Nuclear Dynamics and Chromatin Structure: Implications for Pancreatic Cancer
Abstract
1. Introduction
2. Nuclear Structure
2.1. The Nuclear Envelope
2.2. Chromatin Organization in the Nucleus
2.3. Nuclear Mechanics
3. Mechanisms of Alterations in Nuclear Morphology
4. Alterations in Nuclear Structure and Chromatin Organization in Pancreatic Cancer
5. Unanswered Questions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beale, L.S. Results of the chemical and microscopical examination of solid organs and secretions. Examination of sputum from a case of cancer of the pharynx and the adjacent parts. Arch. Med. 1860, 2, 44–46. [Google Scholar]
- Papanicolaou, G.N. A new procedure for staining vaginal smears. Science 1942, 95, 438–439. [Google Scholar] [CrossRef]
- Papanicolaou, G.N.; Traut, H.F. The diagnostic value of vaginal smears in carcinoma of the uterus 1941. Arch. Pathol. Lab. Med. 1997, 121, 211–224. [Google Scholar]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identi-fied by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Edens, L.J.; White, K.H.; Jevtic, P.; Li, X.; Levy, D.L. Nuclear size regulation: From single cells to development and disease. Trends Cell Biol. 2013, 23, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.; Samuelson, K.; Hanlon, S. Multi-scale organization of the Drosophila melanogaster genome. Genes 2021, 12, 817. [Google Scholar] [CrossRef] [PubMed]
- Walters, A.D.; Bommakanti, A.; Cohen-Fix, O. Shaping the nucleus: Factors and forces. J. Cell. Biochem. 2012, 113, 2813–2821. [Google Scholar] [CrossRef] [PubMed]
- Deolal, P.; Mishra, K. Regulation of diverse nuclear shapes: Pathways working independently, together. Commun. Integr. Biol. 2021, 14, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, P.; Barrales, R.R.; Daga, R.R.; Salas-Pino, S. Nuclear mechanics in the fission yeast. Cells 2019, 8, 1285. [Google Scholar] [CrossRef] [PubMed]
- Harr, J.; Sandoval, A.V.G.; Gasser, S.M. Histones and histone modifications in perinuclear chromatin anchoring: From yeast to man. EMBO Rep. 2016, 17, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.; Witkin, K.L.; Cohen-Fix, O. Sizing up the nucleus: Nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009, 122, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Jevtić, P.; Levy, L.D. Elucidating nuclear size control in the xenopus model system. Vet. Glas. 2018, 72, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wesley, C.C.; Mishra, S.; Levy, D.L. Organelle size scaling over embryonic development. Wiley Interdiscip. Rev. Dev. Biol. 2020, 9, e375. [Google Scholar] [CrossRef]
- Denais, C.; Lammerding, J. Nuclear mechanics in cancer. Cancer Biol. Nucl. Envel. 2014, 773, 435–470. [Google Scholar] [CrossRef]
- Preston, C.C.; Faustino, R.S. Nuclear envelope regulation of oncogenic processes: Roles in pancreatic cancer. Epigenomes 2018, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Burla, R.; La Torre, M.; Maccaroni, K.; Verni, F.; Giunta, S.; Saggio, I. Interplay of the nuclear envelope with chromatin in physiolo-gy and pathology. Nucleus 2020, 11, 205–218. [Google Scholar] [CrossRef]
- Shevelyov, Y.Y.; Ulianov, S.V. The nuclear lamina as an organizer of chromosome architecture. Cells 2019, 8, 136. [Google Scholar] [CrossRef]
- Shin, J.-Y.; Dauer, W.T.; Worman, H.J. Lamina-associated polypeptide 1: Protein interactions and tissue-selective functions. Semin. Cell Dev. Biol. 2014, 29, 164–168. [Google Scholar] [CrossRef][Green Version]
- Brachner, A.; Foisner, R. Evolvement of LEM proteins as chromatin tethers at the nuclear periphery. Biochem. Soc. Trans. 2011, 39, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- Janin, A.; Gache, V. Nesprins and lamins in health and diseases of cardiac and skeletal muscles. Front. Physiol. 2018, 9, 1277. [Google Scholar] [CrossRef] [PubMed]
- Bouzid, T.; Kim, E.; Riehl, B.D.; Esfahani, A.M.; Rosenbohm, J.; Yang, R.; Duan, B.; Lim, J.Y. The LINC complex, mechanotransduction, and mesenchymal stem cell function and fate. J. Biol. Eng. 2019, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Qu, R.; Ouang, J.; Dai, J. A Glance at the nuclear envelope spectrin repeat protein 3. BioMed Res. Int. 2019, 2019, 1651805. [Google Scholar] [CrossRef]
- Taranum, S.; Sur, I.; Muller, R.; Lu, W.; Rashmi, R.N.; Munck, M.; Neumann, S.; Karakesisoglou, I.; Noegel, A.A. Cytoskeletal interac-tions at the nuclear envelope mediated by nesprins. Int. J. Cell Biol. 2012, 2012, 736524. [Google Scholar] [CrossRef][Green Version]
- Hao, H.; Starr, D.A. SUN/KASH interactions facilitate force transmission across the nuclear envelope. Nucleus 2019, 10, 73–80. [Google Scholar] [CrossRef]
- Gerace, L.; Tapia, O. Messages from the voices within: Regulation of signaling by proteins of the nuclear lamina. Curr. Opin. Cell Biol. 2018, 52, 14–21. [Google Scholar] [CrossRef]
- Janota, C.S.; Calero-Cuenca, F.J.; Gomes, E.R. The role of the cell nucleus in mechanotransduction. Curr. Opin. Cell Biol. 2020, 63, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Vlcek, S.; Foisner, R. A-type lamin networks in light of laminopathic diseases. Biochim. Biophys. Acta BBA Bioenerg. 2007, 1773, 661–674. [Google Scholar] [CrossRef]
- Barton, L.; Soshnev, A.; Geyer, P.K. Networking in the nucleus: A spotlight on LEM-domain proteins. Curr. Opin. Cell Biol. 2015, 34, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Berk, J.M.; Tifft, E.K.; Wilson, K.L. The nuclear envelope LEM-domain protein emerin. Nucleus 2013, 4, 298–314. [Google Scholar] [CrossRef]
- Brachner, A.; Foisner, R. Lamina-associated polypeptide (LAP)2α and other LEM proteins in cancer biology. Cancer Biol. Nucl. Envel. 2014, 773, 143–163. [Google Scholar] [CrossRef]
- Lee, K.K.; Wilson, K.L. All in the family: Evidence for four new LEM-domain proteins Lem2 (NET-25), Lem3, Lem4 and Lem5 in the human genome. Symp. Soc. Exp. Biol. 2004, 2004, 329–339. [Google Scholar]
- De Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear lamins: Thin filaments with major functions. Trends Cell Biol. 2018, 28, 34–45. [Google Scholar] [CrossRef]
- Naetar, N.; Ferraioli, S.; Foisner, R. Lamins in the nuclear interior—Life outside the lamina. J. Cell Sci. 2017, 130, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Poleshko, A.; Katz, R.A. Specifying peripheral heterochromatin during nuclear lamina reassembly. Nucleus 2014, 5, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.D.; Guan, T.; Gerace, L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol. Cell. Biol. 2009, 29, 5718–5728. [Google Scholar] [CrossRef] [PubMed]
- Zlopasa, L.; Brachner, A.; Foisner, R. Nucleo-cytoplasmic shuttling of the endonuclease ankyrin repeats and LEM domain-containing protein 1 (Ankle1) is mediated by canonical nuclear export- and nuclear import signals. BMC Cell Biol. 2016, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Elkhatib, R.A.; Paci, M.; Boissier, R.; Longepied, G.; Auguste, Y.; Achard, V.; Bourgeois, P.; Levy, N.; Branger, N.; Mitchell, M.J.; et al. LEM-domain proteins are lost during human spermiogenesis but BAF and BAF-L persist. Reproduction 2017, 154, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Brachner, A.; Reipert, S.; Foisner, R.; Gotzmann, J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 2005, 118, 5797–5810. [Google Scholar] [CrossRef]
- Thanisch, K.; Song, C.; Engelkamp, D.; Koch, J.; Wang, A.; Hallberg, E.; Foisner, R.; Leonhardt, H.; Stewart, C.L.; Joffe, B.; et al. Nuclear envelope localization of LEMD2 is developmentally dynamic and lamin A/C dependent yet insufficient for hetero-chromatin tethering. Differentiation 2017, 94, 58–70. [Google Scholar] [CrossRef]
- Koch, A.J.; Holaska, J.M. Emerin in health and disease. Semin. Cell Dev. Biol. 2014, 29, 95–106. [Google Scholar] [CrossRef]
- Dubińska-Magiera, M.; Kozioł, K.; Machowska, M.; Piekarowicz, K.; Filipczak, D.; Rzepecki, R. Emerin is required for proper nucleus reassembly after mitosis: Implications for new pathogenetic mechanisms for laminopathies detected in EDMD1 patients. Cells 2019, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Dorner, D.; Vlcek, S.; Foeger, N.; Gajewski, A.; Makolm, C.; Gotzmann, J.; Hutchison, C.J.; Foisner, R. Lamina-associated polypeptide 2α regulates cell cycle progression and differentiation via the retinoblastoma–E2F pathway. J. Cell Biol. 2006, 173, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, E.V.; Smith, K.; Reddy, K.L. The shifting shape of genomes: Dynamics of heterochromatin interactions at the nuclear lamina. Curr. Opin. Genet. Dev. 2021, 67, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Shaklai, S.; Amariglio, N.; Rechavi, G.; Simon, A.J. Gene silencing at the nuclear periphery. FEBS J. 2007, 274, 1383–1392. [Google Scholar] [CrossRef]
