Impaired Skeletal Muscle Development and Regeneration in Transglutaminase 2 Knockout Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Experimental Animals
2.3. Cardiotoxin-Induced Muscle Injury Model
2.4. Histological Examination of Various Muscles
2.5. Immunofluorescent Staining
2.6. In Vivo Assessment of Muscle Force
2.7. Ex Vivo Assessment of Muscle Force
2.8. Isolation of Muscle-Derived CD45+ Leukocytes
2.9. Gene Expression Analysis
2.10. Quantification of Intramuscular Immune and Satellite Cells by Flow Cytometry
2.11. C2C12 Cell Culture and Differentiation
2.12. In Vitro Phagocytosis Assay
2.13. Western Blot Analysis
2.14. Quantification of Necrotic Areas
2.15. Statistical Analysis
3. Results
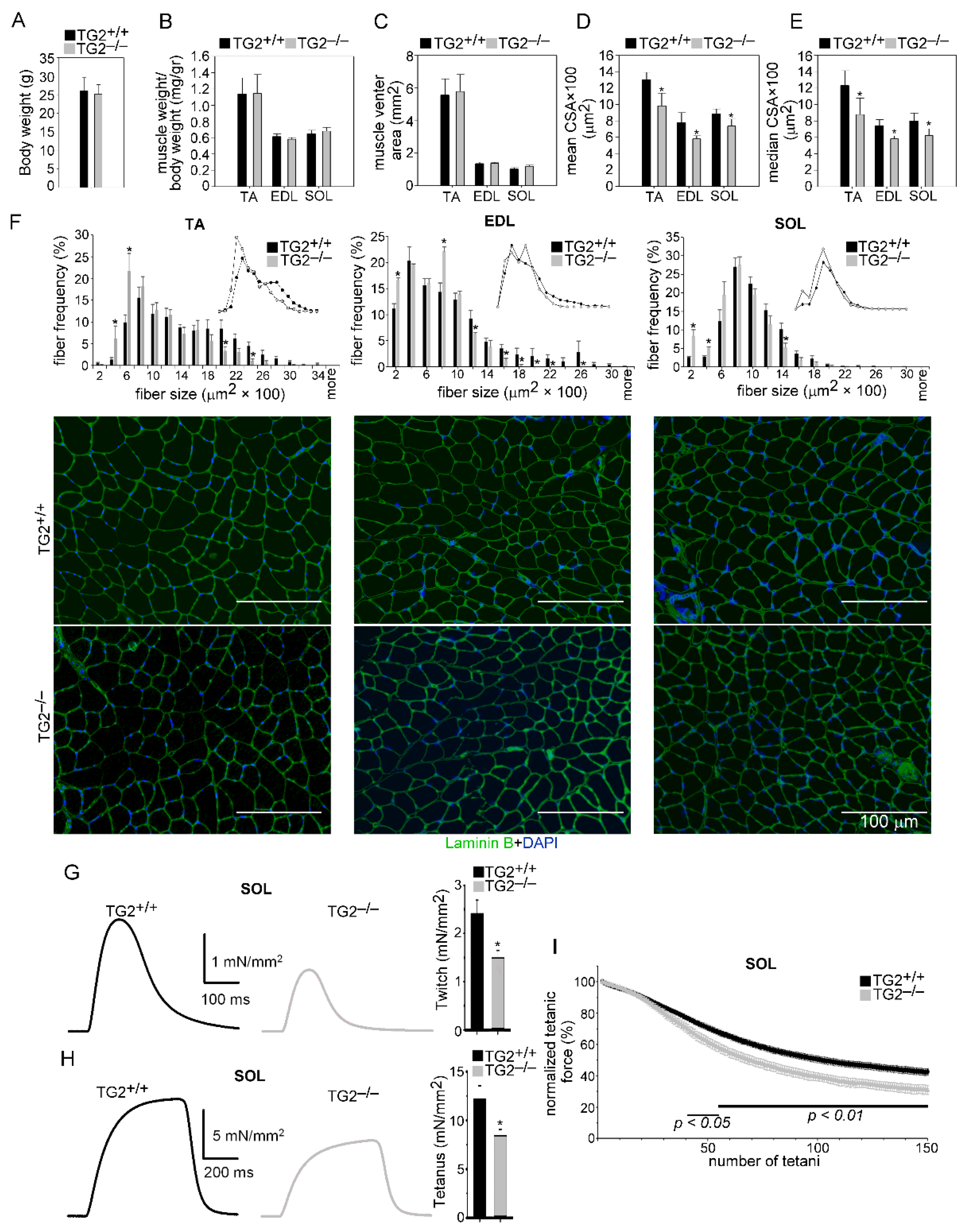
3.1. TG2 Deficiency Impairs Skeletal Muscle Differentiation and Function
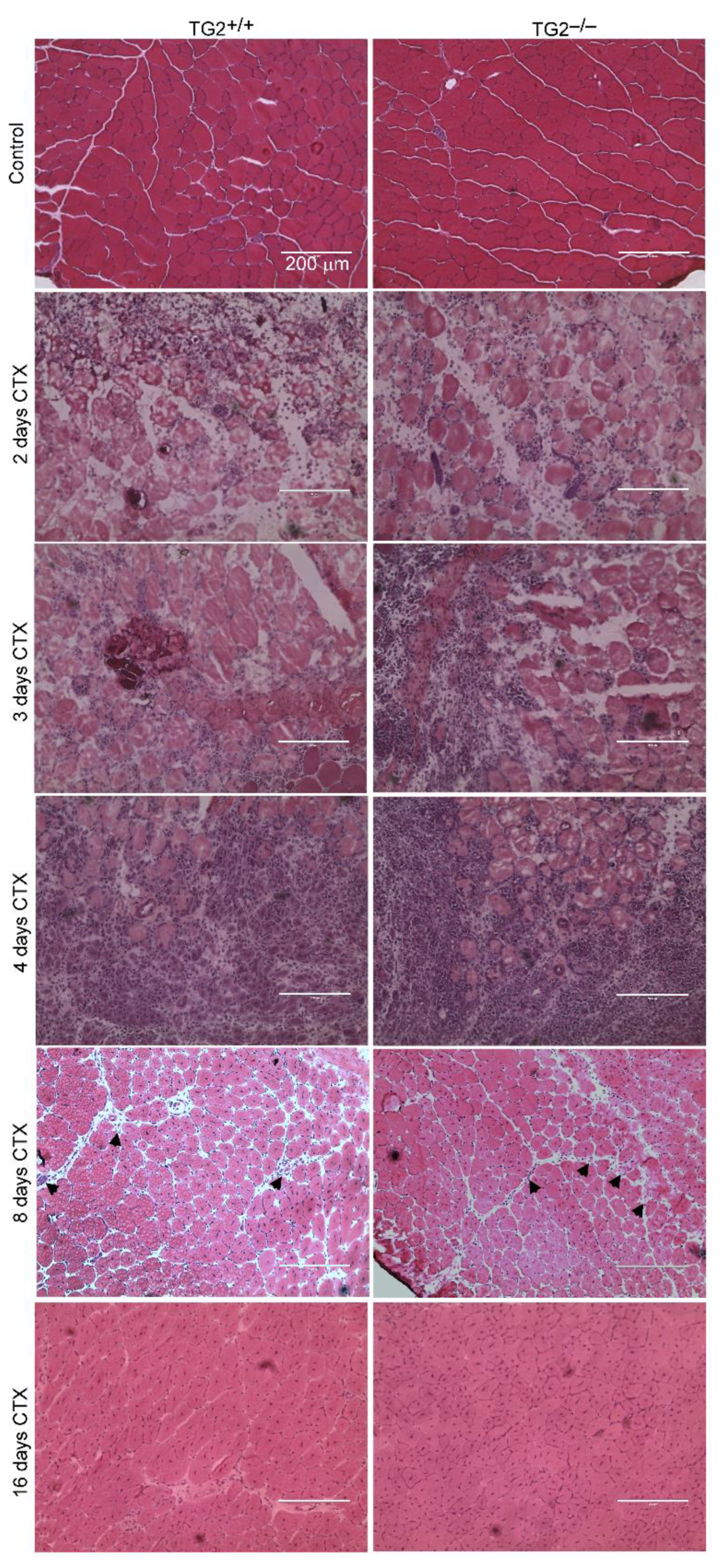
3.2. Normal Histological Appearance of Regenerating TA Muscles in the Absence of TG2
3.3. TG2 Deficiency Impairs TA Muscle Regeneration
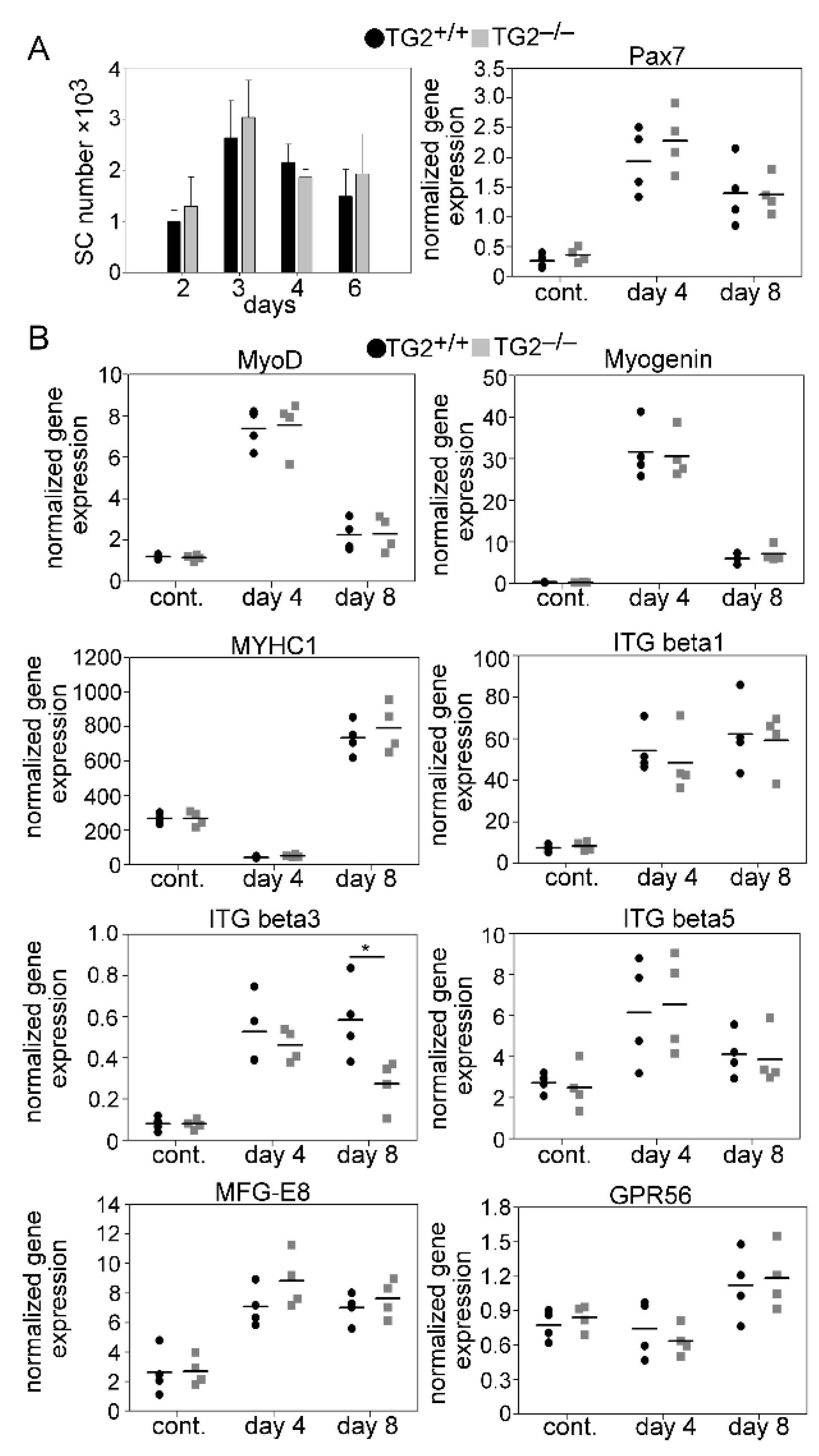
3.4. Normal Proliferation and Differentiation of Satellite Cells (SCs) after Injury in the Absence of TG2
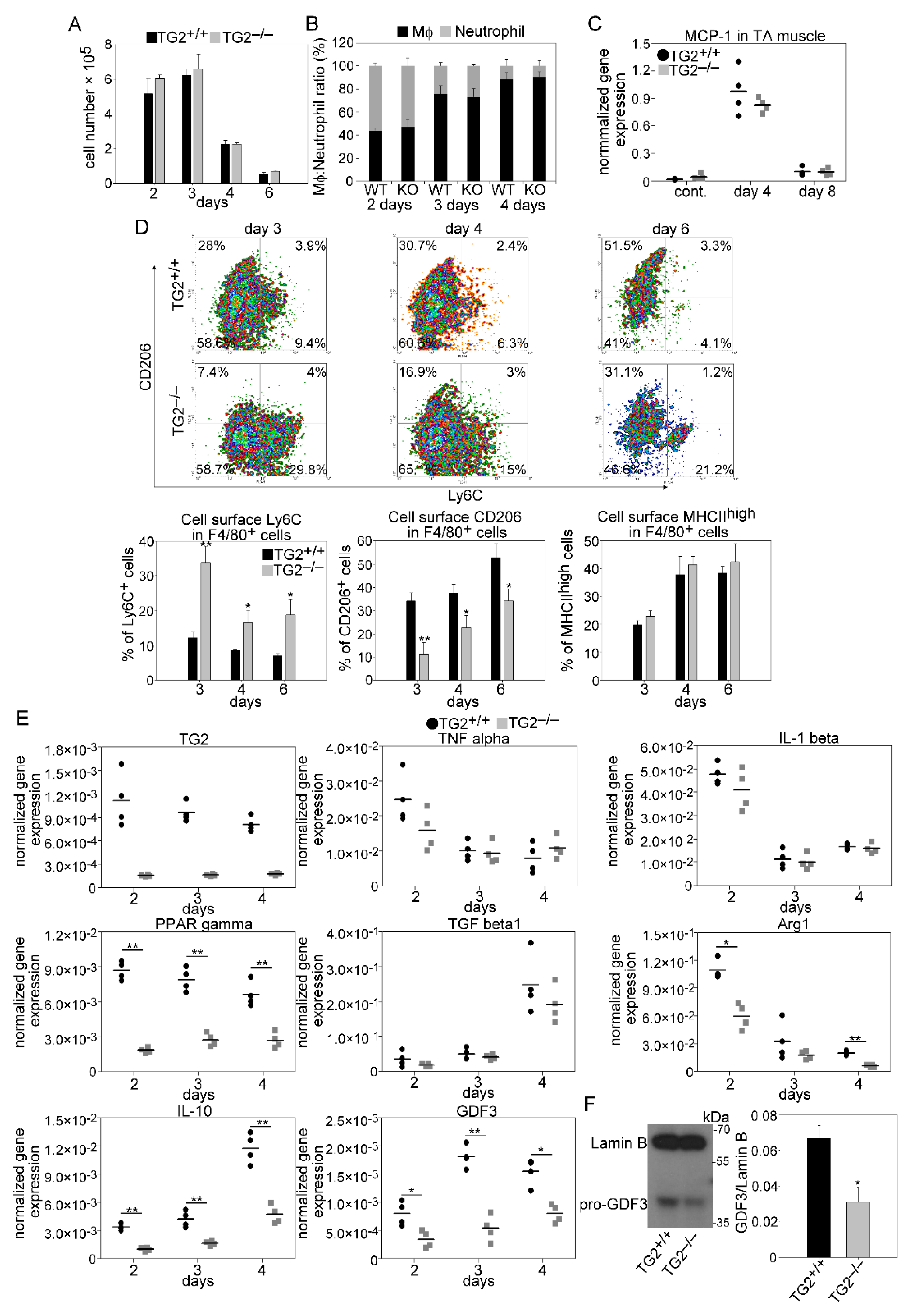
3.5. Unaltered Recruitment of Mϕs and Neutrophils after Injury in the Absence of TG2
3.6. Impaired M1/M2 Phenotypic Switch in TG2 Null Macrophages during the Muscle Regeneration Process
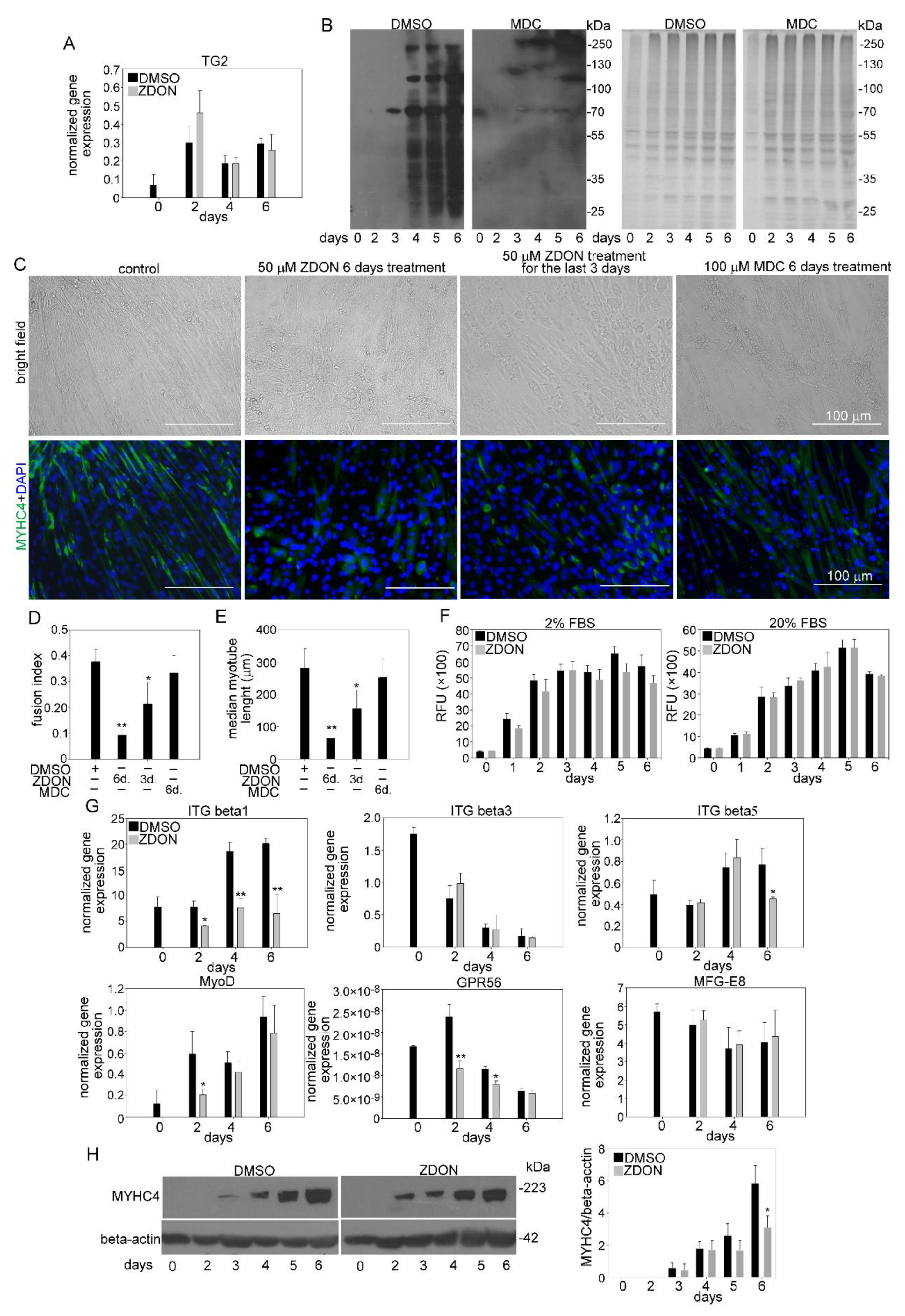
3.7. TG2 Promotes Myoblast Fusion without Using Its Crosslinking Activity
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baghdadi, M.B.; Tajbakhsh, S. Regulation and phylogeny of skeletal muscle regeneration. Dev. Biol. 2018, 433, 200–209. [Google Scholar] [CrossRef]
- Yang, W.; Hu, P. Skeletal muscle regeneration is modulated by inflammation. J. Orthop. Translat. 2018, 13, 25–32. [Google Scholar] [CrossRef]
- Bazgir, B.; Fathi, R.; Rezazadeh Valojerdi, M.; Mozdziak, P.; Asgari, A. Satellite Cells Contribution to Exercise Mediated Muscle Hypertrophy and Repair. Cell J. 2017, 18, 473–484. [Google Scholar]
