Drosophila Dendritic Arborisation Neurons: Fantastic Actin Dynamics and Where to Find Them
Abstract
:1. Introduction
2. The da Neurons of the Drosophila Larva
3. Dendrite Differentiation: Noninvasive Long Term In vivo Imaging Identifies Distinct Phases
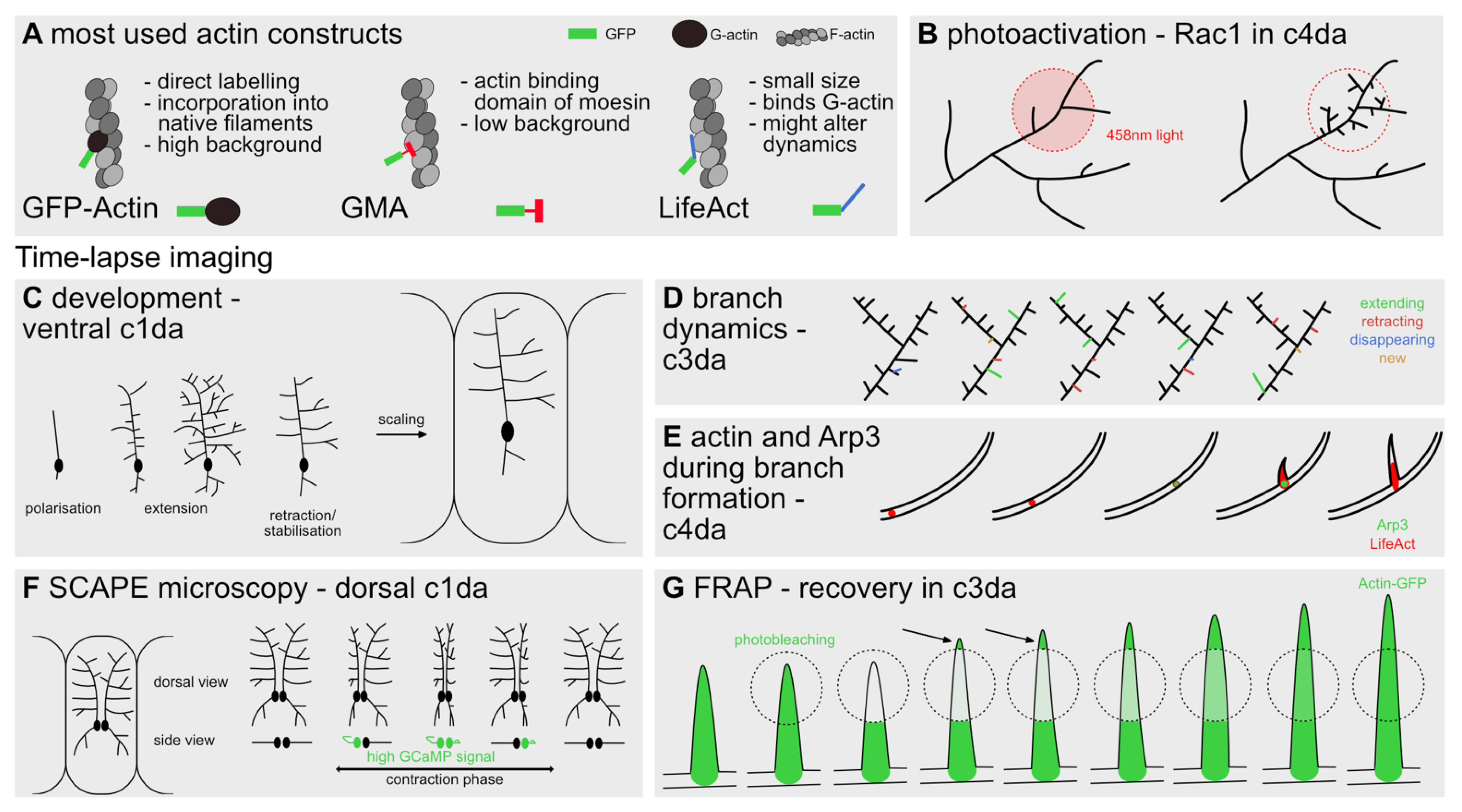
4. Tools to Visualize and Manipulate Actin Dynamics
5. Dendrite Branching
6. Dendrite Extension and Stabilization
7. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jan, Y.-N.; Jan, L.Y. Branching out: Mechanisms of dendritic arborization. Nat. Rev. Neurosci. 2010, 11, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Jan, Y.N.; Jan, L.Y. Dendrites. Genes Dev. 2001, 15, 2627–2641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirazi, P.; Papoutsi, A. Illuminating dendritic function with computational models. Nat. Rev. Neurosci. 2020, 21, 303–321. [Google Scholar] [CrossRef] [PubMed]
- Sidiropoulou, K.; Pissadaki, E.K.; Poirazi, P. Inside the brain of a neuron. EMBO Rep. 2006, 7, 886–892. [Google Scholar] [CrossRef]
- Inberg, S.; Meledin, A.; Kravtsov, V.; Iosilevskii, Y.; Oren-Suissa, M.; Podbilewicz, B. Lessons from Worm Dendritic Patterning. Annu. Rev. Neurosci. 2019, 42, 365–383. [Google Scholar] [CrossRef] [Green Version]
- Albeg, A.; Smith, C.J.; Chatzigeorgiou, M.; Feitelson, D.G.; Hall, D.H.; Schafer, W.R.; Miller, D.M.; Treinin, M. C. elegans multi-dendritic sensory neurons: Morphology and function. Mol. Cell. Neurosci. 2011, 46, 308–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujishima, K.; Kawabata Galbraith, K.; Kengaku, M. Dendritic Self-Avoidance and Morphological Development of Cerebellar Purkinje Cells. Cerebellum 2018, 17, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Grueber, W.B.; Jan, L.Y.; Jan, Y.N. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 2002, 129, 2867–2878. [Google Scholar] [CrossRef]
- Ting, C.-Y.; McQueen, P.G.; Pandya, N.; Lin, T.-Y.; Yang, M.; Reddy, O.V.; O’Connor, M.B.; McAuliffe, M.; Lee, C.-H. Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron 2014, 81, 830–846. [Google Scholar] [CrossRef] [Green Version]
- Vonhoff, F.; Duch, C. Tiling among stereotyped dendritic branches in an identified Drosophila motoneuron. J. Comp. Neurol. 2010, 518, 2169–2185. [Google Scholar] [CrossRef] [Green Version]
- Cuntz, H.; Mathy, A.; Häusser, M. A scaling law derived from optimal dendritic wiring. Proc. Natl. Acad. Sci. USA 2012, 109, 11014–11018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira Castro, A.; Baltruschat, L.; Stürner, T.; Bahrami, A.; Jedlicka, P.; Tavosanis, G.; Cuntz, H. Achieving functional neuronal dendrite structure through sequential stochastic growth and retraction. Elife 2020, 9, e60920. [Google Scholar] [CrossRef] [PubMed]
- Stürner, T.; Castro, A.F.; Philipps, M.; Cuntz, H.; Tavosanis, G. The branching code: A model of actin-driven dendrite arborisation. BioRxiv 2020. [Google Scholar] [CrossRef]
- Nanda, S.; Das, R.; Bhattacharjee, S.; Cox, D.N.; Ascoli, G.A. Morphological determinants of dendritic arborization neurons in Drosophila larva. Brain Struct. Funct. 2018, 223, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Palavalli, A.; Tizón-Escamilla, N.; Rupprecht, J.-F.; Lecuit, T. Deterministic and stochastic rules of branching govern dendrite morphogenesis of sensory neurons. Curr. Biol. 2021, 31, 459–472. [Google Scholar] [CrossRef]