- Somech, R.; Shaklai, S.; Geller, O.; Amariglio, N.; Simon, A.J.; Rechavi, G.; Gal-Yam, E.N. The nuclear-envelope protein and transcrip-tional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005, 118, 4017–4025. [Google Scholar] [CrossRef] [PubMed]
- Mirza, A.N.; Gonzalez, F.; Ha, S.K.; Oro, A.E. The Sky’s the LEMit: New insights into nuclear structure regulation of transcription factor activity. Curr. Opin. Cell Biol. 2020, 68, 173–180. [Google Scholar] [CrossRef]
- Mirza, A.N.; McKellar, S.; Urman, N.M.; Brown, A.S.; Hollmig, T.; Aasi, S.Z.; Oro, A.E. LAP2 proteins chaperone GLI1 movement between the lamina and chromatin to regulate transcription. Cell 2019, 176, 198–212e15. [Google Scholar] [CrossRef]
- Zullo, J.M.; Demarco, I.A.; Pique-Regi, R.; Gaffney, D.; Epstein, C.B.; Spooner, C.J.; Luperchio, T.; Bernstein, B.E.; Pritchard, J.K.; Reddy, K.L.; et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell 2012, 149, 1474–1487. [Google Scholar] [CrossRef]
- Chambers, D.M.; Moretti, L.; Zhang, J.J.; Cooper, S.W.; Chambers, D.M.; Santangelo, P.J.; Barker, T.H. LEM domain-containing protein 3 antagonizes TGFbeta-SMAD2/3 signaling in a stiffness-dependent manner in both the nucleus and cytosol. J. Biol. Chem. 2018, 293, 15867. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.; Basilio, J.; Gotzmann, J.; Brachner, A.; Foisner, R. Comparative interactome analysis of emerin, MAN1 and LEM2 re-veals a unique role for LEM2 in nucleotide excision repair. Cells 2020, 9, 463. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Lajoie, D.; Chen, O.S.; Von Appen, A.; Ladinsky, M.S.; Redd, M.J.; Nikolova, L.; Bjorkman, P.J.; Sundquist, W.I.; Ullman, K.S.; et al. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. bioRxiv 2016, 114, 049312. [Google Scholar] [CrossRef] [PubMed]
- Gatta, A.T.; Olmos, Y.; Stoten, C.L.; Chen, Q.; Rosenthal, P.B.; Carlton, J.G. Author response: CDK1 controls CHMP7-dependent nuclear envelope reformation. Elife 2021, 2021, 10. [Google Scholar] [CrossRef]
- Asencio, C.; Davidson, I.F.; Santarella-Mellwig, R.; Ly-Hartig, T.B.; Mall, M.; Wallenfang, M.R.; Mattaj, I.W.; Gorjanacz, M. Coordina-tion of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell 2012, 150, 122–135. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Ashtiani, Z.O.; Golian, B.S.; Hasheminasab, S.-M.; Modarressi, M.H. Expression of two testis-specific genes, SPATA19 and LEMD1, in prostate cancer. Arch. Med. Res. 2010, 41, 195–200. [Google Scholar] [CrossRef]
- Sasahira, T.; Kurihara, M.; Nakashima, C.; Kirita, T.; Kuniyasu, H. LEM domain containing 1 promotes oral squamous cell carci-noma invasion and endothelial transmigration. Br. J. Cancer 2016, 115, 52–58. [Google Scholar] [CrossRef]
- Takeda, R.; Hirohashi, Y.; Shen, M.; Wang, L.; Ogawa, T.; Murai, A.; Yamamoto, E.; Kubo, T.; Nakatsugawa, M.; Kanaseki, T.; et al. Identification and functional analysis of variants of a cancer/testis antigen LEMD1 in colorectal cancer stem-like cells. Biochem. Biophys. Res. Commun. 2017, 485, 651–657. [Google Scholar] [CrossRef]
- Xu, M.; Lin, B.; Zheng, D.; Wen, J.; Hu, W.; Li, C.; Zhang, X.; Zhang, X.; Qu, J. LEM domain containing 1 promotes thyroid cancer cell proliferation and migration by activating the Wnt/beta-catenin signaling pathway and epithelial-mesenchymal transition. Oncol. Lett. 2021, 21, 442. [Google Scholar] [CrossRef]
- Brachner, A.; Braun, J.; Ghodgaonkar, M.; Castor, D.; Zlopasa, L.; Ehrlich, V.; Jiricny, J.; Gotzmann, J.; Knasmuller, S.; Foisner, R. The endonuclease Ankle1 requires its LEM and GIY-YIG motifs for DNA cleavage in vivo. J. Cell Sci. 2012, 125, 1048–1057. [Google Scholar] [CrossRef]
- Hong, Y.; Sonneville, R.; Wang, B.; Scheidt, V.; Meier, B.; Woglar, A.; Demetriou, S.; Labib, K.; Jantsch, V.; Gartner, A. LEM-3 is a mid-body-tethered DNA nuclease that resolves chromatin bridges during late mitosis. Nat. Commun. 2018, 9, 728. [Google Scholar] [CrossRef]
- Song, J.; Freeman, A.D.; Knebel, A.; Gartner, A.; Lilley, D.M. Human ANKLE1 is a nuclease specific for branched DNA. J. Mol. Biol. 2020, 432, 5825–5834. [Google Scholar] [CrossRef]
- Snyers, L.; Erhart, R.; Laffer, S.; Pusch, O.; Weipoltshammer, K.; Schofer, C. LEM4/ANKLE-2 deficiency impairs post-mitotic re-localization of BAF, LAP2alpha and LaminA to the nucleus, causes nuclear envelope instability in telophase and leads to hy-perploidy in HeLa cells. Eur. J. Cell Biol. 2018, 97, 63–74. [Google Scholar] [CrossRef]
- Gao, A.; Sun, T.; Ma, G.; Cao, J.; Hu, Q.; Chen, L.; Wang, Y.; Wang, Q.; Sun, J.; Wu, R.; et al. LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERalpha pathway. Nat. Commun. 2018, 9, 4180. [Google Scholar] [CrossRef]
- Kaufmann, T.; Kukolj, E.; Brachner, A.; Beltzung, E.; Bruno, M.; Kostrhon, S.; Opravil, S.; Hudecz, O.; Mechtler, K.; Warren, G.; et al. SIRT2 regulates nuclear envelope reassembly via ANKLE2 deacetylation. J. Cell Sci. 2016, 129, 4607–4621. [Google Scholar] [CrossRef] [PubMed]
- Ulbert, S.; Antonin, W.; Platani, M.; Mattaj, I.W. The inner nuclear membrane protein Lem2 is critical for normal nuclear enve-lope morphology. FEBS Lett. 2006, 580, 6435–6441. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shoshan, S.O.; Simon, A.J.; Jacob-Hirsch, J.; Shaklai, S.; Paz-Yaacov, N.; Amariglio, N.; Rechavi, G.; Trakhtenbrot, L. Induction of polyploidy by nuclear fusion mechanism upon decreased expression of the nuclear envelope protein LAP2beta in the hu-man osteosarcoma cell line U2OS. Mol. Cytogenet. 2014, 7, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Coppede, F. Mutations involved in premature-ageing syndromes. Appl. Clin. Genet. 2021, 14, 279–295. [Google Scholar] [CrossRef]
- Goldman, R.D.; Shumaker, D.K.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Gordon, L.B.; Gruenbaum, Y.; Khuon, S.; Mendez, M.; Varga, R.; et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson—Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8963–8968. [Google Scholar] [CrossRef]
- Paradisi, M.; McClintock, D.; Boguslavsky, R.L.; Pedicelli, C.; Worman, H.J.; Djabali, K. Dermal fibroblasts in Hutchinson-Gilford progeria syndrome with the lamin A G608G mutation have dysmorphic nuclei and are hypersensitive to heat stress. BMC Cell Biol. 2005, 6, 27. [Google Scholar] [CrossRef]
- Navarro, C.L.; De Sandre-Giovannoli, A.; Bernard, R.; Boccaccio, I.; Boyer, A.; Geneviève, D.; Hadj-Rabia, S.; Gaudy-Marqueste, C.; Smitt, H.S.; Vabres, P.; et al. Lamin A and ZMPSTE24 (FACE-1) defects cause nuclear disorganization and identify restrictive dermopathy as a lethal neonatal laminopathy. Hum. Mol. Genet. 2004, 13, 2493–2503. [Google Scholar] [CrossRef]
- Sullivan, T.; Escalante-Alcalde, D.; Bhatt, H.; Anver, M.; Bhat, N.; Nagashima, K.; Stewart, C.L.; Burke, B. Loss of A-type lamin ex-pression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999, 147, 913–920. [Google Scholar] [CrossRef]
- Lammerding, J.; Hsiao, J.; Schulze, P.C.; Kozlov, S.; Stewart, C.L.; Lee, R.T. Abnormal nuclear shape and impaired mechanotrans-duction in emerin-deficient cells. J. Cell Biol. 2005, 170, 781–791. [Google Scholar] [CrossRef]
- Lammerding, J.; Fong, L.G.; Ji, J.Y.; Reue, K.; Stewart, C.L.; Young, S.; Lee, R.T. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006, 281, 25768–25780. [Google Scholar] [CrossRef] [PubMed]
- Vergnes, L.; Peterfy, M.; Bergo, M.O.; Young, S.; Reue, K. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA 2004, 101, 10428–10433. [Google Scholar] [CrossRef]
- Coffinier, C.; Jung, H.J.; Nobumori, C.; Chang, S.; Tu, Y.; Barnes, R.H.; Yoshinaga, Y.; de Jong, P.J.; Vergnes, L.; Reue, K.; et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol. Cell. 2011, 22, 4683–4693. [Google Scholar] [CrossRef]
- Chen, N.Y.; Yang, Y.; Weston, T.A.; Belling, J.N.; Heizer, P.; Tu, Y.; Kim, P.; Edillo, L.; Jonas, S.J.; Weiss, P.S.; et al. An ab-sence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc. Natl. Acad. Sci. USA 2019, 116, 25870–25879. [Google Scholar] [CrossRef] [PubMed]
- Jevtić, P.; Edens, L.J.; Li, X.; Nguyen, T.; Chen, P.; Levy, D.L. Concentration-dependent effects of nuclear lamins on nuclear size in xenopus and mammalian cells. J. Biol. Chem. 2015, 290, 27557–27571. [Google Scholar] [CrossRef] [PubMed]