- Chazaud, B. Inflammation and skeletal muscle regeneration: Leave it to the macrophages. Trends Immunol. 2020, 6, 481–492. [Google Scholar] [CrossRef]
- Deng, B.; Wehling-Henricks, M.; Villalta, S.A.; Wang, Y.; Tidball, J.G. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J. Immunol. 2012, 189, 3669–3680. [Google Scholar] [CrossRef]
- Giannakis, N.; Sansbury, B.E.; Patsalos, A.; Hays, T.T.; Riley, C.O.; Han, X.; Spite, M.; Nagy, L. Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration. Nat. Immunol. 2019, 20, 626–636. [Google Scholar] [CrossRef]
- Varga, T.; Mounier, R.; Patsalos, A.; Gogolák, P.; Peloquin, M.; Horvath, A.; Pap, A.; Daniel, B.; Nagy, G.; Pintye, E.; et al. Macrophage PPARγ, a Lipid Activated Transcription Factor Controls the Growth Factor GDF3 and Skeletal Muscle Regeneration. Immunity 2016, 45, 1038–1051. [Google Scholar] [CrossRef]
- Melendez, J.; Sieiro, D.; Salgado, D.; Morin, V.; Dejardin, M.J.; Zhou, C.; Mullen, A.C.; Marcelle, C. TGFβ signalling acts as a molecular brake of myoblast fusion. Nat. Commun. 2021, 12, 749. [Google Scholar] [CrossRef]
- Gundemir, S.; Colak, G.; Tucholski, J.; Johnson, G.V. Transglutaminase 2: A molecular Swiss army knife. Biochim. Biophys. Acta 2012, 1823, 406–419. [Google Scholar] [CrossRef]
- Xu, L.; Begum, S.; Hearn, J.D.; Hynes, R.O. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2006, 103, 9023–9028. [Google Scholar] [CrossRef]
- Akimov, S.S.; Krylov, D.; Fleischman, L.F.; Belkin, A.M. Tissue Transglutaminase Is an Integrin-Binding Adhesion Coreceptor for Fibronectin. J. Cell Biol. 2000, 148, 825–838. [Google Scholar] [CrossRef]
- Herman, J.F.; Mangala, L.S.; Mehta, K. Implications of increased tissue transglutaminase (TG2) expression in drug-resistant breast cancer (MCF-7) cells. Oncogene 2006, 25, 3049–3058. [Google Scholar] [CrossRef]
- Tóth, B.; Garabuczi, E.; Sarang, Z.; Vereb, G.; Vámosi, G.; Aeschlimann, D.; Blaskó, B.; Bécsi, B.; Erdõdi, F.; Lacy-Hulbert, A.; et al. Transglutaminase 2 is needed for the formation of an efficient phagocyte portal in macrophages engulfing apoptotic cells. J. Immunol. 2009, 182, 2084–2092. [Google Scholar] [CrossRef]