- Das, R.; Bhattacharjee, S.; Patel, A.A.; Harris, J.M.; Bhattacharya, S.; Letcher, J.M.; Clark, S.G.; Nanda, S.; Iyer, E.P.R.; Ascoli, G.A.; et al. Dendritic Cytoskeletal Architecture Is Modulated by Combinatorial Transcriptional Regulation in Drosophila melanogaster. Genetics 2017, 207, 1401–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corty, M.M.; Tam, J.; Grueber, W.B. Dendritic diversification through transcription factor-mediated suppression of alternative morphologies. Development 2016, 143, 1351–1362. [Google Scholar] [CrossRef] [Green Version]
- Parrish, J.Z.; Kim, M.D.; Jan, L.Y.; Jan, Y.N. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006, 20, 820–835. [Google Scholar] [CrossRef]
- Ziegler, A.B.; Thiele, C.; Tenedini, F.; Richard, M.; Leyendecker, P.; Hoermann, A.; Soba, P.; Tavosanis, G. Cell-Autonomous Control of Neuronal Dendrite Expansion via the Fatty Acid Synthesis Regulator SREBP. Cell Rep. 2017, 21, 3346–3353. [Google Scholar] [CrossRef] [Green Version]
- Yin, C.; Peterman, E.; Rasmussen, J.P.; Parrish, J.Z. Transparent touch: Insights from model systems on epidermal control of somatosensory innervation. Front. Cell Neurosci. 2021, 15, 680345. [Google Scholar] [CrossRef]
- Yang, W.-K.; Chien, C.-T. Beyond being innervated: The epidermis actively shapes sensory dendritic patterning. Open Biol. 2019, 9, 180257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.-Y.; Chen, P.-J.; Yu, H.-H.; Hsu, C.-P.; Lee, C.-H. Extrinsic factors regulating dendritic patterning. Front. Cell Neurosci. 2020, 14, 622808. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.; Li, Y.; Resseguie, M.; Brenman, J.E. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J. Neurosci. 2005, 25, 8878–8888. [Google Scholar] [CrossRef] [PubMed]
- Konietzny, A.; Bär, J.; Mikhaylova, M. Dendritic actin cytoskeleton: Structure, functions, and regulations. Front. Cell Neurosci. 2017, 11, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drees, F.; Gertler, F.B. Ena/VASP: Proteins at the tip of the nervous system. Curr. Opin. Neurobiol. 2008, 18, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Nithianandam, V.; Chien, C.-T. Actin blobs prefigure dendrite branching sites. J. Cell Biol. 2018, 217, 3731–3746. [Google Scholar] [CrossRef]
- Stürner, T.; Tatarnikova, A.; Mueller, J.; Schaffran, B.; Cuntz, H.; Zhang, Y.; Nemethova, M.; Bogdan, S.; Small, V.; Tavosanis, G. Transient localization of the Arp2/3 complex initiates neuronal dendrite branching in vivo. Development 2019, 146, dev171397. [Google Scholar] [CrossRef] [Green Version]
- Koleske, A.J. Molecular mechanisms of dendrite stability. Nat. Rev. Neurosci. 2013, 14, 536–550. [Google Scholar] [CrossRef]
- Yoong, L.-F.; Lim, H.-K.; Tran, H.; Lackner, S.; Zheng, Z.; Hong, P.; Moore, A.W. Atypical myosin tunes dendrite arbor subdivision. Neuron 2020, 106, 452–467.e8. [Google Scholar] [CrossRef]
- Lefebvre, J.L. Molecular mechanisms that mediate dendrite morphogenesis. Curr. Top. Dev. Biol. 2021, 142, 233–282. [Google Scholar] [CrossRef]
- Grueber, W.B.; Jan, L.Y.; Jan, Y.N. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 2003, 112, 805–818. [Google Scholar] [CrossRef] [Green Version]
