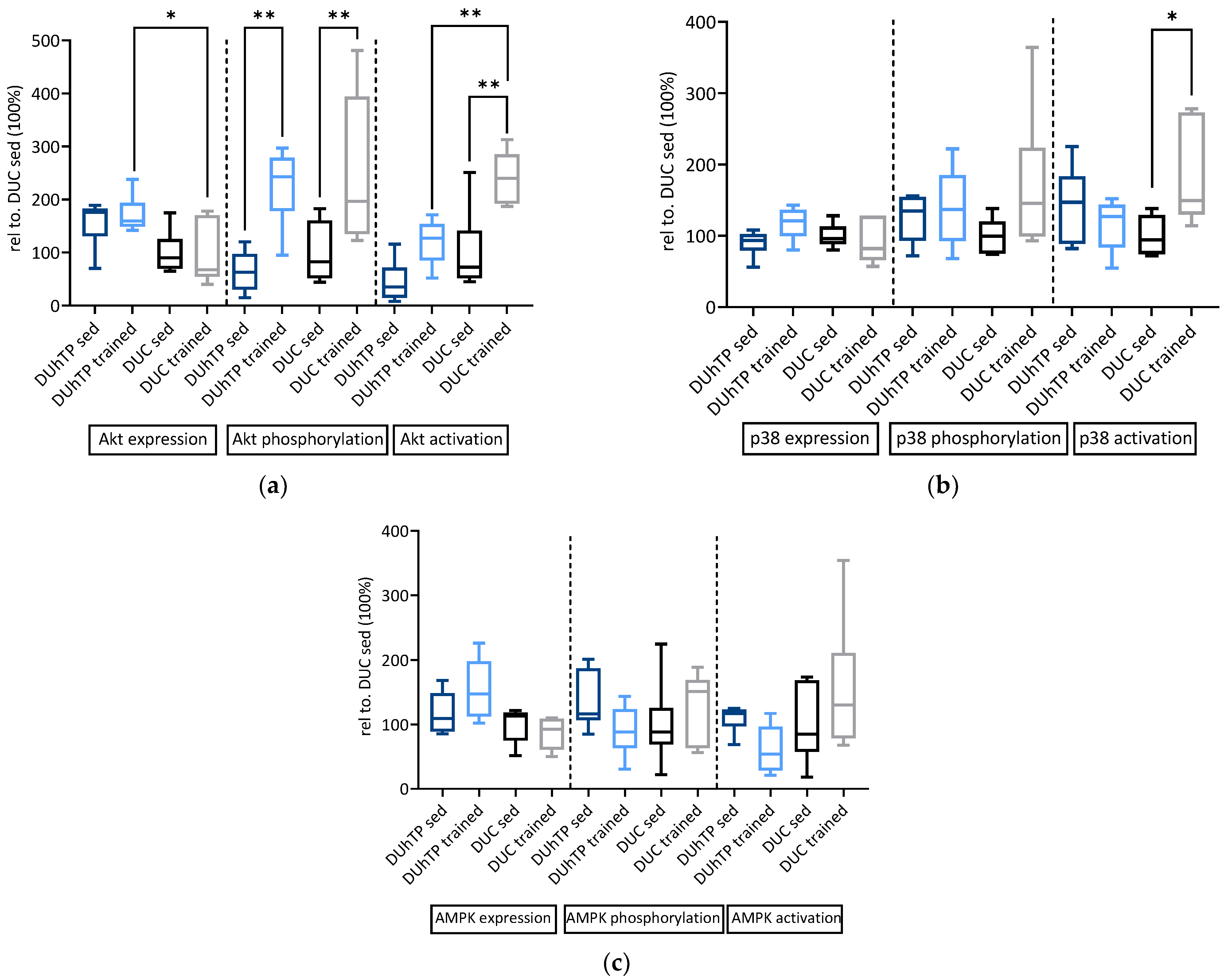
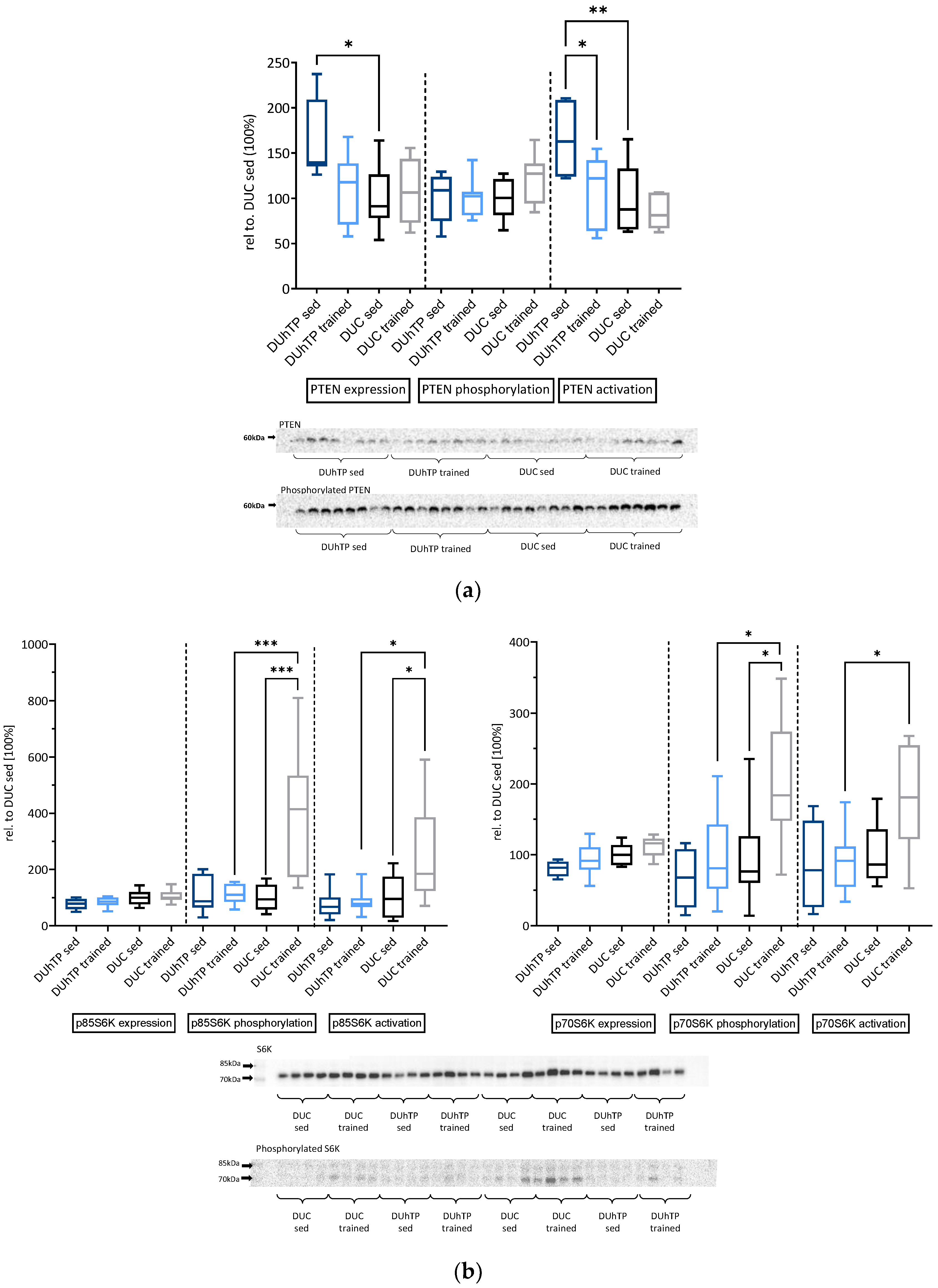
3.3. Effects of Phenotype Selection and Training on Intracellular Signal Transduction in the Muscle
4. Discussion
4.1. Regulation of the Somatotropic Axis
4.2. Exercise-Related Activation of mTORC1 and mTORC2 in Muscle of DUC but Not of DUhTP Mice
4.3. Effects of Phenotype-Selection on mRNA Expression Related to Metabolic Cell Signaling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, J.; Glick, S.M.; Yalow, R.S.; Berson, S.A. Secretion of human growth hormone: Physiologic and experimental modification. Metabolism 1963, 12, 577–579. [Google Scholar]
- Wideman, L.; Weltman, J.Y.; Hartman, M.L.; Veldhuis, J.D.; Weltman, A. Growth hormone release during acute and chronic aerobic and resistance exercise: Recent findings. Sports Med. 2002, 32, 987–1004. [Google Scholar] [CrossRef]
- Liu, W.; Bretz, F.; Böhning, D.; Holt, R.; Böhning, W.; Guha, N.; Sönksen, P.; Cowan, D. Comparison of normal distribution-based and nonparametric decision limits on the GH-2000 score for detecting growth hormone misuse (doping) in sport. Biom. J. 2021, 63, 187–200. [Google Scholar] [CrossRef]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 Mediate Assembly of the Translation Preinitiation Complex through Dynamic Protein Interchange and Ordered Phosphorylation Events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frystyk, J. Exercise and the growth hormone-insulin-like growth factor axis. Med. Sci. Sports Exerc. 2010, 42, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, S.; Moromisato, D.Y.; Moromisato, M.Y.; Woods, J.; Brasel, J.A.; Leroith, D.; Roberts, C.T., Jr.; Cooper, D.M. Effect of training and growth hormone suppression on insulin-like growth factor I mRNA in young rats. J. Appl. Physiol. 1994, 76, 2204–2209. [Google Scholar] [CrossRef] [PubMed]
- Spangenburg, E.E.; Le Roith, D.; Ward, C.W.; Bodine, S.C. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. J. Physiol. 2008, 586, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Brenmoehl, J.; Walz, C.; Spitschak, M.; Wirthgen, E.; Walz, M.; Langhammer, M.; Tuchscherer, A.; Naumann, R.; Hoeflich, A. Partial phenotype conversion and differential trait response to conditions of husbandry in mice. J. Comp. Physiol. B 2018, 188, 527–539. [Google Scholar] [CrossRef] [Green Version]
- Walz, C.; Brenmoehl, J.; Trakooljul, N.; Noce, A.; Caffier, C.; Ohde, D.; Langhammer, M.; Wimmers, K.; Ponsuksili, S.; Hoeflich, A. Control of Protein and Energy Metabolism in the Pituitary Gland in Response to Three-Week Running Training in Adult Male Mice. Cells 2021, 10, 736. [Google Scholar] [CrossRef]
- Walz, M.; Höflich, C.; Walz, C.; Ohde, D.; Brenmoehl, J.; Sawitzky, M.; Vernunft, A.; Zettl, U.K.; Holtze, S.; Hildebrandt, T.B.; et al. Development of a Sensitive Bioassay for the Analysis of IGF-Related Activation of AKT/mTOR Signaling in Biological Matrices. Cells 2021, 10, 482. [Google Scholar] [CrossRef]
- Schaufele, F.; West, B.L.; Reudelhuber, T. Somatotroph- and lactotroph-specific interactions with the homeobox protein binding sites in the rat growth hormone gene promoter. Nucleic Acids Res. 1990, 18, 5235–5243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, L.E.; Wondisford, F.E.; Radovick, S. Role of Pit-1 in the gene expression of growth hormone, prolactin, and thyrotropin. Endocrinol. Metab. Clin. North. Am. 1996, 25, 523–540. [Google Scholar] [CrossRef]