- Shimojima, M.; Yuasa, S.; Motoda, C.; Yozu, G.; Nagai, T.; Ito, S.; Lachmann, M.; Kashimura, S.; Takei, M.; Kusumoto, D.; et al. Emerin plays a crucial role in nuclear invagination and in the nuclear calcium transient. Sci. Rep. 2017, 7, 44312. [Google Scholar] [CrossRef]
- Reis-Sobreiro, M.; Chen, J.F.; Novitskaya, T.; You, S.; Morley, S.; Steadman, K.; Gill, N.K.; Eskaros, A.; Rotinen, M.; Chu, C.Y.; et al. Emerin de-regulation links nuclear shape instability to metastatic potential. Cancer Res. 2018, 78, 6086–6097. [Google Scholar] [CrossRef]
- Kituyi, S.; Edkins, A.L. Hop/STIP1 depletion alters nuclear structure via depletion of nuclear structural protein emerin. Biochem. Biophys. Res. Commun. 2018, 507, 503–509. [Google Scholar] [CrossRef]
- Liddane, A.G.; McNamara, C.A.; Campbell, M.C.; Mercier, I.; Holaska, J.M. Defects in emerin-nucleoskeleton binding disrupt nuclear structure and promote breast cancer cell motility and metastasis. Mol. Cancer Res. 2021, 19, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Fichtman, B.; Zagairy, F.; Biran, N.; Barsheshet, Y.; Chervinsky, E.; Ben Neriah, Z.; Shaag, A.; Assa, M.; Elpeleg, O.; Harel, A.; et al. Combined loss of LAP1B and LAP1C results in an early onset multisystemic nuclear envelopathy. Nat. Commun. 2019, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Lessel, I.; Chen, M.J.; Luttgen, S.; Arndt, F.; Fuchs, S.; Meien, S.; Thiele, H.; Jones, J.R.; Shaw, B.R.; Crossman, D.K.; et al. Two novel cases further expand the phenotype of TOR1AIP1-associated nuclear envelopathies. Hum. Genet. 2020, 139, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.-Y.; Méndez-López, I.; Hong, M.; Wang, Y.; Tanji, K.; Wu, W.; Shugol, L.; Krauss, R.S.; Dauer, W.T.; Worman, H.J. Lamina-associated polypeptide 1 is dispensable for embryonic myogenesis but required for postnatal skeletal muscle growth. Hum. Mol. Genet. 2016, 26, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Kutay, U.; Luithle, N.; De Bos, J.U.; Hovius, R.; Maslennikova, D.; Lewis, R.T.; Ungricht, R.; Fierz, B. Torsin ATPases influence chromatin interaction of the Torsin regulator LAP1. Elife 2020, 9, e63614. [Google Scholar] [CrossRef]
- Naismith, T.V.; Dalal, S.; Hanson, P.I. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J. Biol. Chem. 2009, 284, 27866–27874. [Google Scholar] [CrossRef]
- Galant, D.; Gaborit, B.; Desgrouas, C.; Abdesselam, I.; Bernard, M.; Levy, N.; Merono, F.; Coirault, C.; Roll, P.; Lagarde, A.; et al. A Heterozygous ZMPSTE24 mutation associated with severe metabolic syndrome, ectopic fat accumulation, and dilated cardiomyopathy. Cells 2016, 5, 21. [Google Scholar] [CrossRef]
- Toth, J.I.; Yang, S.H.; Qiao, X.; Beigneux, A.P.; Gelb, M.H.; Moulson, C.L.; Miner, J.H.; Young, S.G.; Fong, L.G. Blocking protein farnesyl-transferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. USA 2005, 102, 12873–12878. [Google Scholar] [CrossRef]
- Zhou, C.; Li, C.; Zhou, B.; Sun, H.; Koullourou, V.; Holt, I.; Puckelwartz, M.J.; Warren, D.T.; Hayward, R.; Lin, Z.; et al. Novel nesprin-1 mutations associated with dilated cardiomyo-pathy cause nuclear envelope disruption and defects in myogenesis. Hum. Mol. Genet. 2017, 26, 2258–2276. [Google Scholar] [CrossRef]
- Zhang, Q.; Bethmann, C.; Worth, N.F.; Davies, J.D.; Wasner, C.; Feuer, A.; Ragnauth, C.D.; Yi, Q.; Mellad, J.A.; Warren, D.T.; et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery–Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007, 16, 2816–2833. [Google Scholar] [CrossRef]
- King, S.J.; Nowak, K.; Suryavanshi, N.; Holt, I.; Shanahan, C.M.; Ridley, A.J. Nesprin-1 and nesprin-2 regulate endothelial cell shape and migration. Cytoskeleton 2014, 71, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Meinke, P.; Mattioli, E.; Haque, F.; Antoku, S.; Columbaro, M.; Straatman, K.R.; Worman, H.J.; Gundersen, G.G.; Lattanzi, G.; Wehnert, M.; et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014, 10, e1004605. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.P.; Williams, J.; Leung, E.; Sanders, J.; Eddy, R.; Castracane, J.; Oktay, M.H.; Entenberg, D.; Condeelis, J.S. SUN-MKL1 cross-talk regulates nuclear deformation and fast motility of breast carcinoma cells in fibrillar ECM microenvironment. Cells 2021, 10, 1549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Pante, N.; Misteli, T.; Elsagga, M.; Crisp, M.; Hodzic, D.; Burke, B.; Roux, K. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 2007, 178, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Donahue, D.A.; Amraoui, S.; di Nunzio, F.; Kieffer, C.; Porrot, F.; Opp, S.; Diaz-Griffero, F.; Casartelli, N.; Schwartz, O. SUN2 Overexpression Deforms Nuclear Shape and Inhibits HIV. J. Virol. 2016, 90, 4199–4214. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, R.E.; Kim, C.E.; Dauer, W. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 2005, 48, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Laudermilch, E.; Tsai, P.L.; Graham, M.; Turner, E.; Zhao, C.; Schlieker, C. Dissecting torsin/cofactor function at the nuclear enve-lope: A genetic study. Mol. Biol. Cell. 2016, 27, 3964–3971. [Google Scholar] [CrossRef]
- Vander Heyden, A.B.; Naismith, T.V.; Snapp, E.L.; Hodzic, D.; Hanson, P.I. LULL1 retargets TorsinA to the nuclear envelope reveal-ing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell. 2009, 20, 2661–2672. [Google Scholar] [CrossRef]
- Imbalzano, K.M.; Cohet, N.; Wu, Q.; Underwood, J.M.; Imbalzano, A.N.; Nickerson, J.A. Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS ONE 2013, 8, e55628. [Google Scholar] [CrossRef]
- Hill, D.A.; Chiosea, S.; Jamaluddin, S.; Roy, K.; Fischer, A.; Boyd, D.D.; Nickerson, J.A.; Imbalzano, A.N. Inducible changes in cell size and attachment area due to expression of a mutant SWI/SNF chromatin remodeling enzyme. J. Cell Sci. 2004, 117, 5847–5854. [Google Scholar] [CrossRef][Green Version]
- Somsuan, K.; Peerapen, P.; Boonmark, W.; Plumworasawat, I.; Samol, R.; Sakulsak, N.; Thongboonkerd, V. ARID1A knockdown triggers epithelial-mesenchymal transition and carcinogenesis features of renal cells: Role in renal cell carcinoma. FASEB J. 2019, 33, 12226–12239. [Google Scholar] [CrossRef]
- Boyle, S.; Flyamer, I.M.; Williamson, I.; Sengupta, D.; Bickmore, W.A.; Illingworth, R.S. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev. 2020, 34, 931–949. [Google Scholar] [CrossRef]
- Karoutas, A.; Szymanski, W.; Rausch, T.; Guhathakurta, S.; Rog-Zielinska, E.A.; Peyronnet, R.; Seyfferth, J.; Chen, H.-R.; De Leeuw, R.; Herquel, B.; et al. The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell Biol. 2019, 21, 1248–1260. [Google Scholar] [CrossRef]
- George, C.M.; Bozler, J.; Nguyen, H.Q.; Bosco, G. Condensins are required for maintenance of nuclear architecture. Cells 2014, 3, 865–882. [Google Scholar] [CrossRef]
- Douet, J.; Corujo, D.; Malinverni, R.; Renauld, J.; Sansoni, V.; Posavec Marjanovic, M.; Cantarino, N.; Valero, V.; Mongelard, F.; Bouvet, P.; et al. MacroH2A histone variants maintain nuclear organization and heterochromatin archi-tecture. J. Cell Sci. 2017, 130, 1570–1582. [Google Scholar]
- Fu, Y.; Lv, P.; Yan, G.; Fan, H.; Cheng, L.; Zhang, F.; Dang, Y.; Wu, H.; Wen, B. MacroH2A1 associates with nuclear lamina and main-tains chromatin architecture in mouse liver cells. Sci. Rep. 2015, 5, 17186. [Google Scholar] [CrossRef]
- Furusawa, T.; Rochman, M.; Taher, L.; Dimitriadis, E.K.; Nagashima, K.; Anderson, S.; Bustin, M. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 2015, 6, 6138. [Google Scholar] [CrossRef] [PubMed]
- Jevtic, P.; Schibler, A.C.; Wesley, C.C.; Pegoraro, G.; Misteli, T.; Levy, D.L. The nucleoporin ELYS regulates nuclear size by control-ling NPC number and nuclear import capacity. EMBO Rep. 2019, 20, e47283. [Google Scholar] [CrossRef] [PubMed]
- Vuorinen, E.M.; Rajala, N.K.; Ihalainen, T.O.; Kallioniemi, A. Depletion of nuclear import protein karyopherin alpha 7 (KPNA7) induces mitotic defects and deformation of nuclei in cancer cells. BMC Cancer 2018, 18, 325. [Google Scholar] [CrossRef] [PubMed]
- Hawryluk-Gara, L.A.; Shibuya, E.K.; Wozniak, R.W. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing Complex. Mol. Biol. Cell 2005, 16, 2382–2394. [Google Scholar] [CrossRef]
- Fahrenkrog, B.; Martinelli, V.; Nilles, N.; Fruhmann, G.; Chatel, G.; Juge, S.; Sauder, U.; Di Giacomo, D.; Mecucci, C.; Schwaller, J. Ex-pression of leukemia-associated Nup98 fusion proteins generates an aberrant nuclear envelope phenotype. PLoS ONE 2016, 11, e0152321. [Google Scholar] [CrossRef][Green Version]