- Jinushi-Nakao, S.; Arvind, R.; Amikura, R.; Kinameri, E.; Liu, A.W.; Moore, A.W. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron 2007, 56, 963–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimura, K.; Satoh, D.; Estes, P.; Crews, S.; Uemura, T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron 2004, 43, 809–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, B.; Kim, J.H.; Yang, L.; McLachlan, I.; Younger, S.; Jan, L.Y.; Jan, Y.N. Differential regulation of dendritic and axonal development by the novel Krüppel-like factor Dar1. J. Neurosci. 2011, 31, 3309–3319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Onishi, M.; Jan, L.Y.; Jan, Y.N. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc. Natl. Acad. Sci. USA 2007, 104, 5199–5204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaadia, R.D.; Li, W.; Voleti, V.; Singhania, A.; Hillman, E.M.C.; Grueber, W.B. Characterization of Proprioceptive System Dynamics in Behaving Drosophila Larvae Using High-Speed Volumetric Microscopy. Curr. Biol. 2019, 29, 935–944.e4. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Gulyanon, S.; Mihovilovic Skanata, M.; Karagyozov, D.; Heckscher, E.S.; Krieg, M.; Tsechpenakis, G.; Gershow, M.; Tracey, W.D. Direction selectivity in drosophila proprioceptors requires the mechanosensory channel tmc. Curr. Biol. 2019, 29, 945–956.e3. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Petersen, M.; Hoyer, N.; Spitzweck, B.; Tenedini, F.; Wang, D.; Gruschka, A.; Burchardt, L.S.; Szpotowicz, E.; Schweizer, M.; et al. Sensory integration and neuromodulatory feedback facilitate Drosophila mechanonociceptive behavior. Nat. Neurosci. 2017, 20, 1085–1095. [Google Scholar] [CrossRef] [Green Version]
- Tsubouchi, A.; Caldwell, J.C.; Tracey, W.D. Dendritic filopodia, Ripped Pocket, NOMPC, and NMDARs contribute to the sense of touch in Drosophila larvae. Curr. Biol. 2012, 22, 2124–2134. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Zhang, W.; He, Y.; Gorczyca, D.; Xiang, Y.; Cheng, L.E.; Meltzer, S.; Jan, L.Y.; Jan, Y.N. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature 2013, 493, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Bellido, R.; Himmel, N.J.; Gutstein, H.B.; Cox, D.N.; Galko, M.J. An assay for chemical nociception in Drosophila larvae. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20190282. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Yuan, Q.; Vogt, N.; Looger, L.L.; Jan, L.Y.; Jan, Y.N. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 2010, 468, 921–926. [Google Scholar] [CrossRef] [Green Version]
- Tracey, W.D.; Wilson, R.I.; Laurent, G.; Benzer, S. painless, a Drosophila gene essential for nociception. Cell 2003, 113, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Gorczyca, D.A.; Younger, S.; Meltzer, S.; Kim, S.E.; Cheng, L.; Song, W.; Lee, H.Y.; Jan, L.Y.; Jan, Y.N. Identification of Ppk26, a DEG/ENaC Channel Functioning with Ppk1 in a Mutually Dependent Manner to Guide Locomotion Behavior in Drosophila. Cell Rep. 2014, 9, 1446–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Wang, Y.; Wang, Q.; Wang, Z. The role of PPK26 in Drosophila larval mechanical nociception. Cell Rep. 2014, 9, 1183–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neely, G.G.; Keene, A.C.; Duchek, P.; Chang, E.C.; Wang, Q.-P.; Aksoy, Y.A.; Rosenzweig, M.; Costigan, M.; Woolf, C.J.; Garrity, P.A.; et al. TrpA1 regulates thermal nociception in Drosophila. PLoS ONE 2011, 6, e24343. [Google Scholar] [CrossRef]