- Fésüs, L.; Szondy, Z. Transglutaminase 2 in the balance of cell death and survival. FEBS Lett. 2005, 579, 3297–3302. [Google Scholar] [CrossRef]
- Johnson, T.S.; Griffin, M.; Thomas, G.L.; Skill, J.; Cox, A.; Yang, B.; Nicholas, B.; Birckbichler, P.J.; Muchaneta-Kubara, C.; El Nahas, A.M. The role of transglutaminase in the rat subtotal nephrectomy model of renal fibrosis. J. Clin. Investig. 1997, 99, 2950–2960. [Google Scholar] [CrossRef]
- Szondy, Z.; Sarang, Z.; Molnar, P.; Nemeth, T.; Piacentini, M.; Mastroberardino, P.G.; Falasca, L.; Aeschlimann, D.; Kovacs, J.; Kiss, I.; et al. Transglutaminase 2-/- mice reveal a phagocytosis-associated crosstalk between macrophages and apoptotic cells. Proc. Natl. Acad. Sci. USA 2003, 100, 7812–7817. [Google Scholar] [CrossRef]
- Haroon, Z.A.; Hettasch, J.M.; Lai, T.S.; Dewhirst, M.W.; Greenberg, C.S. Tissue transglutaminase is expressed, active, and directly involved in rat dermal wound healing and angiogenesis. FASEB J. 1999, 13, 1787–1795. [Google Scholar] [CrossRef]
- Lee, J.; Kim, Y.S.; Choi, D.H.; Bang, M.S.; Han, T.R.; Joh, T.H.; Kim, S.Y. Transglutaminase 2 induces nuclear factor-kappaB activation via a novel pathway in BV-2 microglia. J. Biol. Chem. 2004, 279, 53725–53735. [Google Scholar] [CrossRef]
- Lee, S.K.; Chi, J.G.; Park, S.C.; Chung, S.I. Transient expression of transglutaminase C during prenatal development of human muscles. J. Histochem. Cytochem. 2000, 48, 1565–1574. [Google Scholar] [CrossRef]
- Hand, D.; Campoy, F.J.; Clark, S.; Fisher, A.; Haynes, L.W. Activity and distribution of tissue transglutaminase in association with nerve-muscle synapses. J. Neurochem. 1993, 61, 1064–1072. [Google Scholar] [CrossRef]
- Bersten, A.M.; Ahkong, Q.F.; Hallinan, T.; Nelson, S.J.; Lucy, J.A. Inhibition of the formation of myotubes in vitro by inhibitors of transglutaminase. Biochim. Biophys. Acta. 1983, 762, 429–436. [Google Scholar] [CrossRef]
- Kang, S.J.; Shin, K.S.; Song, W.K.; Ha, D.B.; Chung, C.H.; Kang, M.S. Involvement of Transglutaminase in Myofibril Assembly of Chick Embryonic Myoblasts in Culture. J. Cell Biol. 1995, 130, 1127–1136. [Google Scholar] [CrossRef]