- Kojima, M.; Hosoda, H.; Date, Y.; Nakazato, M.; Matsuo, H.; Kangawa, K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 1999, 402, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Gibney, J.; Healy, M.L.; Sönksen, P.H. The growth hormone/insulin-like growth factor-I axis in exercise and sport. Endocr. Rev. 2007, 28, 603–624. [Google Scholar] [CrossRef] [Green Version]
- de Alcantara Borba, D.; da Silva Alves, E.; Rosa, J.P.P.; Facundo, L.A.; Costa, C.M.A.; Silva, A.C.; Narciso, F.V.; Silva, A.; de Mello, M.T. Can IGF-1 Serum Levels Really be Changed by Acute Physical Exercise? A Systematic Review and Meta-Analysis. J. Phys. Act. Health 2020, 17, 575–584. [Google Scholar] [CrossRef]
- Philp, A.; Hamilton, D.L.; Baar, K. Signals mediating skeletal muscle remodeling by resistance exercise: PI3-kinase independent activation of mTORC1. J. Appl. Physiol. 2011, 110, 561–568. [Google Scholar] [CrossRef] [Green Version]
- Hornberger, T.A.; Stuppard, R.; Conley, K.E.; Fedele, M.J.; Fiorotto, M.L.; Chin, E.R.; Esser, K.A. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem. J. 2004, 380, 795–804. [Google Scholar] [CrossRef]
- Hamilton, D.L.; Philp, A.; MacKenzie, M.G.; Baar, K. A limited role for PI(3,4,5)P3 regulation in controlling skeletal muscle mass in response to resistance exercise. PLoS ONE 2010, 5, e11624. [Google Scholar] [CrossRef] [PubMed]
- Marino, F.E.; Mbambo, Z.; Kortekaas, E.; Wilson, G.; Lambert, M.I.; Noakes, T.D.; Dennis, S.C. Advantages of smaller body mass during distance running in warm, humid environments. Pflug. Arch. 2000, 441, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Brenmoehl, J.; Ohde, D.; Albrecht, E.; Walz, C.; Tuchscherer, A.; Hoeflich, A. Browning of subcutaneous fat and higher surface temperature in response to phenotype selection for advanced endurance exercise performance in male DUhTP mice. J. Physiol. B 2017, 187, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, M.; Moriya, N.; Takemasa, T. Transient activation of mTORC1 signaling in skeletal muscle is independent of Akt1 regulation. Physiol. Rep. 2020, 8, e14599. [Google Scholar] [CrossRef] [PubMed]
- Kotani, T.; Takegaki, J.; Tamura, Y.; Kouzaki, K.; Nakazato, K.; Ishii, N. Repeated bouts of resistance exercise in rats alter mechanistic target of rapamycin complex 1 activity and ribosomal capacity but not muscle protein synthesis. Exp. Physiol. 2021, 106, 1950–1960. [Google Scholar] [CrossRef]
- Solsona, R.; Pavlin, L.; Bernardi, H.; Sanchez, A.M. Molecular Regulation of Skeletal Muscle Growth and Organelle Biosynthesis: Practical Recommendations for Exercise Training. Int. J. Mol. Sci. 2021, 22, 2741. [Google Scholar] [CrossRef]
- Schoenfeld, B.J.; Ogborn, D.; Krieger, J.W. Dose-response relationship between weekly resistance training volume and increases in muscle mass: A systematic review and meta-analysis. J. Sports Sci. 2017, 35, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Alimov, A.P.; McCarthy, J.J. Ribosome Biogenesis is Necessary for Skeletal Muscle Hypertrophy. Exerc. Sport Sci. Rev. 2016, 44, 110–115. [Google Scholar] [CrossRef]
- West, D.W.; Baehr, L.M.; Marcotte, G.R.; Chason, C.M.; Tolento, L.; Gomes, A.V.; Bodine, S.C.; Baar, K. Acute resistance exercise activates rapamycin-sensitive and -insensitive mechanisms that control translational activity and capacity in skeletal muscle. J. Physiol. 2016, 594, 453–468. [Google Scholar] [CrossRef] [Green Version]