- Zhong, L.; Hwang, R.Y.; Tracey, W.D. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr. Biol. 2010, 20, 429–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poe, A.R.; Xu, Y.; Zhang, C.; Lei, J.; Li, K.; Labib, D.; Han, C. Low FoxO expression in Drosophila somatosensory neurons protects dendrite growth under nutrient restriction. Elife 2020, 9, e53351. [Google Scholar] [CrossRef]
- Watanabe, K.; Furumizo, Y.; Usui, T.; Hattori, Y.; Uemura, T. Nutrient-dependent increased dendritic arborization of somatosensory neurons. Genes Cells 2017, 22, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Das, R.; Bhattacharjee, S.; Letcher, J.M.; Harris, J.M.; Nanda, S.; Foldi, I.; Lottes, E.N.; Bobo, H.M.; Grantier, B.D.; Mihály, J.; et al. Formin3 directs dendritic architecture via microtubule regulation and is required for somatosensory nociceptive behavior. Development 2021, 148, dev187609. [Google Scholar] [CrossRef]
- Li, W.; Wang, F.; Menut, L.; Gao, F.-B. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron 2004, 43, 823–834. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Usui, T.; Satoh, D.; Moriyama, S.; Shimono, K.; Itoh, T.; Shirahige, K.; Uemura, T. Sensory-neuron subtype-specific transcriptional programs controlling dendrite morphogenesis: Genome-wide analysis of Abrupt and Knot/Collier. Dev. Cell 2013, 27, 530–544. [Google Scholar] [CrossRef] [Green Version]
- Parrish, J.Z.; Xu, P.; Kim, C.C.; Jan, L.Y.; Jan, Y.N. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron 2009, 63, 788–802. [Google Scholar] [CrossRef] [Green Version]
- Baltruschat, L.; Tavosanis, G.; Cuntz, H. A developmental stretch-and-fill process that optimises dendritic wiring. BioRxiv 2020. [Google Scholar] [CrossRef]
- Melak, M.; Plessner, M.; Grosse, R. Correction: Actin visualization at a glance. J. Cell Sci. 2017, 130, 1688. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Kim, D.; Mun, J.Y. Direct Visualization of Actin Filaments and Actin-Binding Proteins in Neuronal Cells. Front. Cell Dev. Biol. 2020, 8, 588556. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.I.; Frey, D.; Lungu, O.I.; Jaehrig, A.; Schlichting, I.; Kuhlman, B.; Hahn, K.M. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009, 461, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Krueger, D.; Izquierdo, E.; Viswanathan, R.; Hartmann, J.; Pallares Cartes, C.; De Renzis, S. Principles and applications of optogenetics in developmental biology. Development 2019, 146, dev175067. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, J.; Krueger, D.; De Renzis, S. Using optogenetics to tackle systems-level questions of multicellular morphogenesis. Curr. Opin. Cell Biol. 2020, 66, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Bloor, J.W.; Ruiz-Gomez, M.; VijayRaghavan, K.; Kiehart, D.P. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis 2002, 34, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.A.; Demsky, M.; Montague, R.A.; Weymouth, N.; Kiehart, D.P. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev. Biol. 1997, 191, 103–117. [Google Scholar] [CrossRef] [Green Version]
- Riedl, J.; Crevenna, A.H.; Kessenbrock, K.; Yu, J.H.; Neukirchen, D.; Bista, M.; Bradke, F.; Jenne, D.; Holak, T.A.; Werb, Z.; et al. Lifeact: A versatile marker to visualize F-actin. Nat. Methods 2008, 5, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Spracklen, A.J.; Fagan, T.N.; Lovander, K.E.; Tootle, T.L. The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev. Biol. 2014, 393, 209–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belyy, A.; Merino, F.; Sitsel, O.; Raunser, S. Structure of the Lifeact-F-actin complex. PLoS Biol. 2020, 18, e3000925. [Google Scholar] [CrossRef]