- Choi, Y.C.; Kim, T.S.; Kim, S.Y. Increase in transglutaminase 2 in idiopathic inflammatory myopathies. Eur. Neurol. 2004, 51, 10–14. [Google Scholar] [CrossRef]
- De Laurenzi, V.; Melino, G. Gene disruption of tissue transglutaminase. Mol. Cell Biol. 2001, 21, 148–155. [Google Scholar] [CrossRef]
- Al-Zaeed, N.; Budai, Z.; Szondy, Z.; Sarang, Z. TAM kinase signaling is indispensable for proper skeletal muscle regeneration in mice. Cell Death Dis. 2021, 12, 611. [Google Scholar] [CrossRef]
- Sztretye, M.; Singlár, Z.; Szabó, L.; Angya, Á.; Balogh, N.; Vakilzadeh, F.; Szentesi, P.; Dienes, B.; Csernoch, L. Improved Tetanic Force and Mitochondrial Calcium Homeostasis by Astaxanthin Treatment in Mouse Skeletal Muscle. Antioxidants 2020, 9, 98. [Google Scholar] [CrossRef]
- Budai, Z.; Újlaki-Nagy, L.; Kis, N.G.; Antal, M.; Bankó, C.; Szondy, Z.; Sarang, Z. Macrophages engulf apoptotic and primary necrotic thymocytes through similar phosphatidylserine-dependent mechanisms. FEBS Open Bio 2019, 9, 446–456. [Google Scholar] [CrossRef]
- Zhang, J.; Qu, C.; Li, T.; Cui, W.; Wang, X.; Du, J. Phagocytosis mediated by scavenger receptor class BI promotes macrophage transition during skeletal muscle regeneration. J. Biol. Chem. 2019, 294, 15672–15685. [Google Scholar] [CrossRef]
- Rose, D.M.; Sydlaske, A.D.; Agha-Babakhani, A.; Johnson, K.; Terkeltaub, R. Transglutaminase 2 limits murine peritoneal acute gout-like inflammation by regulating macrophage clearance of apoptotic neutrophils. Arthritis. Rheum. 2006, 54, 3363–3371. [Google Scholar] [CrossRef]
- Chikazawa, M.; Shimizu, M.; Yamauchi, Y.; Sato, R. Bridging molecules are secreted from the skeletal muscle and potentially regulate muscle differentiation. Biochem. Biophys. Res. Commun. 2020, 522, 113–120. [Google Scholar] [CrossRef]
- Li, H.; Xu, W.; Ma, Y.; Zhou, S.; Xiao, R. Milk fat globule membrane protein promotes C2C12 cell proliferation through the PI3K/Akt signaling pathway. Int. J. Biol. Macromol. 2018, 114, 1305–1314. [Google Scholar] [CrossRef]