- Csibi, A.; Cornille, K.; Leibovitch, M.P.; Poupon, A.; Tintignac, L.A.; Sanchez, A.M.; Leibovitch, S.A. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS ONE 2010, 5, e8994. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Harris, T.E.; Roth, R.A.; Lawrence, J.C., Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 2007, 282, 20036–20044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 2007, 25, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Civiletto, G.; Dogan, S.A.; Cerutti, R.; Fagiolari, G.; Moggio, M.; Lamperti, C.; Benincá, C.; Viscomi, C.; Zeviani, M. Rapamycin rescues mitochondrial myopathy via coordinated activation of autophagy and lysosomal biogenesis. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Hresko, R.C.; Mueckler, M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 2005, 280, 40406–40416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogasawara, R.; Fujita, S.; Hornberger, T.A.; Kitaoka, Y.; Makanae, Y.; Nakazato, K.; Naokata, I. The role of mTOR signalling in the regulation of skeletal muscle mass in a rodent model of resistance exercise. Sci. Rep. 2016, 6, 31142. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Song, C.; Huang, D.; Narayanan, N.; Qiu, J.; Jia, Z.; Yuan, Z.; Oprescu, S.N.; Roseguini, B.T.; Deng, M.; et al. PTEN Inhibition Ameliorates Muscle Degeneration and Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. 2021, 29, 132–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, Y.; Ma, J.; Wang, W.; Sun, B.; Zheng, T.; Wei, M.; Sun, Y. MicroRNA-20a participates in the aerobic exercise-based prevention of coronary artery disease by targeting PTEN. Biomed. Pharmacother. 2017, 95, 756–763. [Google Scholar] [CrossRef]
- Gao, H.E.; Wu, D.S.; Sun, L.; Yang, L.D.; Qiao, Y.B.; Ma, S.; Wu, Z.J.; Ruan, L.; Li, F.H. Effects of lifelong exercise on age-related body composition, oxidative stress, inflammatory cytokines, and skeletal muscle proteome in rats. Mech. Ageing Dev. 2020, 189, 111262. [Google Scholar] [CrossRef]
- Gao, H.E.; Li, F.H.; Xie, T.; Ma, S.; Qiao, Y.B.; Wu, D.S.; Sun, L. Lifelong Exercise in Age Rats Improves Skeletal Muscle Function and MicroRNA Profile. Med. Sci. Sports Exerc. 2021, 53, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Bengal, E.; Aviram, S.; Hayek, T. p38 MAPK in Glucose Metabolism of Skeletal Muscle: Beneficial or Harmful? Int. J. Mol. Sci. 2020, 21, 6480. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Rhee, J.; St-Pierre, J.; Handschin, C.; Puigserver, P.; Lin, J.; Jäeger, S.; Erdjument-Bromage, H.; Tempst, P.; Spiegelman, B.M. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: Modulation by p38 MAPK. Genes Dev. 2004, 18, 278–289. [Google Scholar] [CrossRef] [Green Version]
- Michael, L.F.; Wu, Z.; Cheatham, R.B.; Puigserver, P.; Adelmant, G.; Lehman, J.J.; Kelly, D.P.; Spiegelman, B.M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. USA 2001, 98, 3820–3825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; New, L.; Kravchenko, V.V.; Kato, Y.; Gram, H.; di Padova, F.; Olson, E.N.; Ulevitch, R.J.; Han, J. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell Biol. 1999, 19, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Fukatsu, Y.; Noguchi, T.; Hosooka, T.; Ogura, T.; Kotani, K.; Abe, T.; Shibakusa, T.; Inoue, K.; Sakai, M.; Tobimatsu, K.; et al. Muscle-specific overexpression of heparin-binding epidermal growth factor-like growth factor increases peripheral glucose disposal and insulin sensitivity. Endocrinology 2009, 150, 2683–2691. [Google Scholar] [CrossRef] [Green Version]