- Westphal, M.; Jungbluth, A.; Heidecker, M.; Mühlbauer, B.; Heizer, C.; Schwartz, J.M.; Marriott, G.; Gerisch, G. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr. Biol. 1997, 7, 176–183. [Google Scholar] [CrossRef]
- Freymuth, P.S.; Fitzsimons, H.L. The ERM protein Moesin is essential for neuronal morphogenesis and long-term memory in Drosophila. Mol. Brain 2017, 10, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanda, S.; Bhattacharjee, S.; Cox, D.N.; Ascoli, G.A. Distinct Relations of Microtubules and Actin Filaments with Dendritic Architecture. iScience 2020, 23, 101865. [Google Scholar] [CrossRef] [PubMed]
- Wolterhoff, N.; Gigengack, U.; Rumpf, S. PP2A phosphatase is required for dendrite pruning via actin regulation in Drosophila. EMBO Rep. 2020, 21, e48870. [Google Scholar] [CrossRef]
- Emoto, K.; He, Y.; Ye, B.; Grueber, W.B.; Adler, P.N.; Jan, L.Y.; Jan, Y.-N. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 2004, 119, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Kanellopoulos, A.K.; Richter, M.; Petersen, M.; Konietzny, A.; Tenedini, F.M.; Hoyer, N.; Cheng, L.; Poon, C.L.C.; Harvey, K.F.; et al. Conserved tao kinase activity regulates dendritic arborization, cytoskeletal dynamics, and sensory function in drosophila. J. Neurosci. 2020, 40, 1819–1833. [Google Scholar] [CrossRef] [PubMed]
- Chazeau, A.; Giannone, G. Organization and dynamics of the actin cytoskeleton during dendritic spine morphological remodeling. Cell Mol. Life Sci. 2016, 73, 3053–3073. [Google Scholar] [CrossRef]
- Koike-Kumagai, M.; Yasunaga, K.; Morikawa, R.; Kanamori, T.; Emoto, K. The target of rapamycin complex 2 controls dendritic tiling of Drosophila sensory neurons through the Tricornered kinase signalling pathway. EMBO J. 2009, 28, 3879–3892. [Google Scholar] [CrossRef] [Green Version]
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Rüegg, M.A.; Hall, A.; Hall, M.N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004, 6, 1122–1128. [Google Scholar] [CrossRef]
- Threadgill, R.; Bobb, K.; Ghosh, A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron 1997, 19, 625–634. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.; Li, W.; Xu, K.; Bogert, B.A.; Su, K.; Gao, F.-B. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 2003, 130, 5543–5552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papayannopoulos, V.; Co, C.; Prehoda, K.E.; Snapper, S.; Taunton, J.; Lim, W.A. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol. Cell 2005, 17, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Padrick, S.B.; Doolittle, L.K.; Brautigam, C.A.; King, D.S.; Rosen, M.K. Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. USA 2011, 108, E472–E479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zallen, J.A.; Cohen, Y.; Hudson, A.M.; Cooley, L.; Wieschaus, E.; Schejter, E.D. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 2002, 156, 689–701. [Google Scholar] [CrossRef] [Green Version]
- Hudson, A.M.; Cooley, L. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 2002, 156, 677–687. [Google Scholar] [CrossRef]
- Korobova, F.; Svitkina, T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell 2010, 21, 165–176. [Google Scholar] [CrossRef] [Green Version]