- Wu, M.P.; Doyle, J.R.; Barry, B.; Beauvais, A.; Rozkalne, A.; Piao, X.; Lawlor, M.W.; Kopin, A.S.; Walsh, C.A.; Gussoni, E. G-protein coupled receptor 56 promotes myoblast fusion through serum response factor- and nuclear factor of activated T-cell-mediated signalling but is not essential for muscle development in vivo. FEBS J. 2013. 280, 6097–6113. [CrossRef]
- Schwander, M.; Leu, M.; Stumm, M.; Dorchies, O.M.; Ruegg, U.T.; Schittny, J.; Muller, U. Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev. Cell 2003, 4, 673–685. [Google Scholar] [CrossRef]
- Sinanan, A.C.; Machell, J.R.; Wynne-Hughes, G.T.; Hunt, N.P.; Lewis, M.P. Alpha v beta 3 and alpha v beta 5 integrins and their role in muscle precursor cell adhesion. Biol. Cell 2008, 100, 465–477. [Google Scholar] [CrossRef]
- Martinez, C.O.; McHale, M.J.; Wells, J.T.; Ochoa, O.; Michalek, J.E.; McManus, L.M.; Shireman, P.K. Regulation of skeletal muscle regeneration by CCR2-activating chemokines is directly related to macrophage recruitment. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 299, R832–R842. [Google Scholar] [CrossRef]
- Yaffe, D.; Saxel, O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977, 270, 725–727. [Google Scholar] [CrossRef]
- Hochreiter-Hufford, A.E.; Lee, C.S.; Kinchen, J.M.; Sokolowski, J.D.; Arandjelovic, S.; Call, J.A.; Klibanov, A.L.; Yan, Z.; Mandell, J.W.; Ravichandran, K.S. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 2013, 497, 263–267. [Google Scholar] [CrossRef]
- McConoughey, S.J.; Basso, M.; Niatsetskaya, Z.V.; Sleiman, S.F.; Smirnova, N.A.; Langley, B.C.; Mahishi, L.; Cooper, A.J.; Antonyak, M.A.; Cerione, R.A.; et al. Inhibition of transglutaminase 2 mitigates transcriptional dysregulation in models of Huntington disease. EMBO Mol. Med. 2010, 2, 349–370. [Google Scholar] [CrossRef]
- Keillor, J.W.; Apperley, K.Y.P.; Akbar, A. Inhibitors of tissue transglutaminase. Trends Pharmacol. Sci. 2015, 36, 32–40. [Google Scholar] [CrossRef]