- Zhou, L.; Panté, N. The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett. 2010, 584, 3013–3020. [Google Scholar] [CrossRef]
- Wiggan, O.; Schroder, B.; Krapf, D.; Bamburg, J.R.; DeLuca, J.G. Cofilin regulates nuclear architecture through a myosin-II de-pendent mechanotransduction module. Sci. Rep. 2017, 7, 40953. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Wagstaff, K.M.; Suarez-Sanchez, R.; Zinker, S.; Jans, D.A.; Cisneros, B. Nuclear localization of the dystrophin-associated protein alpha-dystrobrevin through importin alpha2/beta1 is critical for interaction with the nuclear lami-na/maintenance of nuclear integrity. FASEB J. 2015, 29, 1842–1858. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Almendrala, D.K.; Go, M.M.; Krauss, S.W. Structural protein 4.1R is integrally involved in nuclear envelope protein localization, centrosome–nucleus association and transcriptional signaling. J. Cell Sci. 2011, 124, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Russ, A.; Louderbough, J.M.V.; Zarnescu, D.; Schroeder, J.A. Hugl1 and Hugl2 in mammary epithelial cells: Polarity, proliferation, and differentiation. PLoS ONE 2012, 7, e47734. [Google Scholar] [CrossRef]
- Takaki, T.; Montagner, M.; Serres, M.P.; Le Berre, M.; Russell, M.; Collinson, L.; Szuhai, K.; Howell, M.; Boulton, S.J.; Sahai, E.; et al. Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat. Commun. 2017, 8, 16013. [Google Scholar] [CrossRef] [PubMed]
- Capo-chichi, C.D.; Cai, K.Q.; Testa, J.R.; Godwin, A.K.; Xu, X.X. Loss of GATA6 leads to nuclear deformation and aneuploidy in ovarian cancer. Mol. Cell Biol. 2009, 29, 4766–4777. [Google Scholar] [CrossRef]
- Lee, S.; Ahn, Y.M.; Kim, J.Y.; Cho, Y.E.; Park, J.H. Downregulation of NOP53 ribosome biogenesis factor leads to abnormal nuclear division and chromosomal instability in human cervical cancer cells. Pathol. Oncol. Res. 2020, 26, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.A.; Matsunaga, S.; Uchiyama, S.; Fukui, K. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem. J. 2008, 415, 345–351. [Google Scholar] [CrossRef]
- Lazar, I.; Fabre, B.; Feng, Y.; Khateb, A.; Turko, P.; Gomez, J.M.M.; Frederick, D.T.; Levesque, M.P.; Feld, L.; Zhang, G.; et al. SPANX control of lamin A/C modulates nuclear architecture and promotes melanoma growth. Mol. Cancer Res. 2020, 18, 1560–1573. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, A.; Santarella-Mellwig, R.; Santama, N.; Mattaj, I.W. Transmembrane protein TMEM170A is a newly discovered regulator of ER and nuclear envelope morphogenesis in human cells. J. Cell Sci. 2016, 129, 1552–1565. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, A.; Asaka, M.N.; Matsumoto, K.; Nagata, K. Centrosome maturation requires YB-1 to regulate dynamic instability of microtubules for nucleus reassembly. Sci. Rep. 2015, 5, 8768. [Google Scholar] [CrossRef] [PubMed]
- Olins, A.L.; Rhodes, G.; Welch, D.B.; Zwerger, M.; Olins, D.E. Lamin B receptor: Multi-tasking at the nuclear envelope. Nucleus 2010, 1, 53–70. [Google Scholar] [CrossRef]
- Guarda, A.; Bolognese, F.; Bonapace, I.M.; Badaracco, G. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp. Cell Res. 2009, 315, 1895–1903. [Google Scholar] [CrossRef]
- Hirano, Y.; Hizume, K.; Kimura, H.; Takeyasu, K.; Haraguchi, T.; Hiraoka, Y. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J. Biol. Chem. 2012, 287, 42654–42663. [Google Scholar] [CrossRef]
- Foisner, R.; Gerace, L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 1993, 73, 1267–1279. [Google Scholar] [CrossRef]
- Rampello, A.J.; Prophet, S.M.; Schlieker, C. The role of torsin AAA+ proteins in preserving nuclear envelope integrity and safeguarding against disease. Biomolecules 2020, 10, 468. [Google Scholar] [CrossRef]
- Dunlevy, K.L.; Medvedeva, V.; Wilson, J.E.; Hoque, M.; Pellegrin, T.; Maynard, A.; Kremp, M.M.; Wasserman, J.S.; Poleshko, A.; Katz, R.A. The PRR14 heterochromatin tether encodes modular domains that mediate and regulate nuclear lamina targeting. J. Cell Sci. 2020, 133, jcs240416. [Google Scholar] [CrossRef]
- Poleshko, A.; Mansfield, K.M.; Burlingame, C.C.; Andrake, M.D.; Shah, N.R.; Katz, R.A. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013, 5, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.D.; Martins, F.; Santos, M.; Mueller, T.; da Cruz, E.S.O.A.B.; Rebelo, S. Nuclear accumulation of LAP1:TRF2 complex during DNA damage response uncovers a novel role for LAP1. Cells 2020, 9, 1804. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Domingues, S.C.; Costa, P.; Muller, T.; Galozzi, S.; Marcus, K.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.; Rebelo, S. Identi-fication of a novel human LAP1 isoform that is regulated by protein phosphorylation. PLoS ONE 2014, 9, e113732. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Rebelo, S.; Van Kleeff, P.J.M.; Kim, C.E.; Dauer, W.T.; Fardilha, M.; Silva, O.A.D.C.E.; Silva, E.F.D.C.E. The nuclear envelope protein, LAP1B, is a novel protein phosphatase 1 substrate. PLoS ONE 2013, 8, e76788. [Google Scholar] [CrossRef] [PubMed]
- Serrano, J.B.; Silva, O.A.B.D.C.E.; Rebelo, S. Lamina associated polypeptide 1 (LAP1) interactome and its functional features. Membranes 2016, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Tenga, R.; Medalia, O. Structure and unique mechanical aspects of nuclear lamin filaments. Curr. Opin. Struct. Biol. 2020, 64, 152–159. [Google Scholar] [CrossRef]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.G.T.S.R.D.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, T.; Goldman, R.D.; Medalia, Y.T.M.E.A.D.-G.K.T.S.O. The molecular architecture of lamins in somatic cells. Nat. Cell Biol. 2017, 543, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, A.T.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Gesson, K.; Rescheneder, P.; Skoruppa, M.P.; Von Haeseler, A.; Dechat, T.; Foisner, R. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res. 2016, 26, 462–473. [Google Scholar] [CrossRef]
- Naetar, N.; Korbei, B.; Kozlov, S.; Kerenyi, M.A.; Dorner, D.; Kral, R.; Gotic, I.; Fuchs, P.; Cohen, T.V.; Bittner, R.; et al. Loss of nucleoplasmic LAP2α–lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat. Cell Biol. 2008, 10, 1341–1348. [Google Scholar] [CrossRef]
- Alvarado-Kristensson, M.; Rossello, C.A. The biology of the nuclear envelope and its implications in cancer biology. Int. J. Mol. Sci. 2019, 20, 2586. [Google Scholar] [CrossRef] [PubMed]
- Shimi, T.; Pfleghaar, K.; Kojima, S.-I.; Pack, C.-G.; Solovei, I.; Goldman, A.E.; Adam, S.; Shumaker, D.K.; Kinjo, M.; Cremer, T.; et al. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 2008, 22, 3409–3421. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Chojnowski, A.; Boudier, T.; Lim, J.S.; Ahmed, S.; Ser, Z.; Stewart, C.; Burke, B. A-type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr. Biol. 2016, 26, 2651–2658. [Google Scholar] [CrossRef]
- Elgin, S.C.R. Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 1996, 6, 193–202. [Google Scholar] [CrossRef]
- Hildebrand, E.M.; Dekker, J. Mechanisms and functions of chromosome compartmentalization. Trends Biochem. Sci. 2020, 45, 385–396. [Google Scholar] [CrossRef]
- Bizhanova, A.; Kaufman, P.D. Close to the edge: Heterochromatin at the nucleolar and nuclear peripheries. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194666. [Google Scholar] [CrossRef]
- Politz, J.C.R.; Scalzo, D.; Groudine, M. The redundancy of the mammalian heterochromatic compartment. Curr. Opin. Genet. Dev. 2016, 37, 1–8. [Google Scholar] [CrossRef]
- Vertii, A.; Ou, J.; Yu, J.; Yan, A.; Pagès, H.; Liu, H.; Zhu, L.J.; Kaufman, P.D. Two contrasting classes of nucleolus-associated domains in mouse fibroblast heterochromatin. Genome Res. 2019, 29, 1235–1249. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Hübner, M.R.; Eckersley-Maslin, M.; Spector, D.L. Chromatin organization and transcriptional regulation. Curr. Opin. Genet. Dev. 2013, 23, 89–95. [Google Scholar] [CrossRef]
- Reid, G.; Gallais, R.; Métivier, R. Marking time: The dynamic role of chromatin and covalent modification in transcription. Int. J. Biochem. Cell Biol. 2009, 41, 155–163. [Google Scholar] [CrossRef]
- Espiritu, D.; Gribkova, A.K.; Gupta, S.; Shaytan, A.K.; Panchenko, A.R. Molecular mechanisms of oncogenesis through the lens of nucleosomes and histones. J. Phys. Chem. B 2021, 125, 3963–3976. [Google Scholar] [CrossRef]