- Messi, M.L.; Li, T.; Wang, Z.M.; Marsh, A.P.; Nicklas, B.; Delbono, O. Resistance Training Enhances Skeletal Muscle Innervation without Modifying the Number of Satellite Cells or their Myofiber Association in Obese Older Adults. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 1273–1280. [Google Scholar] [CrossRef]
- Adamovich, Y.; Dandavate, V.; Ezagouri, S.; Manella, G.; Zwighaft, Z.; Sobel, J.; Kuperman, Y.; Golik, M.; Auerbach, A.; Itkin, M.; et al. Clock proteins and training modify exercise capacity in a daytime-dependent manner. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, Y.; Zhang, L.; Wang, S.; Yin, B.; Zhao, S.; Li, X. Synergistic effects of TGFβ2, WNT9a, and FGFR4 signals attenuate satellite cell differentiation during skeletal muscle development. Aging Cell 2018, 17, e12788. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Schweitzer, L.D.; Zoncu, R.; Sabatini, D.M. Ragulator Is a GEF for the Rag GTPases that Signal Amino Acid Levels to mTORC1. Cell 2012, 150, 1196–1208. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.S.; Jiang, B.; Li, M.; Zhu, M.; Peng, Y.; Zhang, Y.L.; Wu, Y.Q.; Li, T.Y.; Liang, Y.; Lu, Z.; et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014, 20, 526–540. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, H.M.; Maarbjerg, S.J.; Crane, J.D.; Jeppesen, J.; Jørgensen, S.B.; Schertzer, J.D.; Shyroka, O.; Kiens, B.; van Denderen, B.J.; Tarnopolsky, M.A.; et al. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc. Natl. Acad. Sci. USA 2011, 108, 16092–16097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costford, S.R.; Kavaslar, N.; Ahituv, N.; Chaudhry, S.N.; Schackwitz, W.S.; Dent, R.; Pennacchio, L.A.; McPherson, R.; Harper, M.E. Gain-of-function R225W mutation in human AMPKgamma(3) causing increased glycogen and decreased triglyceride in skeletal muscle. PLoS ONE 2007, 2, e903. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.R.; Michell, B.J.; van Denderen, B.J.; Watt, M.J.; Carey, A.L.; Fam, B.C.; Andrikopoulos, S.; Proietto, J.; Görgün, C.Z.; Carling, D.; et al. Tumor necrosis factor alpha-induced skeletal muscle insulin resistance involves suppression of AMP-kinase signaling. Cell Metab. 2006, 4, 465–474. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Xu, Z.; Zhang, L.; Liu, J.; Feng, J.; Wang, X.; Shan, T.; Wang, Y. Muscle-specific deletion of Prkaa1 enhances skeletal muscle lipid accumulation in mice fed a high-fat diet. J. Physiol. Biochem. 2018, 74, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Dali-Youcef, N.; Lagouge, M.; Froelich, S.; Koehl, C.; Schoonjans, K.; Auwerx, J. Sirtuins: The ‘magnificent seven’, function, metabolism and longevity. Ann. Med. 2007, 39, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Hooshmand-Moghadam, B.; Eskandari, M.; Golestani, F.; Rezae, S.; Mahmoudi, N.; Gaeini, A.A. The effect of 12-week resistance exercise training on serum levels of cellular aging process parameters in elderly men. Exp. Gerontol. 2020, 141, 111090. [Google Scholar] [CrossRef] [PubMed]
- Brenmoehl, J.; Hoeflich, A. Dual control of mitochondrial biogenesis by sirtuin 1 and sirtuin 3. Mitochondrion 2013, 13, 755–761. [Google Scholar] [CrossRef] [PubMed]