- Ruggiero, L.; Sarang, Z.; Szondy, Z.; Finnemann, S.C.; Szondy, Z.; Finnemann, S.C. αvβ5 integrin-dependent diurnal phagocytosis of shed photoreceptor outer segments by RPE cells is independent of the integrin coreceptor transglutaminase-2. Adv. Exp. Med. Biol. 2012, 723, 731–737. [Google Scholar]
- Feinberg, H.; Jégouzo, S.A.F.; Lasanajak, Y.; Smith, D.F.; Drickamer, K.; Weis, W.I.; Taylor, M.E. Structural analysis of carbohydrate binding by the macrophage mannose receptor CD206. J. Biol. Chem. 2021, 296, 100368. [Google Scholar] [CrossRef]
- Garg, M.; Johri, S.; Sagar, S.; Mundhada, A.; Agrawal, A.; Prabir, R.; Chakraborty, K. Cardiolipin-mediated PPARγ S112 phosphorylation impairs IL-10 production and inflammation resolution during bacterial pneumonia. Cell Rep. 2021, 34, 108736. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, C.; Yao, Q.; Qian, L.; Liu, J.; Xie, X.; Ma, W.; Nie, X.; Lai, B.; Xiao, L.; et al. Procyanidin B2 Activates PPARγ to Induce M2 Polarization in Mouse Macrophages. Front. Immunol. 2019, 10, 1895. [Google Scholar] [CrossRef]
- Rochlin, K.; Yu, S.; Roy, S.; Baylies, M.K. Myoblast fusion: When it takes more to make one. Dev. Biol. 2010, 341, 66–83. [Google Scholar] [CrossRef]
- Doherty, K.R.; Cave, A.; Davis, D.B.; Delmonte, A.J.; Posey, A.; Earley, J.U.; Hadhazy, M.; McNally, A.M. Normal myoblast fusion requires myoferlin. Development 2005, 132, 5565–5575. [Google Scholar] [CrossRef]
- Park, S.Y.; Yun, Y.; Lim, J.S.; Kim, M.J.; Kim, S.Y.; Kim, J.E.; Kim, I.S. Stabilin-2 modulates the efficiency of myoblast fusion during myogenic differentiation and muscle regeneration. Nat. Commun. 2016, 7, 10871. [Google Scholar] [CrossRef]
- Rozo, M.; Li, L.; Fan, C.M. Targeting β1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nat. Med. 2016, 22, 889–896. [Google Scholar] [CrossRef]
- Liu, H.; Niu, A.; Chen, S.E.; Li, Y.P. Beta3-integrin mediates satellite cell differentiation in regenerating mouse muscle. FASEB J. 2011, 25, 1914–1921. [Google Scholar] [CrossRef]