- Kurumizaka, H.; Kujirai, T.; Takizawa, Y. Contributions of histone variants in nucleosome structure and function. J. Mol. Biol. 2021, 433, 166678. [Google Scholar] [CrossRef]
- Chen, Q.W.; Zhu, X.Y.; Li, Y.Y.; Meng, Z.Q. Epigenetic regulation and cancer (Review). Oncol. Rep. 2013, 31, 523–532. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Espada, J.; Esteller, M. Epigenetic control of nuclear architecture. Cell. Mol. Life Sci. 2007, 64, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Madakashira, B.P.; Sadler, K.C. DNA methylation, nuclear organization, and cancer. Front. Genet. 2017, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Pombo, A.; Dillon, N. Three-dimensional genome architecture: Players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015, 16, 245–257. [Google Scholar] [CrossRef]
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M.; Hubner, B.; Silahtaroglu, A.; Hendzel, M.; Lanctot, C.; Strickfaden, H.; Cremer, C. The interchromatin compartment participates in the structural and functional organization of the cell nucleus. Bioessays 2020, 42, e1900132. [Google Scholar] [CrossRef] [PubMed]
- Finn, E.H.; Misteli, T. Molecular basis and biological function of variability in spatial genome organization. Science 2019, 365, eaaw9498. [Google Scholar] [CrossRef] [PubMed]
- Boveri, T. Die blastomerenkerne von ascaris megalocephala. Arch für Zellforsch 1909, 3, 181. [Google Scholar]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef] [PubMed]
- Lichter, P.; Cremer, T.; Borden, J.; Manuelidis, L.; Ward, D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Qual. Life Res. 1988, 80, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.K.; Vitalini, M.W.; Wallrath, L.L. Nuclear organization: Taking a position on gene expression. Curr. Opin. Cell Biol. 2011, 23, 354–359. [Google Scholar] [CrossRef]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef]
- Szabo, Q.; Bantignies, F.; Cavalli, G. Principles of genome folding into topologically associating domains. Sci. Adv. 2019, 5, eaaw1668. [Google Scholar] [CrossRef]
- Razin, S.V.; Ulianov, S.V. Gene functioning and storage within a folded genome. Cell. Mol. Biol. Lett. 2017, 22, 18. [Google Scholar] [CrossRef]
- Alpsoy, A.; Sood, S.; Dykhuizen, E. At the crossroad of gene regulation and genome organization: Potential roles for ATP-dependent chromatin remodelers in the regulation of CTCF-mediated 3D architecture. Biology 2021, 10, 272. [Google Scholar] [CrossRef]
- Gonzalez-Sandoval, A.; Gasser, S.M. On TADs and LADs: Spatial control over gene expression. Trends Genet. 2016, 32, 485–495. [Google Scholar] [CrossRef]
- Sikorska, N.; Sexton, T. Defining functionally relevant spatial chromatin domains: It is a TAD complicated. J. Mol. Biol. 2020, 432, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.S.; Huntley, M.H.; Durand, N.C.; Stamenova, E.K.; Bochkov, I.D.; Robinson, J.T.; Sanborn, A.L.; Machol, I.; Omer, A.D.; Lander, E.S.; et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Wutz, G.; Várnai, C.; Nagasaka, K.; Cisneros, D.A.; Stocsits, R.R.; Tang, W.; Schoenfelder, S.; Jessberger, G.; Muhar, M.; Hossain, M.J.; et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017, 36, 3573–3599. [Google Scholar] [CrossRef]
- Van Steensel, B.; Belmont, A.S. Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila melano-gaster genome at the nuclear lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Amendola, M.; van Steensel, B. Nuclear lamins are not required for lamina-associated domain organization in mouse embry-onic stem cells. EMBO Rep. 2015, 16, 610–617. [Google Scholar] [CrossRef]
- Reguant, L.P.; Blanco, E.; Galan, S.; Le Dily, F.; Cuartero, Y.; Serra-Bardenys, G.; Di Carlo, V.; Iturbide, A.; Cebrià-Costa, J.P.; Nonell, L.; et al. Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun. 2018, 9, 3420. [Google Scholar] [CrossRef]
- Smith, K.S.; Liu, L.L.; Ganesan, S.; Michor, F.; De, S. Nuclear topology modulates the mutational landscapes of cancer genomes. Nat. Struct. Mol. Biol. 2017, 24, 1000–1006. [Google Scholar] [CrossRef]
- Lenain, C.; de Graaf, C.A.; Pagie, L.; Visser, N.L.; de Haas, M.; de Vries, S.S.; Peric-Hupkes, D.; van Steensel, B.; Peeper, D.S. Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res. 2017, 27, 1634–1644. [Google Scholar] [CrossRef] [PubMed]
- Meuleman, W.; Peric-Hupkes, D.; Kind, J.; Beaudry, J.-B.; Pagie, L.; Kellis, M.; Reinders, M.; Wessels, L.; Van Steensel, B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2012, 23, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Wu, H.; Shinkai, Y.A.; Irizarry, R.; Feinberg, A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009, 41, 246–250. [Google Scholar] [CrossRef]
- McDonald, O.G.; Li, X.; Saunders, T.; Tryggvadottir, R.; Mentch, S.J.; Warmoes, M.O.; Word, A.E.; Carrer, A.; Salz, T.H.; Natsume, S.; et al. Epige-nomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 2017, 49, 367–376. [Google Scholar] [CrossRef]
- McDonald, O.G.; Wu, H.; Timp, W.; Doi, A.; Feinberg, A.P. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat. Struct. Mol. Biol. 2011, 18, 867–874. [Google Scholar] [CrossRef]
- Khan, Z.S.; Santos, J.M.; Hussain, F. Aggressive prostate cancer cell nuclei have reduced stiffness. Biomicrofluidics 2018, 12, 014102. [Google Scholar] [CrossRef]
- Deville, S.S.; Cordes, N. The extracellular, cellular, and nuclear stiffness, a trinity in the cancer resistome—A Review. Front. Oncol. 2019, 9, 1376. [Google Scholar] [CrossRef]
- Roberts, A.B.; Zhang, J.; Raj Singh, V.; Nikolic, M.; Moeendarbary, E.; Kamm, R.D.; So, P.T.C.; Scarcelli, G. Tumor cell nuclei soften dur-ing transendothelial migration. J. Biomech. 2021, 121, 110400. [Google Scholar] [CrossRef]
- Nguyen, A.V.; Nyberg, K.D.; Scott, M.B.; Welsh, A.M.; Nguyen, A.H.; Wu, N.; Hohlbauch, S.V.; Geisse, N.A.; Gibb, E.A.; Robertson, A.G.; et al. Stiffness of pancreatic cancer cells is associated with increased invasive potential. Integr. Biol. 2016, 8, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2005, 172, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Ketema, M.; Wilhelmsen, K.; Kuikman, I.; Janssen, H.; Hodzic, D.; Sonnenberg, A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci. 2007, 120, 3384–3394. [Google Scholar] [CrossRef]
- Lu, W.; Schneider, M.; Neumann, S.; Jaeger, V.-M.; Taranum, S.; Munck, M.; Cartwright, S.; Richardson, C.; Carthew, J.R.; Noh, K.; et al. Nesprin interchain associations control nuclear size. Cell. Mol. Life Sci. 2012, 69, 3493–3509. [Google Scholar] [CrossRef]
- Roux, K.J.; Crisp, M.L.; Liu, Q.; Kim, D.; Kozlov, S.; Stewart, C.L.; Burke, B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA 2009, 106, 2194–2199. [Google Scholar] [CrossRef]
- Svitkina, T.M.; Verkhovsky, A.B.; Borisy, G.G. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol. 1996, 135, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Hieda, M. Implications for diverse functions of the LINC complexes based on the structure. Cells 2017, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Uzer, G.; Rubin, C.T.; Rubin, J. Cell mechanosensitivity is enabled by the LINC nuclear complex. Curr. Mol. Biol. Rep. 2016, 2, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Thakar, K.; May, C.K.; Rogers, A.; Carroll, C.W. Opposing roles for distinct LINC complexes in regulation of the small GTPase RhoA. Mol. Biol. Cell 2017, 28, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Maizels, Y.; Gerlitz, G. Shaping of interphase chromosomes by the microtubule network. FEBS J. 2015, 282, 3500–3524. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.W. Nuclear pore complex tethers to the cytoskeleton. Semin. Cell Dev. Biol. 2017, 68, 52–58. [Google Scholar] [CrossRef]
- Guilluy, C.; Osborne, L.D.; Van Landeghem, L.; Sharek, L.; Superfine, R.; Garcia-Mata, R.; Burridge, K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol. 2014, 16, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Liu, P.Z.; Banigan, E.J.; Almassalha, L.M.; Backman, V.; Adam, S.A.; Goldman, R.D.; Marko, J.F. Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol. Biol. Cell. 2018, 29, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Wintner, O.; Hirsch-Attas, N.; Schlossberg, M.; Brofman, F.; Friedman, R.; Kupervaser, M.; Kitsberg, D.; Buxboim, A. A unified linear viscoelastic model of the cell nucleus defines the mechanical contributions of lamins and chromatin. Adv. Sci. 2020, 7, 1901222. [Google Scholar] [CrossRef] [PubMed]