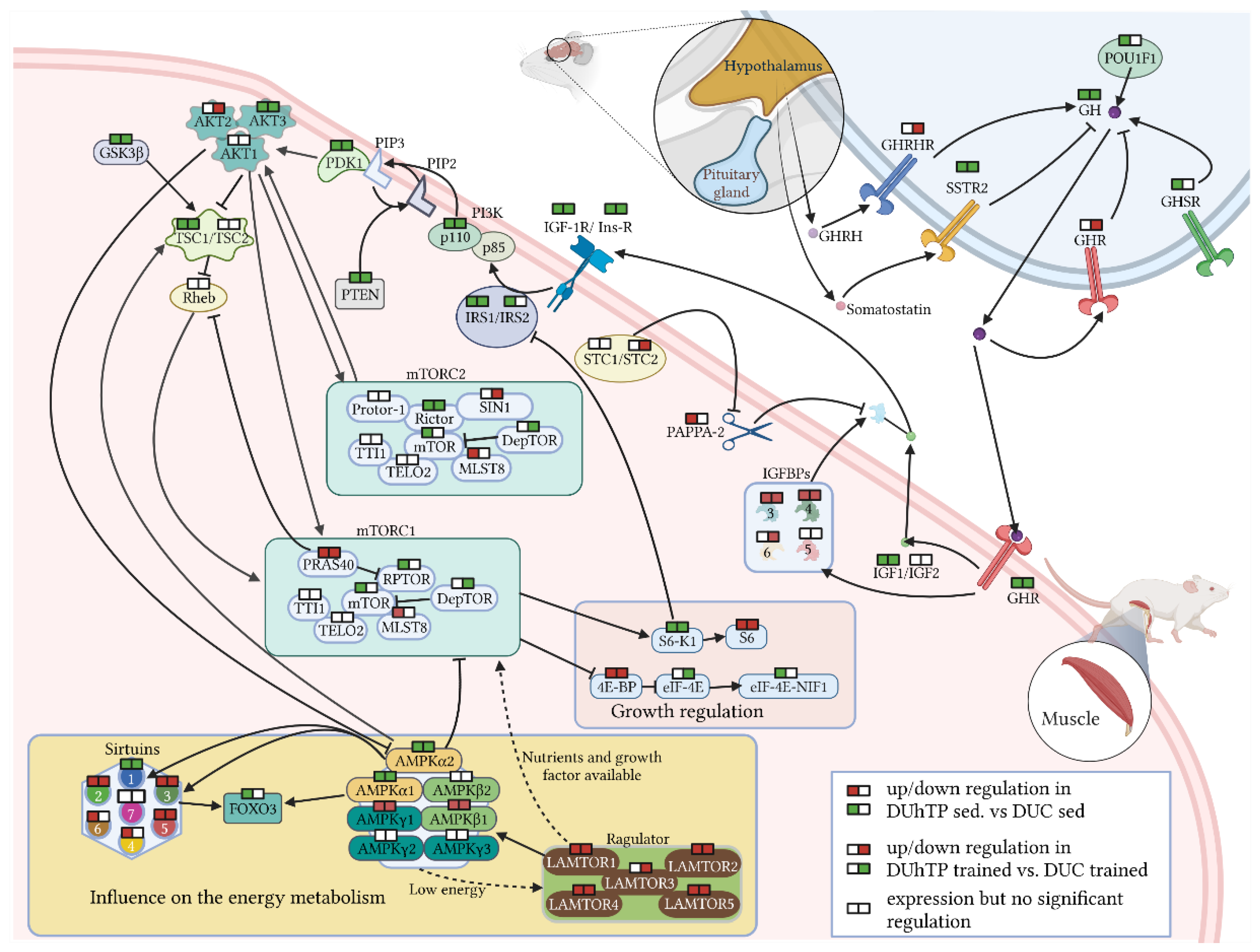
| Signaling Pathway | Comparison Parameters | Expression in the Pituitary Gland | Expression in Skeletal Muscle | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | DUhTP vs. DUC | DUC | DUhTP | DUhTP vs. DUC | DUC | DUhTP | ||||
| Sed | Trained | Trained vs. Sed | Sed | Trained | Trained vs. Sed | |||||
| GH axis | Pou1f1 | log2FC | −0.378 | −0.067 | −0.339 | −0.028 | ||||
| FDR | 0.002 | 0.650 | 0.027 | 1.000 | ||||||
| Gh | log2FC | −0.440 | −0.724 | 0.378 | 0.095 | |||||
| FDR | 0.066 | 0.000 | 0.158 | 1.000 | ||||||
| Ghsr | log2FC | −0.813 | −0.134 | −0.683 | −0.004 | |||||
| FDR | 0.016 | 0.744 | 0.082 | 1.000 | ||||||
| Sstr1 | log2FC | 0.676 | −0.084 | 0.862 | 0.102 | |||||
| FDR | 0.107 | 0.861 | 0.053 | 1.000 | ||||||
| Sstr2 | log2FC | −1.367 | −1.653 | −0.287 | −0.573 | |||||
| FDR | 0.004 | 0.000 | 0.668 | 1.000 | ||||||
| Ghrhr | log2FC | −0.144 | 0.493 | −0.176 | 0.462 | |||||
| FDR | 0.545 | 0.002 | 0.427 | 0.087 | ||||||
| Ghr | log2FC | −0.014 | 0.397 | −0.190 | 0.221 | −0.291 | −0.557 | −0.097 | −0.363 | |
| FDR | 0.977 | 0.073 | 0.526 | 1.000 | 0.034 | 0.000 | 0.859 | 0.025 | ||
| IGF system | Igf1 | log2FC | −0.075 | 0.299 | −0.563 | −0.189 | −0.882 | −1.722 | 0.098 | −0.742 |
| FDR | 0.906 | 0.426 | 0.179 | 1.000 | 0.000 | 0.000 | 0.914 | 0.002 | ||
| Igf2 | log2FC | −4.277 | −0.280 | −3.217 | 0.780 | −0.332 | −0.711 | 1.017 | 0.638 | |
| FDR | 0.000 | 0.843 | 0.021 | 1.000 | 0.569 | 0.138 | 0.303 | 0.363 | ||
| Igf1r | log2FC | −0.302 | 0.117 | −0.209 | 0.211 | −0.480 | −0.279 | −0.173 | 0.029 | |
| FDR | 0.069 | 0.487 | 0.267 | 0.989 | 0.000 | 0.046 | 0.670 | 0.913 | ||
| Igf2r | log2FC | −0.249 | 0.109 | −0.307 | 0.050 | −0.717 | −0.625 | 0.013 | 0.105 | |
| FDR | 0.086 | 0.450 | 0.049 | 1.000 | 0.000 | 0.000 | 0.987 | 0.565 | ||
| Irs1 | log2FC | 0.164 | 0.394 | −0.207 | 0.022 | −0.826 | −1.161 | −0.021 | −0.357 | |
| FDR | 0.579 | 0.054 | 0.445 | 1.000 | 0.000 | 0.000 | 0.985 | 0.091 | ||
| Irs2 | log2FC | −0.334 | −0.132 | −0.068 | 0.134 | −1.119 | −0.331 | −0.236 | 0.552 | |
| FDR | 0.271 | 0.648 | 0.868 | 1.000 | 0.037 | 0.562 | 0.920 | 0.474 | ||
| Insr | log2FC | −0.157 | 0.208 | −0.204 | 0.161 | −0.604 | −0.724 | −0.114 | −0.235 | |
| FDR | 0.389 | 0.147 | 0.242 | 1.000 | 0.000 | 0.000 | 0.740 | 0.080 | ||
| Igfbp2 | log2FC | −1.494 | −0.943 | −0.678 | −0.127 | |||||
| FDR | 0.001 | 0.032 | 0.220 | 1.000 | ||||||
| Igfbp3 | log2FC | −0.204 | −0.106 | −0.133 | −0.035 | 0.364 | 0.495 | −0.394 | −0.263 | |
| FDR | 0.318 | 0.570 | 0.547 | 1.000 | 0.053 | 0.006 | 0.302 | 0.295 | ||
| Igfbp4 | log2FC | 0.368 | −0.238 | 0.527 | −0.079 | 0.211 | 0.459 | 0.066 | 0.314 | |
| FDR | 0.220 | 0.378 | 0.080 | 1.000 | 0.094 | 0.000 | 0.899 | 0.030 | ||
| Igfbp5 | log2FC | −0.194 | 0.000 | −0.175 | 0.019 | −0.215 | −0.107 | −0.326 | −0.218 | |
| FDR | 0.305 | 1.000 | 0.367 | 1.000 | 0.273 | 0.589 | 0.419 | 0.399 | ||
| Igfbp6 | log2FC | −0.565 | −0.525 | −0.149 | −0.109 | 0.174 | 0.582 | −0.041 | 0.367 | |
| FDR | 0.395 | 0.344 | 0.859 | 1.000 | 0.289 | 0.000 | 0.955 | 0.043 | ||
| Igfbp7 | log2FC | −0.153 | −0.577 | 0.233 | −0.191 | 0.179 | 0.307 | 0.015 | 0.142 | |
| FDR | 0.479 | 0.000 | 0.236 | 1.000 | 0.141 | 0.006 | 0.985 | 0.387 | ||
| Pappa2 | log2FC | −0.759 | −0.566 | −0.090 | 0.103 | −0.233 | −0.341 | 0.560 | 0.453 | |
| FDR | 0.000 | 0.006 | 0.792 | 1.000 | 0.660 | 0.470 | 0.659 | 0.493 | ||