- Blaschuk, K.L.; Guérin, C.; Holland, P.C. Myoblast alpha v beta3 integrin levels are controlled by transcriptional regulation of expression of the beta3 subunit and down-regulation of beta3 subunit expression is required for skeletal muscle cell differentiation. Dev. Biol. 1997, 184, 266–277. [Google Scholar] [CrossRef][Green Version]
- Kitakaze, T.; Yoshikawa, M.; Kobayashi, Y.; Kimura, N.; Goshima, N.; Ishikawa, T.; Ogata, Y.; Yamashita, Y.; Ashida, H.; Harada, N.; et al. Extracellular transglutaminase 2 induces myotube hypertrophy through G protein-coupled receptor 56. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118563. [Google Scholar] [CrossRef]
- Augusto, V.; Padovani, C.R.; Campos, G.E.R. Skeletal muscle fiber types in C57BL6J mice. J. Morphol. Sci. 2004, 21, 89–94. [Google Scholar]
- Westerblad, H.; Bruton, J.D.; Katz, A. Skeletal muscle: Energy metabolism, fiber types, fatigue and adaptability. Exp. Cell Res. 2010, 316, 3093–3099. [Google Scholar] [CrossRef]
- Szondy, Z.; Mastroberardino, P.G.; Váradi, J.; Farrace, M.G.; Nagy, N.; Bak, I.; Viti, I.; Wieckowski, M.R.; Melino, G.; Rizzuto, R.; et al. Tissue transglutaminase (TG2) protects cardiomyocytes against ischemia/reperfusion injury by regulating ATP synthesis. Cell Death Differ. 2006, 13, 1827–1829. [Google Scholar] [CrossRef]
- Battaglia, G.; Farrace, M.G.; Mastroberardino, P.G.; Viti, I.; Fimia, G.M.; van Beeumen, J.; Devreese, B.; Melino, G.; Molinaro, G.; Busceti, C.L.; et al. Transglutaminase 2 ablation leads to defective function of mitochondrial respiratory complex I affecting neuronal vulnerability in experimental models of extrapyramidal disorders. J. Neurochem. 2007, 100, 36–49. [Google Scholar] [CrossRef]


| TWITCH | TETANUS | |||
|---|---|---|---|---|
| TG2+/+ | TG2−/− | TG2+/+ | TG2−/− | |
| Number of animals | 8 | 7 | 8 | 7 |
| Number of muscles | 14 | 11 | 14 | 11 |
| Muscle weight (mg) | 17.41 ± 0.91 | 16.61 ± 0.55 | - | - |
| Force (mN/mm2) | 1.75 ± 0.24 | 1.47 ± 0.12 | 8.37 ± 1.31 | 7.61 ± 0.63 |
| TTP (ms) | 32.7 ± 1.0 | 31.0 ± 0.6 | 186.9 ± 7.4 | 186.7 ± 5.0 |
| HRT (ms) | 26.5 ± 1.4 | 24.5 ± 1.4 | 74.4 ± 8.2 | 75.7 ± 5.1 |
| Duration (ms) | 205.6 ± 33.3 | 181.2 ± 25.8 | 344.6 ± 8.8 | 343.0 ± 5.2 |