- Janin, A.; Bauer, D.; Ratti, F.; Millat, G.; Mejat, A. Nuclear envelopathies: A complex LINC between nuclear envelope and pa-thology. Orphanet. J. Rare Dis. 2017, 12, 147. [Google Scholar] [CrossRef]
- Marcelot, A.; Worman, H.J.; Zinn-Justin, S. Protein structural and mechanistic basis of progeroid laminopathies. FEBS J. 2021, 288, 2757–2772. [Google Scholar] [CrossRef]
- Stiekema, M.; Van Zandvoort, M.A.M.J.; Ramaekers, F.C.S.; Broers, J.L.V. Structural and mechanical aberrations of the nuclear lamina in disease. Cells 2020, 9, 1884. [Google Scholar] [CrossRef]
- Tatli, M.; Medalia, O. Insight into the functional organization of nuclear lamins in health and disease. Curr. Opin. Cell Biol. 2018, 54, 72–79. [Google Scholar] [CrossRef]
- Wong, X.; Stewart, C.L. The laminopathies and the insights they provide into the structural and Functional organization of the nucleus. Annu. Rev. Genom. Hum. Genet 2020, 21, 263–288. [Google Scholar] [CrossRef]
- Jahn, D.; Schramm, S.; Schnolzer, M.; Heilmann, C.J.; de Koster, C.G.; Schutz, W.; Benavente, R.; Alsheimer, M. A truncated lamin A in the Lmna -/- mouse line: Implications for the understanding of laminopathies. Nucleus 2012, 3, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Tariq, Z.; Zhang, H.; Chia-Liu, A.; Shen, Y.; Gete, Y.; Xiong, Z.-M.; Tocheny, C.; Campanello, L.; Wu, D.; Losert, W.; et al. Lamin A and microtubules collaborate to maintain nuclear morphology. Nucleus 2017, 8, 433–446. [Google Scholar] [CrossRef]
- Smith, E.R.; Meng, Y.; Moore, R.; Tse, J.D.; Xu, A.G.; Xu, X.-X. Nuclear envelope structural proteins facilitate nuclear shape changes accompanying embryonic differentiation and fidelity of gene expression. BMC Cell Biol. 2017, 18, 8. [Google Scholar] [CrossRef]
- Coffinier, C.; Chang, S.Y.; Nobumori, C.; Tu, Y.; Farber, E.A.; Toth, J.I.; Fong, L.G.; Young, S. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. USA 2010, 107, 5076–5081. [Google Scholar] [CrossRef]
- Jevtić, P.; Edens, L.J.; Vuković, L.D.; Levy, D.L. Sizing and shaping the nucleus: Mechanisms and significance. Curr. Opin. Cell Biol. 2014, 28, 16–27. [Google Scholar] [CrossRef]
- Tamashunas, A.C.; Tocco, V.J.; Matthews, J.; Zhang, Q.; Atanasova, K.R.; Paschall, L.; Pathak, S.; Ratnayake, R.; Stephens, A.D.; Luesch, H.; et al. High-throughput gene screen reveals modulators of nuclear shape. Mol. Biol. Cell 2020, 31, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Bergo, M.O.; Gavino, B.; Ross, J.; Schmidt, W.K.; Hong, C.; Kendall, L.V.; Mohr, A.; Meta, M.; Genant, H.; Jiang, Y.; et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. USA 2002, 99, 13049–13054. [Google Scholar] [CrossRef] [PubMed]
- Pendas, A.M.; Zhou, Z.; Cadiñanos, J.; Freije, J.M.; Wang, J.; Hultenby, K.; Astudillo, A.; Wernerson, A.; Rodríguez, F.; Tryggvason, K.; et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice. Nat. Genet. 2002, 31, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-J.; Meers, O.; Buschbeck, M.; Heidel, F. The role of MacroH2A histone variants in cancer. Cancers 2021, 13, 3003. [Google Scholar] [CrossRef] [PubMed]
- Cantarino, N.; Douet, J.; Buschbeck, M. MacroH2A—An epigenetic regulator of cancer. Cancer Lett. 2013, 336, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Giallongo, S.; Re, O.L.; Vinciguerra, M. Macro histone variants: Emerging rheostats of gastrointestinal cancers. Cancers 2019, 11, 676. [Google Scholar] [CrossRef]
- Lavigne, A.-C.; Castells, M.; Mermet, J.; Kocanova, S.; Dalvai, M.; Bystricky, K. Increased macroH2A1.1 expression correlates with poor survival of triple-negative breast cancer patients. PLoS ONE 2014, 9, e98930. [Google Scholar] [CrossRef]
- Yang, G.; Yao, Y.; Wu, D.; Guo, H.; Zhou, S.; Sun, D.; Guo, X.; Zheng, T.; Wang, J.; Zhang, S.; et al. Upregulated mH2A1 serves as an unfavorable prognostic indicator and promotes the progress of hepa-tocellular carcinoma (HCC). Life Sci. 2020, 263, 118576. [Google Scholar] [CrossRef]
- Rochman, M.; Postnikov, Y.; Correll, S.; Malicet, C.; Wincovitch, S.; Karpova, T.S.; McNally, J.G.; Wu, X.; Bubunenko, N.A.; Grigoryev, S.; et al. The interaction of NSBP1/HMGN5 with nucleosomes in euchromatin counteracts linker histone-mediated chroma-tin compaction and modulates transcription. Mol. Cell. 2009, 35, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschenes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [PubMed]
- Biedzinski, S.; Agsu, G.; Vianay, B.; Delord, M.; Blanchoin, L.; Larghero, J.; Faivre, L.; Thery, M.; Brunet, S. Microtubules control nucle-ar shape and gene expression during early stages of hematopoietic differentiation. EMBO J. 2020, 39, e103957. [Google Scholar] [CrossRef] [PubMed]
- Heffler, J.; Shah, P.P.; Robison, P.; Phyo, S.; Veliz, K.; Uchida, K.; Bogush, A.; Rhoades, J.; Jain, R.; Prosser, B.L. A balance between intermediate filaments and microtubules maintains nuclear architecture in the cardiomyocyte. Circ. Res. 2020, 126, e10–e26. [Google Scholar] [CrossRef]
- Timme, S.; Schmitt, E.; Stein, S.; Schwarz-Finsterle, J.; Wagner, J.; Walch, A.; Werner, M.; Hausmann, M.; Wiech, T. Nuclear position and shape deformation of chromosome 8 territories in pancreatic ductal adenocarcinoma. Anal. Cell Pathol. 2011, 34, 21–33. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Du, Y.; Kong, X.; Li, Z.; Jia, Z.; Cui, J.; Gao, J.; Wang, G.; Xie, K. Lamin B1 is a novel therapeutic target of betulinic acid in pancreatic cancer. Clin. Cancer Res. 2013, 19, 4651–4661. [Google Scholar] [CrossRef]
- Kim, H.-J.; Hwang, S.-H.; Han, M.-E.; Baek, S.; Sim, H.-E.; Yoon, S.; Baek, S.-Y.; Kim, B.-S.; Kim, J.-H.; Kim, S.-Y.; et al. LAP2 Is Widely Overexpressed in Diverse Digestive Tract Cancers and Regulates Motility of Cancer Cells. PLOS ONE 2012, 7, e39482. [Google Scholar] [CrossRef]
- Trotter, K.W.; Archer, T.K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 2008, 6, e004. [Google Scholar] [CrossRef]
- Schick, S.; Rendeiro, A.F.; Runggatscher, K.; Ringler, A.; Boidol, B.; Hinkel, M.; Majek, P.; Vulliard, L.; Penz, T.; Parapatics, K.; et al. Systematic characterization of BAF mutations pro-vides insights into intracomplex synthetic lethalities in human cancers. Nat. Genet. 2019, 51, 1399–1410. [Google Scholar] [CrossRef]
- Shain, A.H.; Giacomini, C.P.; Matsukuma, K.; Karikari, C.A.; Bashyam, M.D.; Hidalgo, M.; Maitra, A.; Pollack, J.R. Convergent structur-al alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive com-plex in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E252–E259. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Marquez-Vilendrer, S.B.; Thompson, K.; Lu, L.; Reisman, D. Mechanism of BRG1 silencing in primary cancers. Oncotarget 2016, 7, 56153–56169. [Google Scholar] [CrossRef]
- Von Figura, G.; Fukuda, A.; Roy, N.; Liku, M.E.; Morris Iv, J.P.; Kim, G.E.; Russ, H.A.; Firpo, M.A.; Mulvihill, S.J.; Dawson, D.W.; et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mu-cinous neoplasm and pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2014, 16, 255–267. [Google Scholar] [CrossRef]
- Tsuda, M.; Fukuda, A.; Roy, N.; Hiramatsu, Y.; Leonhardt, L.; Kakiuchi, N.; Hoyer, K.; Ogawa, S.; Goto, N.; Ikuta, K.; et al. The BRG1/SOX9 axis is critical for acinar cell–derived pancreatic tumorigenesis. J. Clin. Investig. 2018, 128, 3475–3489. [Google Scholar] [CrossRef]
- Laurila, E.; Savinainen, K.; Kuuselo, R.; Karhu, R.; Kallioniemi, A. Characterization of the 7q21-q22 amplicon identifies ARPC1A, a subunit of the Arp2/3 complex, as a regulator of cell migration and invasion in pancreatic cancer. Genes Chromosom. Cancer 2009, 48, 330–339. [Google Scholar] [CrossRef]
- Laurila, E.; Vuorinen, E.; Savinainen, K.; Rauhala, H.; Kallioniemi, A. KPNA7, a nuclear transport receptor, promotes malignant properties of pancreatic cancer cells in vitro. Exp. Cell Res. 2014, 322, 159–167. [Google Scholar] [CrossRef]
- Network CGAR. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017, 32, 185–203. [Google Scholar] [CrossRef]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarci-noma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Lomberk, G.; Blum, Y.; Nicolle, R.; Nair, A.; Gaonkar, K.S.; Marisa, L.; Mathison, A.; Sun, Z.; Yan, H.; Elarouci, N.; et al. Distinct epige-netic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat. Commun. 2018, 9, 1978. [Google Scholar] [CrossRef]