| Stc1 | log2FC | −0.247 | −0.003 | −0.481 | −0.237 | −0.810 | −0.627 | −0.308 | −0.124 | |
| FDR | 0.536 | 0.993 | 0.173 | 1.000 | 0.059 | 0.153 | 0.850 | 0.877 | ||
| Stc2 | log2FC | 0.511 | 0.667 | 0.289 | 0.445 | 0.550 | 0.595 | 0.240 | 0.285 | |
| FDR | 0.256 | 0.064 | 0.566 | 1.000 | 0.088 | 0.049 | 0.850 | 0.525 | ||
| Slc2a4 | log2FC | 0.178 | −0.129 | 0.276 | −0.031 | 0.227 | 0.362 | 0.134 | 0.268 | |
| FDR | 0.732 | 0.751 | 0.530 | 1.000 | 0.033 | 0.000 | 0.654 | 0.033 | ||
| Functional Group | Comparison Parameters | Expression in the Pituitary Gland | Expression in Skeletal Muscle | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | DUhTP vs. DUC | DUC | DUhTP | DUhTP vs. DUC | DUC | DUhTP | ||||
| Sed | Trained | Trained vs. Sed | Sed | Trained | Trained vs. Sed | |||||
| Hormonal control of mTORC activity | Akt1 | log2FC | −0.033 | −0.180 | 0.088 | −0.059 | 0.125 | 0.199 | 0.082 | 0.156 |
| FDR | 0.834 | 0.049 | 0.476 | 1.000 | 0.368 | 0.116 | 0.877 | 0.384 | ||
| Akt2 | log2FC | −0.113 | −0.138 | −0.016 | −0.042 | 0.079 | 0.254 | −0.028 | 0.146 | |
| FDR | 0.395 | 0.190 | 0.929 | 1.000 | 0.496 | 0.010 | 0.954 | 0.292 | ||
| Akt3 | log2FC | −0.005 | 0.227 | −0.070 | 0.162 | −0.668 | −0.467 | −0.076 | 0.124 | |
| FDR | 0.983 | 0.060 | 0.695 | 1.000 | 0.000 | 0.000 | 0.881 | 0.508 | ||
| Gsk3b | log2FC | −0.239 | 0.002 | −0.267 | −0.026 | −0.684 | −0.642 | −0.189 | −0.147 | |
| FDR | 0.157 | 0.991 | 0.133 | 1.000 | 0.000 | 0.000 | 0.506 | 0.383 | ||
| Bmal1 | log2FC | 0.332 | −0.064 | −0.444 | −0.840 | −0.165 | −0.650 | −0.755 | −1.240 | |
| FDR | 0.288 | 0.845 | 0.153 | 0.012 | 0.684 | 0.055 | 0.269 | 0.001 | ||
| Clock | log2FC | −0.093 | 0.153 | −0.350 | −0.105 | −0.539 | −1.026 | −0.509 | −0.997 | |
| FDR | 0.713 | 0.386 | 0.074 | 1.000 | 0.000 | 0.000 | 0.012 | 0.000 | ||
| Mtor | log2FC | −0.106 | 0.159 | −0.207 | 0.058 | −0.330 | 0.010 | −0.142 | 0.198 | |
| FDR | 0.556 | 0.233 | 0.189 | 1.000 | 0.005 | 0.944 | 0.687 | 0.205 | ||
| Pik3ca | log2FC | −0.140 | 0.123 | −0.203 | 0.060 | −0.532 | −0.618 | −0.303 | −0.389 | |
| FDR | 0.378 | 0.355 | 0.180 | 1.000 | 0.000 | 0.000 | 0.111 | 0.003 | ||
| Pik3cb | log2FC | −0.092 | 0.034 | −0.086 | 0.041 | −0.400 | −0.527 | −0.005 | −0.133 | |
| FDR | 0.704 | 0.867 | 0.706 | 1.000 | 0.011 | 0.000 | 0.994 | 0.589 | ||
| Pik3c2a | log2FC | −0.131 | 0.156 | −0.296 | −0.009 | −0.986 | −1.328 | −0.240 | −0.582 | |
| FDR | 0.604 | 0.413 | 0.168 | 1.000 | 0.000 | 0.000 | 0.678 | 0.020 | ||
| Pdpk1 | log2FC | −0.140 | 0.072 | −0.163 | 0.049 | −0.797 | −0.771 | −0.016 | 0.010 | |
| FDR | 0.508 | 0.700 | 0.413 | 1.000 | 0.000 | 0.000 | 0.980 | 0.968 | ||
| Tsc1 | log2FC | 0.019 | 0.279 | −0.163 | 0.097 | −0.557 | −0.447 | 0.008 | 0.119 | |
| FDR | 0.946 | 0.038 | 0.352 | 1.000 | 0.000 | 0.000 | 0.991 | 0.441 | ||
| Tsc2 | log2FC | −0.254 | −0.046 | −0.143 | 0.065 | −0.182 | 0.047 | 0.048 | 0.277 | |
| FDR | 0.008 | 0.688 | 0.223 | 1.000 | 0.091 | 0.691 | 0.919 | 0.025 | ||
| Rheb | log2FC | −0.054 | −0.304 | 0.198 | −0.052 | 0.094 | −0.028 | 0.061 | −0.061 | |
| FDR | 0.710 | 0.000 | 0.077 | 1.000 | 0.388 | 0.805 | 0.881 | 0.701 | ||
| Pten | log2FC | −0.186 | 0.243 | −0.361 | 0.069 | −0.678 | −1.056 | 0.017 | −0.361 | |
| FDR | 0.404 | 0.166 | 0.083 | 1.000 | 0.000 | 0.000 | 0.981 | 0.009 | ||
| Rictor | log2FC | 0.008 | 0.347 | −0.227 | 0.112 | −0.550 | −0.772 | −0.257 | −0.478 | |