| Muscle venter area (mm2) | 1.36 ± 0.08 | 1.37 ± 0.06 | - | - |
| Fatigue at 50 (%) | - | - | 33.8 ± 1.8 | 37.2 ± 2.3 |
| Fatigue at 100 (%) | - | - | 63.7 ± 1.8 | 63.6 ± 2.1 |
| Fatigue at 150 (%) | - | - | 72.9 ± 1.8 | 76.4 ± 1.8 |
| TWITCH | TETANUS | |||
|---|---|---|---|---|
| TG2+/+ | TG2−/− | TG2+/+ | TG2−/− | |
| Number of animals | 8 | 7 | 8 | 7 |
| Number of muscles | 13 | 11 | 13 | 11 |
| Muscle weight (mg) | 18.32 ± 1.24 | 19.48 ± 1.31 | - | - |
| Force (mN/mm2) | 2.41 ± 0.28 | 1.49 ± 0.16 * | 12.18 ± 1.36 | 8.42 ± 0.66 * |
| TTP (ms) | 77.9 ± 3.8 | 77.1 ± 1.6 | 520.4 ± 1.4 | 522.9 ± 1.1 |
| HRT (ms) | 73.5 ± 5.6 | 72.4 ± 5.1 | 106.9 ± 3.8 | 105.7 ± 3.4 |
| Duration (ms) | 314.2 ± 24.4 | 301.4 ± 17.9 | 766.4 ± 10.0 | 771.5 ± 18.1 |
| Muscle venter area (mm2) | 1.02 ± 0.09 | 1.18 ± 0.08 | - | - |
| Fatigue at 50 (%) | - | - | 29.3 ± 0.5 | 38.4 ± 3.0 * |
| Fatigue at 100 (%) | - | - | 49.6 ± 0.6 | 60.7 ± 2.5 ** |
| Fatigue at 150 (%) | - | - | 57.5 ± 0.6 | 69.1 ± 2.9 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budai, Z.; Al-Zaeed, N.; Szentesi, P.; Halász, H.; Csernoch, L.; Szondy, Z.; Sarang, Z. Impaired Skeletal Muscle Development and Regeneration in Transglutaminase 2 Knockout Mice. Cells 2021, 10, 3089. https://doi.org/10.3390/cells10113089
Budai Z, Al-Zaeed N, Szentesi P, Halász H, Csernoch L, Szondy Z, Sarang Z. Impaired Skeletal Muscle Development and Regeneration in Transglutaminase 2 Knockout Mice. Cells. 2021; 10(11):3089. https://doi.org/10.3390/cells10113089
Chicago/Turabian StyleBudai, Zsófia, Nour Al-Zaeed, Péter Szentesi, Hajnalka Halász, László Csernoch, Zsuzsa Szondy, and Zsolt Sarang. 2021. "Impaired Skeletal Muscle Development and Regeneration in Transglutaminase 2 Knockout Mice" Cells 10, no. 11: 3089. https://doi.org/10.3390/cells10113089
APA StyleBudai, Z., Al-Zaeed, N., Szentesi, P., Halász, H., Csernoch, L., Szondy, Z., & Sarang, Z. (2021). Impaired Skeletal Muscle Development and Regeneration in Transglutaminase 2 Knockout Mice. Cells, 10(11), 3089. https://doi.org/10.3390/cells10113089