- Martinez-Useros, J.; Martin-Galan, M.; Garcia-Foncillas, J. The match between molecular subtypes, histology and microenvi-ronment of pancreatic cancer and its relevance for chemoresistance. Cancers 2021, 13, 322. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual micro-dissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Lenkiewicz, E.; Malasi, S.; Hogenson, T.L.; Flores, L.F.; Barham, W.; Phillips, W.J.; Roesler, A.S.; Chambers, K.R.; Rajbhandari, N.; Hayashi, A.; et al. Genomic and epigenomic landscaping defines new therapeutic targets for adenosquamous carcinoma of the pancreas. Cancer Res. 2020, 80, 4324–4334. [Google Scholar] [CrossRef] [PubMed]
- Espinet, E.; Gu, Z.; Imbusch, C.D.; Giese, N.; Buscher, M.; Safavi, M.; Weisenburger, S.; Klein, C.; Vogel, V.; Falcone, M.; et al. Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell-of-origin. Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell of origin. Cancer Discov. 2021, 11, 638–659. [Google Scholar] [PubMed]
- Berman, B.P.; Weisenberger, D.J.; Aman, J.F.; Hinoue, T.; Ramjan, Z.; Liu, Y.; Noushmehr, H.; Lange, C.P.; van Dijk, C.M.; Tollenaar, R.A.; et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat. Genet. 2011, 44, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Digel, W.; Lubbert, M. DNA methylation disturbances as novel therapeutic target in lung cancer: Preclinical and clinical re-sults. Crit. Rev. Oncol. Hematol. 2005, 55, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nat. Cell Biol. 1983, 301, 89–92. [Google Scholar] [CrossRef]
- Martinelli, P.; Cañamero, M.; Del Pozo, N.; Madriles, F.; Zapata, A.G.; Real, F.X. Gata6is required for complete acinar differentiation and maintenance of the exocrine pancreas in adult mice. Gut 2012, 62, 1481–1488. [Google Scholar] [CrossRef]
- O’Kane, G.M.; Grünwald, B.T.; Jang, G.-H.; Masoomian, M.; Picardo, S.; Grant, R.C.; Denroche, R.E.; Zhang, A.; Wang, Y.; Lam, B.; et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin. Cancer Res. 2020, 26, 4901–4910. [Google Scholar] [CrossRef]
- Patil, S.; Steuber, B.; Kopp, W.; Kari, V.; Urbach, L.; Wang, X.; Küffer, S.; Bohnenberger, H.; Spyropoulou, D.; Zhang, Z.; et al. EZH2 regulates pancreatic cancer subtype identity and tumor progression via transcriptional repression of GATA6. Cancer Res. 2020, 80, 4620–4632. [Google Scholar] [CrossRef]
- Eyres, M.; Lanfredini, S.; Xu, H.; Burns, A.; Blake, A.; Willenbrock, F.; Goldin, R.; Hughes, D.; Hughes, S.; Thapa, A.; et al. TET2 drives 5hmc marking of GATA6 and epigenetically defines pancreatic ductal adenocarcinoma transcriptional subtypes. Gastroenterology 2021, 161, 653–668e16. [Google Scholar] [CrossRef]
- Melamed, P.; Yosefzon, Y.; David, C.; Tsukerman, A.; Pnueli, L. Tet enzymes, variants, and differential effects on function. front. Cell Dev. Biol. 2018, 6, 22. [Google Scholar] [CrossRef]
- Diao, Z.; Ji, Q.; Wu, Z.; Zhang, W.; Cai, Y.; Wang, Z.; Hu, J.; Liu, Z.; Wang, Q.; Bi, S.; et al. SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 2021, 49, 4203–4219. [Google Scholar] [CrossRef] [PubMed]
- Tocco, V.J.; Li, Y.; Christopher, K.G.; Matthews, J.H.; Aggarwal, V.; Paschall, L.; Luesch, H.; Licht, J.D.; Dickinson, R.B.; Lele, T.P. The nucleus is irreversibly shaped by motion of cell boundaries in cancer and non-cancer cells. J. Cell Physiol. 2018, 233, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Stephens, A.D.; Banigan, E.; Marko, J.F. Chromatin’s physical properties shape the nucleus and its functions. Curr. Opin. Cell Biol. 2019, 58, 76–84. [Google Scholar] [CrossRef]
- Smith, E.R.; Capo-Chichi, C.D.; Xu, X.X. Defective nuclear lamina in aneuploidy and carcinogenesis. Front. Oncol. 2018, 8, 529. [Google Scholar] [CrossRef]
- Santos, M.; Costa, P.; Martins, F.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.; Rebelo, S. LAP1 is a crucial protein for the mainte-nance of the nuclear envelope structure and cell cycle progression. Mol. Cell Biochem. 2015, 399, 143–153. [Google Scholar] [CrossRef]
- Von Appen, A.; LaJoie, D.; Johnson, I.E.; Trnka, M.J.; Pick, S.M.; Burlingame, A.L.; Ullman, K.S.; Frost, A. LEM2 phase separation pro-motes ESCRT-mediated nuclear envelope reformation. Nature 2020, 582, 115–118. [Google Scholar] [CrossRef]
- Mukherjee, R.N.; Chen, P.; Levy, D.L. Recent advances in understanding nuclear size and shape. Nucleus 2016, 7, 167–186. [Google Scholar] [CrossRef]
- Alhudiri, I.M.; Nolan, C.C.; Ellis, I.; Elzagheid, A.; Rakha, E.A.; Green, A.R.; Chapman, C.J. Expression of lamin A/C in early-stage breast cancer and its prognostic value. Breast Cancer Res. Treat. 2019, 174, 661–668. [Google Scholar] [CrossRef]
- Capo-chichi, C.D.; Cai, K.Q.; Smedberg, J.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.-X. Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin. J. Cancer 2011, 30, 415–425. [Google Scholar] [CrossRef]
- Matsumoto, A.; Hieda, M.; Yokoyama, Y.; Nishioka, Y.; Yoshidome, K.; Tsujimoto, M.; Matsuura, N. Global loss of a nuclear lamina component, lamin A/C, and LINC complex components SUN 1, SUN 2, and nesprin-2 in breast cancer. Cancer Med. 2015, 4, 1547–1557. [Google Scholar] [CrossRef]
- Wazir, U.; Ahmed, M.H.; Bridger, J.; Harvey, A.; Jiang, W.; Sharma, A.K.; Mokbel, K. The clinicopathological significance of lamin A/C, lamin B1 and lamin B receptor mRNA expression in human breast cancer. Cell. Mol. Biol. Lett. 2013, 18, 595–611. [Google Scholar] [CrossRef]
- Abdalla, F.B.E.; Markus, R.; Buhmeida, A.; Boder, J.; Syrjänen, K.; Collan, Y. Estrogen receptor, progesterone receptor, and nuclear size features in female breast cancer in Libya: Correlation with clinical features and survival. Anticancer Res. 2012, 32, 3485–3493. [Google Scholar]
- Hayward, M.K.; Louise Jones, J.; Hall, A.; King, L.; Ironside, A.J.; Nelson, A.C.; Shelley Hwang, E.; Weaver, V.M. Derivation of a nuclear heterogeneity image index to grade DCIS. Comput. Struct. Biotechnol. J. 2020, 18, 4063–4070. [Google Scholar] [CrossRef]
- Kalhan, S.; Dubey, S.; Sharma, S.; Dudani, S.; Preeti; Dixit, M. Significance of nuclear morphometry in cytological aspirates of breast masses. J. Cytol. 2010, 27, 16–21. [Google Scholar] [CrossRef]
- Capo-chichi, C.D.; Cai, K.Q.; Simpkins, F.; Ganjei-Azar, P.; Godwin, A.K.; Xu, X.X. Nuclear envelope structural defects cause chromo-somal numerical instability and aneuploidy in ovarian cancer. BMC Med. 2011, 9, 28. [Google Scholar] [CrossRef]
- Irianto, J.; Pfeifer, C.R.; Ivanovska, I.L.; Swift, J.; Discher, D.E. Nuclear lamins in cancer. Cell. Mol. Bioeng. 2016, 9, 258–267. [Google Scholar] [CrossRef]
- Lochs, S.J.A.; Kefalopoulou, S.; Kind, J. Lamina associated domains and gene regulation in development and cancer. Cells 2019, 8, 271. [Google Scholar] [CrossRef]
- Briand, N.; Collas, P. Laminopathy-causing lamin A mutations reconfigure lamina-associated domains and local spatial chromatin conformation. Nucleus 2018, 9, 216–226. [Google Scholar] [CrossRef]
- Chang, L.; Li, M.; Shao, S.; Li, C.; Ai, S.; Xue, B.; Hou, Y.; Zhang, Y.; Li, R.; Fan, X.; et al. Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell 2020, 1–23. [Google Scholar] [CrossRef]
- Coste Pradas, J.; Auguste, G.; Matkovich, S.J.; Lombardi, R.; Chen, S.N.; Garnett, T.; Chamberlain, K.; Riyad, J.M.; Weber, T.; Singh, S.K.; et al. Identification of genes and pathways regulated by lamin A in heart. J. Am. Heart Assoc. 2020, 9, e015690. [Google Scholar] [CrossRef]
- Galiová, G.; Bártová, E.; Raška, I.; Krejčí, J.; Kozubek, S. Chromatin changes induced by lamin A/C deficiency and the histone deacetylase inhibitor trichostatin A. Eur. J. Cell Biol. 2008, 87, 291–303. [Google Scholar] [CrossRef]
- Malhas, A.; Lee, C.F.; Sanders, R.; Saunders, N.J.; Vaux, D.J. Defects in lamin B1 expression or processing affect interphase chromo-some position and gene expression. J. Cell Biol. 2007, 176, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Reichart, B.; Klafke, R.; Dreger, C.; Kruger, E.; Motsch, I.; Ewald, A.; Schafer, J.; Reichmann, H.; Muller, C.R.; Dabauvalle, M.C. Expression and localization of nuclear proteins in autosomal-dominant Emery-Dreifuss muscular dystrophy with LMNA R377H mutation. BMC Cell Biol. 2004, 5, 12. [Google Scholar] [CrossRef]
- Singh, M.; Hunt, C.R.; Pandita, R.K.; Kumar, R.; Yang, C.R.; Horikoshi, N.; Bachoo, R.; Serag, S.; Story, M.D.; Shay, J.W.; et al. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell Biol. 2013, 33, 1210–1222. [Google Scholar] [CrossRef]
- Chen, N.Y.; Kim, P.H.; Fong, L.G.; Young, S.G. Nuclear membrane ruptures, cell death, and tissue damage in the setting of nuclear lamin deficiencies. Nucleus 2020, 11, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.-H.; Factor, R.E.; Ullman, K.S. The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer 2012, 12, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Dubik, N.; Mai, S. Lamin A/C: Function in normal and tumor cells. Cancers 2020, 12, 3688. [Google Scholar] [CrossRef]