| FDR | 0.980 | 0.015 | 0.215 | 1.000 | 0.004 | 0.000 | 0.630 | 0.044 | ||
| Rptor | log2FC | −0.127 | −0.011 | −0.072 | 0.044 | −0.264 | −0.058 | 0.047 | 0.252 | |
| FDR | 0.283 | 0.932 | 0.585 | 1.000 | 0.008 | 0.599 | 0.919 | 0.036 | ||
| Deptor | log2FC | 0.178 | 0.582 | −0.122 | 0.281 | −0.120 | −0.529 | 0.150 | −0.258 | |
| FDR | 0.576 | 0.006 | 0.709 | 1.000 | 0.501 | 0.000 | 0.783 | 0.217 | ||
| Rps6kb1 | log2FC | −0.113 | 0.151 | −0.184 | 0.080 | −0.341 | −0.786 | −0.033 | −0.479 | |
| FDR | 0.583 | 0.325 | 0.308 | 1.000 | 0.013 | 0.000 | 0.962 | 0.002 | ||
| Rps6 | log2FC | −0.088 | −0.451 | 0.375 | 0.011 | 0.368 | 0.529 | 0.048 | 0.209 | |
| FDR | 0.696 | 0.001 | 0.034 | 1.000 | 0.000 | 0.000 | 0.908 | 0.069 | ||
| Mlst8 | log2FC | −0.002 | −0.200 | 0.076 | −0.122 | 0.274 | 0.169 | 0.104 | −0.001 | |
| FDR | 0.994 | 0.090 | 0.645 | 1.000 | 0.020 | 0.154 | 0.808 | 0.996 | ||
| Mapkap1 | log2FC | −0.008 | −0.196 | 0.063 | −0.126 | 0.095 | 0.155 | 0.074 | 0.135 | |
| FDR | 0.976 | 0.096 | 0.714 | 1.000 | 0.334 | 0.080 | 0.823 | 0.259 | ||
| Akt1s1 | log2FC | −0.035 | −0.225 | 0.232 | 0.042 | 0.406 | 0.629 | −0.101 | 0.122 | |
| FDR | 0.900 | 0.145 | 0.215 | 1.000 | 0.000 | 0.000 | 0.813 | 0.472 | ||
| Telo2 | log2FC | −0.230 | −0.352 | −0.009 | −0.131 | 0.043 | −0.054 | 0.088 | −0.010 | |
| FDR | 0.085 | 0.002 | 0.971 | 1.000 | 0.805 | 0.738 | 0.883 | 0.973 | ||
| Eif4ebp1 | log2FC | −0.194 | −0.213 | 0.199 | 0.180 | 0.563 | 0.862 | −0.048 | 0.251 | |
| FDR | 0.651 | 0.506 | 0.615 | 1.000 | 0.000 | 0.000 | 0.944 | 0.186 | ||
| Eif4enif1 | log2FC | −0.004 | 0.086 | −0.083 | 0.007 | −0.219 | −0.099 | −0.065 | 0.056 | |
| FDR | 0.982 | 0.390 | 0.493 | 1.000 | 0.021 | 0.318 | 0.866 | 0.722 | ||
| Foxo3 | log2FC | −0.018 | 0.194 | −0.142 | 0.070 | −0.388 | 0.209 | −0.015 | 0.582 | |
| FDR | 0.952 | 0.195 | 0.460 | 1.000 | 0.002 | 0.093 | 0.987 | 0.000 | ||
| Signaling Pathway Members | Comparison Parameters | Expression in the Pituitary Gland | Expression in Skeletal Muscle | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene ID | DUhTP vs. DUC | DUC | DUhTP | DUhTP vs. DUC | DUC | DUhTP | ||||
| Sed | Trained | Trained vs. Sed | Sed | Trained | Trained vs. Sed | |||||
| Lamtors | Lamtor1 | log2FC | 0.035 | −0.265 | 0.237 | −0.063 | 0.470 | 0.525 | 0.131 | 0.186 |
| FDR | 0.887 | 0.049 | 0.159 | 1.000 | 0.000 | 0.000 | 0.593 | 0.109 | ||
| Lamtor2 | log2FC | −0.097 | −0.640 | 0.427 | −0.116 | 0.377 | 0.331 | −0.029 | −0.076 | |
| FDR | 0.710 | 0.000 | 0.033 | 1.000 | 0.000 | 0.000 | 0.951 | 0.611 | ||
| Lamtor3 | log2FC | 0.086 | −0.049 | 0.022 | −0.112 | 0.191 | 0.238 | −0.077 | −0.030 | |
| FDR | 0.571 | 0.708 | 0.904 | 1.000 | 0.068 | 0.017 | 0.850 | 0.868 | ||
| Lamtor4 | log2FC | 0.090 | −0.395 | 0.416 | −0.069 | 0.490 | 0.486 | 0.014 | 0.010 | |
| FDR | 0.751 | 0.021 | 0.050 | 1.000 | 0.000 | 0.000 | 0.983 | 0.959 | ||
| Lamtor5 | log2FC | 0.019 | −0.097 | 0.199 | 0.084 | 0.446 | 0.399 | 0.069 | 0.023 | |
| FDR | 0.955 | 0.609 | 0.329 | 1.000 | 0.000 | 0.000 | 0.876 | 0.905 | ||
| Sirtuins | Sirt1 | log2FC | −0.247 | 0.060 | −0.258 | 0.049 | −0.554 | −0.899 | 0.040 | −0.305 |
| FDR | 0.216 | 0.771 | 0.207 | 1.000 | 0.000 | 0.000 | 0.956 | 0.128 | ||
| Sirt2 | log2FC | 0.021 | 0.002 | 0.043 | 0.024 | 0.260 | 0.384 | 0.011 | 0.134 | |
| FDR | 0.896 | 0.988 | 0.739 | 1.000 | 0.002 | 0.000 | 0.986 | 0.249 | ||
| Sirt3 | log2FC | 0.205 | 0.074 | 0.166 | 0.034 | 0.480 | 0.454 | 0.106 | 0.080 | |