- Jansen, S.; Collins, A.; Yang, C.; Rebowski, G.; Svitkina, T.; Dominguez, R. Mechanism of actin filament bundling by fascin. J. Biol. Chem. 2011, 286, 30087–30096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, J.; Delandre, C.; Zhang, Y.; Förstner, F.; Moore, A.W.; Tavosanis, G. Fascin controls neuronal class-specific dendrite arbor morphology. Development 2012, 139, 2999–3009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, A.J.; Millard, T.H.; Evans, I.R.; Wood, W. Ena orchestrates remodelling within the actin cytoskeleton to drive robust Drosophila macrophage chemotaxis. J. Cell Sci. 2019, 132, jcs224618. [Google Scholar] [CrossRef] [Green Version]
- Dimitrova, S.; Reissaus, A.; Tavosanis, G. Slit and Robo regulate dendrite branching and elongation of space-filling neurons in Drosophila. Dev. Biol. 2008, 324, 18–30. [Google Scholar] [CrossRef] [Green Version]
- D’Este, E.; Kamin, D.; Göttfert, F.; El-Hady, A.; Hell, S.W. STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep. 2015, 10, 1246–1251. [Google Scholar] [CrossRef] [Green Version]
- Avery, A.W.; Thomas, D.D.; Hays, T.S. β-III-spectrin spinocerebellar ataxia type 5 mutation reveals a dominant cytoskeletal mechanism that underlies dendritic arborization. Proc. Natl. Acad. Sci. USA 2017, 114, E9376–E9385. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, T.; Ou, Y.; Li, S.; Giniger, E.; van Meyel, D.J. Dendrite architecture organized by transcriptional control of the F-actin nucleator Spire. Development 2014, 141, 650–660. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, M.E.; Hilgert, S.; Bedrossian, A.; Mullins, R.D.; Kerkhoff, E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 2007, 179, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Dahlgaard, K.; Raposo, A.A.S.F.; Niccoli, T.; St Johnston, D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 2007, 13, 539–553. [Google Scholar] [CrossRef] [Green Version]
- Bradley, A.O.; Vizcarra, C.L.; Bailey, H.M.; Quinlan, M.E. Spire stimulates nucleation by Cappuccino and binds both ends of actin filaments. Mol. Biol. Cell 2020, 31, 273–286. [Google Scholar] [CrossRef]
- Qu, Y.; Hahn, I.; Webb, S.E.D.; Pearce, S.P.; Prokop, A. Periodic actin structures in neuronal axons are required to maintain microtubules. Mol. Biol. Cell 2017, 28, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.; Kovar, D.R. Internetwork competition for monomers governs actin cytoskeleton organization. Nat. Rev. Mol. Cell Biol. 2016, 17, 799–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavosanis, G. Dendrite enlightenment. Curr. Opin. Neurobiol. 2021, 69, 222–230. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kilo, L.; Stürner, T.; Tavosanis, G.; Ziegler, A.B. Drosophila Dendritic Arborisation Neurons: Fantastic Actin Dynamics and Where to Find Them. Cells 2021, 10, 2777. https://doi.org/10.3390/cells10102777
Kilo L, Stürner T, Tavosanis G, Ziegler AB. Drosophila Dendritic Arborisation Neurons: Fantastic Actin Dynamics and Where to Find Them. Cells. 2021; 10(10):2777. https://doi.org/10.3390/cells10102777
Chicago/Turabian StyleKilo, Lukas, Tomke Stürner, Gaia Tavosanis, and Anna B. Ziegler. 2021. "Drosophila Dendritic Arborisation Neurons: Fantastic Actin Dynamics and Where to Find Them" Cells 10, no. 10: 2777. https://doi.org/10.3390/cells10102777