- Fischer, E.G. Nuclear Morphology and the Biology of Cancer Cells. Acta Cytol. 2020, 64, 511–519. [Google Scholar] [CrossRef]
- Maynard, S.; Keijzers, G.; Akbari, M.; Ben Ezra, M.; Hall, A.; Morevati, M.; Scheibye-Knudsen, M.; Gonzalo, S.; Bartek, J.; Bohr, V.A. Lamin A/C promotes DNA base excision repair. Nucleic Acids Res. 2019, 47, 11709–11728. [Google Scholar] [CrossRef] [PubMed]
- Cenni, V.; Squarzoni, S.; Loi, M.; Mattioli, E.; Lattanzi, G.; Capanni, C. Emerin phosphorylation during the early phase of the oxidative stress response influences emerin-BAF interaction and BAF nuclear localization. Cells 2020, 9, 1415. [Google Scholar] [CrossRef]
- Gong, G.; Chen, P.; Li, L.; Tan, H.; Zhou, J.; Zhou, Y.; Yang, X.; Wu, X. Loss of lamin A but not lamin C expression in epithelial ovarian cancer cells is associated with metastasis and poor prognosis. Pathol. Res. Pract. 2015, 211, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wu, L.; Weng, D.; Xu, D.; Geng, J.; Zhao, F. Reduced expression of lamin A/C correlates with poor histological differentia-tion and prognosis in primary gastric carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 8. [Google Scholar] [CrossRef]
- Maciejowski, J.; Hatch, E.M. Nuclear membrane rupture and its consequences. Annu. Rev. Cell Dev. Biol. 2020, 36, 85–114. [Google Scholar] [CrossRef]
- Capo-Chichi, C.D.; Yeasky, T.M.; Smith, E.R.; Xu, X.X. Nuclear envelope structural defect underlies the main cause of aneuploidy in ovarian carcinogenesis. BMC Cell Biol. 2016, 17, 37. [Google Scholar] [CrossRef]
- Kong, L.; Schäfer, G.; Bu, H.; Zhang, Y.; Zhang, Y.; Klocker, H. Lamin A/C protein is overexpressed in tissue-invading prostate cancer and promotes prostate cancer cell growth, migration and invasion through the PI3K/AKT/PTEN pathway. Carcinogenesis 2012, 33, 751–759. [Google Scholar] [CrossRef]
- Liu, H.; Li, D.; Zhou, L.; Kan, S.; He, G.; Zhou, K.; Wang, L.; Chen, M.; Shu, W. LMNA functions as an oncogene in hepatocellular carcinoma by regulating the proliferation and migration ability. J. Cell Mol. Med. 2020, 24, 12008–12019. [Google Scholar] [CrossRef]
- Lautscham, L.A.; Kammerer, C.; Lange, J.R.; Kolb, T.; Mark, C.; Schilling, A.; Strissel, P.L.; Strick, R.; Gluth, C.; Rowat, A.C.; et al. Migration in confined 3D environments is determined by a combination of adhesiveness, nuclear volume, contractility, and cell stiffness. Biophys. J. 2015, 109, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Rizzotto, A.; Schirmer, E.C. Breaking the scale: How disrupting the karyoplasmic ratio gives cancer cells an advantage for metastatic invasion. Biochem. Soc. Trans. 2017, 45, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, R.N.; Salle, J.; Dmitrieff, S.; Nelson, K.M.; Oakey, J.; Minc, N.; Levy, D.L. The perinuclear ER scales nuclear size independently of cell size in early embryos. Dev. Cell. 2020, 54, 395–409 e7. [Google Scholar] [CrossRef]
- Ting, D.T.; Lipson, D.; Paul, S.; Brannigan, B.W.; Akhavanfard, S.; Coffman, E.J.; Contino, G.; Deshpande, V.; Iafrate, A.J.; Letovsky, S.; et al. Aberrant overexpression of satellite repeats in pancreatic and other epi-thelial cancers. Science 2011, 331, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Cournac, A.; Koszul, R.; Mozziconacci, J. The 3D folding of metazoan genomes correlates with the association of similar repeti-tive elements. Nucleic Acids Res. 2016, 44, 245–255. [Google Scholar] [CrossRef]
- Mangiavacchi, A.; Liu, P.; Della Valle, F.; Orlando, V. New insights into the functional role of retrotransposon dynamics in mammalian somatic cells. Cell. Mol. Life Sci. 2021, 78, 5245–5256. [Google Scholar] [CrossRef] [PubMed]
- Van de Werken, H.J.G.; Haan, J.C.; Feodorova, Y.; Bijos, D.; Weuts, A.; Theunis, K.; Holwerda, S.J.B.; Meuleman, W.; Pagie, L.; Thanisch, K.; et al. Small chromosomal regions posi-tion themselves autonomously according to their chromatin class. Genome Res. 2017, 27, 922–933. [Google Scholar] [CrossRef] [PubMed]
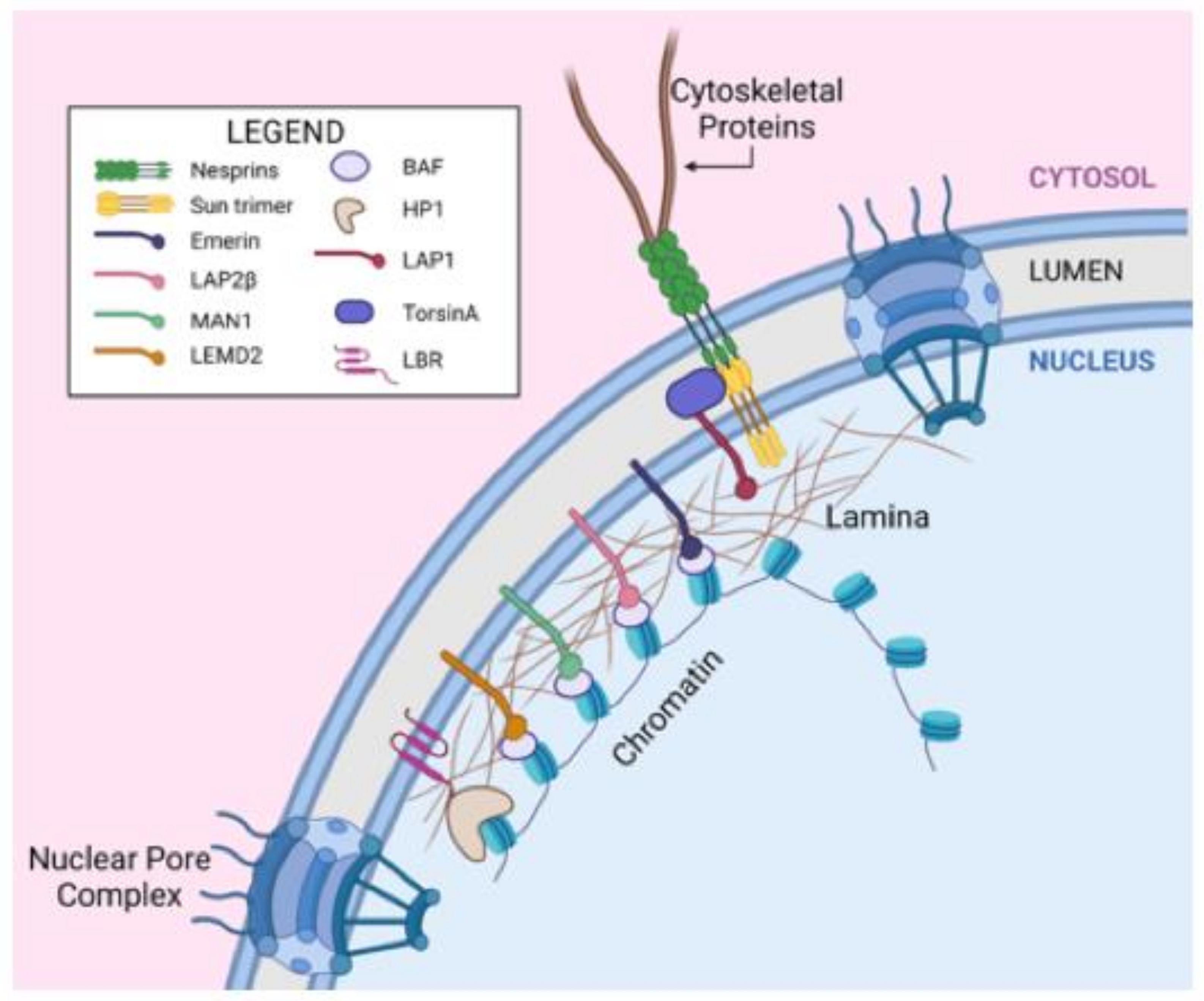
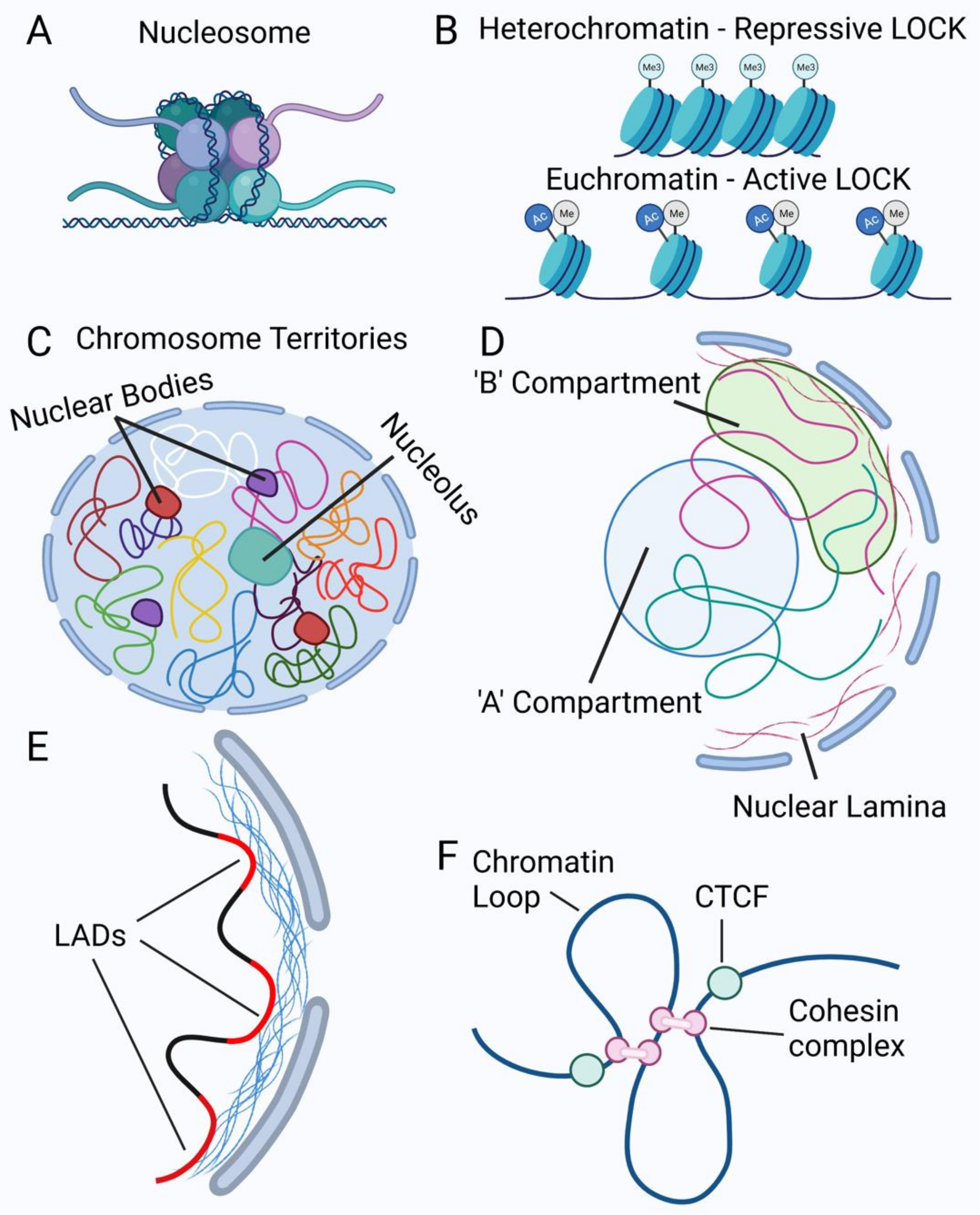

| Protein Alteration | Nuclear Changes | References |
|---|---|---|
| Lamins and Associated Proteins | ||
| Lamin A mutation | Lobulations | [68,69,70,71] |
| Lamin A truncation | Increased area, blebs, lobulations, aneuploidy | [72,73,74] |
| Lamin B1 truncation | Blebs | [74,75,76] |
| Lamin B2 deletion | Elongation | [76] |
| Lamin B2 deletion | Ruptures | [77] |
| Lamin A, B1 or B2 depletion | Decreased area | [78] |
| Lamin A, B1 or B2 overexpression | Increased area | [78] |
| Emerin mutation | Increased area | [79] |
| Emerin deletion | Increased area | [73] |
| Emerin depletion | Increased area, lobulations, blebs | [79,80] |
| Emerin depletion | Reduced area, invaginations | [81] |
| Emerin overexpression | Increased nuclear area | [82] |
| LEM2 depletion | Lobulations | [66] |
| LAP1 mutation | cytoplasmic channels, lobulations, invaginations | [83,84] |
| LAP1 deletion | Ruffled | [85] |
| LAP1 overexpression | Lobulations | [86] |
| LAP1C overexpression | Invaginations | [87] |
| LAP2b depletion | Increased area, hyperploidy | [67] |
| ANKLE2 depletion | Lobulations, increased area, hyperploidy | [55,63,65] |
| ZMPSTE24 mutation | Lobulations | [88,89] |
| LINC Complex Proteins | ||
| Nesprin 1 mutation | Lobulations | [90] |
| Nesprin 1 or Nesprin 2 depletion | Lobulations, increased area | [91,92] |
| SUN1 mutation | Enhance blebs in Lamin A mutant cells | [93] |
| SUN1/SUN2 depletion | Lobulations | [94,95] |
| SUN2 overexpression | Lobulations | [96] |
| Torsin deletion | Intraluminal blebs | [97,98] |
| Torsin 1 overexpression | Blebs, invaginations | [99] |
| Chromatin Enzymes | ||
| BRG1 depletion | Lobulations | [100] |
| BRG1-ATPase deficient | Increased area | [101] |
| ARID1A | Increased area | [102] |
| RING1B depletion | Increased area, hyperploidy | [103] |
| MOF deletion | Blebs, micronuclei | [104] |
| NCAPH2 or NCAPD3 depletion | Lobulations, increased area | [105] |
| SMC2 depletion | Lobulations | [105] |
| Nucleosome Proteins | ||
| mH2A1 and mH2A2 deletion | Lobulations, blebs, increased area | [106,107] |
| HMGN5 overexpression | Blebs | [108] |
| Nuclear Pore-Related Proteins | ||
| ELYS depletion | Decreased size | [109] |
| KPNA7 depletion | Lobulations | [110] |
| NUP53 depletion | Lobulations | [111] |
| NUP98 depletion | Lobulations | [111,112] |
| NUP153 depletion | Lobulations, invaginations | [113] |
| Cytoskeletal-Associated Proteins | ||
| Cofilin and ADF depletion | Lobulations | [114] |
| DIAPH3 depletion | Lobulations | [80] |
| a-dystrobrevin depletion | Lobulations, blebs, septa | [115] |
| EPB41 depletion | Blebs, lobulations | [116] |
| LLGL1 or LLGL2 depletion | Increased area | [117] |
| PPP1R12A or PPP1CB depletion | Lobulations, blebs | [118] |
| Other | ||
| GATA6 decrease | Larger size, lobulations, aneuploidy | [119] |
| NOP53 depletion | Lobulations | [120] |
| Nucleophosmin depletion | Lobulations | [121] |
| SIRT2 depletion | Increased area | [65] |
| SPANX depletion | Increased area, lobulations | [122] |
| STIP1 depletion | Reduced size, invaginations | [81] |
| TMEM170A depletion | Increased area, lobulations | [123] |
| YBX1 depletion | Lobulations | [124] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flores, L.F.; Tader, B.R.; Tolosa, E.J.; Sigafoos, A.N.; Marks, D.L.; Fernandez-Zapico, M.E. Nuclear Dynamics and Chromatin Structure: Implications for Pancreatic Cancer. Cells 2021, 10, 2624. https://doi.org/10.3390/cells10102624
Flores LF, Tader BR, Tolosa EJ, Sigafoos AN, Marks DL, Fernandez-Zapico ME. Nuclear Dynamics and Chromatin Structure: Implications for Pancreatic Cancer. Cells. 2021; 10(10):2624. https://doi.org/10.3390/cells10102624
Chicago/Turabian StyleFlores, Luis F., Brooke R. Tader, Ezequiel J. Tolosa, Ashley N. Sigafoos, David L. Marks, and Martin E. Fernandez-Zapico. 2021. "Nuclear Dynamics and Chromatin Structure: Implications for Pancreatic Cancer" Cells 10, no. 10: 2624. https://doi.org/10.3390/cells10102624
APA StyleFlores, L. F., Tader, B. R., Tolosa, E. J., Sigafoos, A. N., Marks, D. L., & Fernandez-Zapico, M. E. (2021). Nuclear Dynamics and Chromatin Structure: Implications for Pancreatic Cancer. Cells, 10(10), 2624. https://doi.org/10.3390/cells10102624