| FDR | 0.105 | 0.567 | 0.228 | 1.000 | 0.000 | 0.000 | 0.839 | 0.718 | ||
| Sirt4 | log2FC | −0.098 | −0.028 | −0.025 | 0.045 | 0.297 | 0.134 | 0.191 | 0.029 | |
| FDR | 0.567 | 0.858 | 0.904 | 1.000 | 0.028 | 0.333 | 0.591 | 0.905 | ||
| Sirt5 | log2FC | 0.064 | 0.239 | 0.044 | 0.219 | 0.570 | 1.177 | −0.093 | 0.514 | |
| FDR | 0.836 | 0.196 | 0.884 | 1.000 | 0.001 | 0.000 | 0.909 | 0.012 | ||
| Sirt6 | log2FC | 0.041 | −0.136 | 0.084 | −0.092 | 0.337 | 0.285 | 0.035 | −0.017 | |
| FDR | 0.876 | 0.395 | 0.692 | 1.000 | 0.041 | 0.078 | 0.968 | 0.959 | ||
| Sirt7 | log2FC | −0.227 | −0.339 | 0.097 | −0.016 | −0.047 | 0.104 | 0.025 | 0.177 | |
| FDR | 0.090 | 0.002 | 0.549 | 1.000 | 0.810 | 0.541 | 0.978 | 0.437 | ||
| AMPK subunits | Prkaa1 | log2FC | −0.046 | 0.133 | −0.202 | −0.024 | −0.680 | −0.844 | −0.105 | −0.269 |
| FDR | 0.842 | 0.364 | 0.224 | 1.000 | 0.000 | 0.000 | 0.818 | 0.099 | ||
| Prkaa2 | log2FC | 0.051 | 0.351 | −0.203 | 0.096 | −0.672 | −0.885 | −0.084 | −0.297 | |
| FDR | 0.872 | 0.046 | 0.372 | 1.000 | 0.000 | 0.000 | 0.881 | 0.080 | ||
| Prkab1 | log2FC | 0.066 | 0.028 | 0.083 | 0.045 | 0.355 | 0.315 | −0.037 | −0.077 | |
| FDR | 0.720 | 0.852 | 0.610 | 1.000 | 0.010 | 0.021 | 0.957 | 0.736 | ||
| Prkab2 | log2FC | 0.077 | 0.277 | −0.121 | 0.079 | −0.111 | −0.101 | 0.031 | 0.041 | |
| FDR | 0.762 | 0.077 | 0.575 | 1.000 | 0.577 | 0.592 | 0.974 | 0.897 | ||
| Prkag1 | log2FC | −0.091 | −0.286 | 0.186 | −0.009 | 0.437 | 0.527 | 0.026 | 0.115 | |
| FDR | 0.569 | 0.007 | 0.177 | 1.000 | 0.000 | 0.000 | 0.960 | 0.434 | ||
| Prkag2 | log2FC | 0.029 | 0.099 | −0.186 | −0.115 | 0.244 | 0.217 | 0.077 | 0.049 | |
| FDR | 0.883 | 0.405 | 0.167 | 1.000 | 0.150 | 0.188 | 0.917 | 0.864 | ||
| Prkag3 | log2FC | 0.297 | −0.043 | 0.330 | −0.010 | |||||
| FDR | 0.163 | 0.856 | 0.498 | 0.981 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brenmoehl, J.; Walz, C.; Caffier, C.; Brosig, E.; Walz, M.; Ohde, D.; Trakooljul, N.; Langhammer, M.; Ponsuksili, S.; Wimmers, K.; et al. Central Suppression of the GH/IGF Axis and Abrogation of Exercise-Related mTORC1/2 Activation in the Muscle of Phenotype-Selected Male Marathon Mice (DUhTP). Cells 2021, 10, 3418. https://doi.org/10.3390/cells10123418
Brenmoehl J, Walz C, Caffier C, Brosig E, Walz M, Ohde D, Trakooljul N, Langhammer M, Ponsuksili S, Wimmers K, et al. Central Suppression of the GH/IGF Axis and Abrogation of Exercise-Related mTORC1/2 Activation in the Muscle of Phenotype-Selected Male Marathon Mice (DUhTP). Cells. 2021; 10(12):3418. https://doi.org/10.3390/cells10123418
Chicago/Turabian StyleBrenmoehl, Julia, Christina Walz, Caroline Caffier, Elli Brosig, Michael Walz, Daniela Ohde, Nares Trakooljul, Martina Langhammer, Siriluck Ponsuksili, Klaus Wimmers, and et al. 2021. "Central Suppression of the GH/IGF Axis and Abrogation of Exercise-Related mTORC1/2 Activation in the Muscle of Phenotype-Selected Male Marathon Mice (DUhTP)" Cells 10, no. 12: 3418. https://doi.org/10.3390/cells10123418
APA StyleBrenmoehl, J., Walz, C., Caffier, C., Brosig, E., Walz, M., Ohde, D., Trakooljul, N., Langhammer, M., Ponsuksili, S., Wimmers, K., Zettl, U. K., & Hoeflich, A. (2021). Central Suppression of the GH/IGF Axis and Abrogation of Exercise-Related mTORC1/2 Activation in the Muscle of Phenotype-Selected Male Marathon Mice (DUhTP). Cells, 10(12), 3418. https://doi.org/10.3390/cells10123418









