Terc Gene Cluster Variants Predict Liver Telomere Length in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experiment 1
2.1.1. Experiment 1: Overview
2.1.2. Experiment 1: Subjects
2.1.3. Experiment 1: Liver Dissection and DNA Extraction
2.1.4. Experiment 1: Telomere Length Quantification
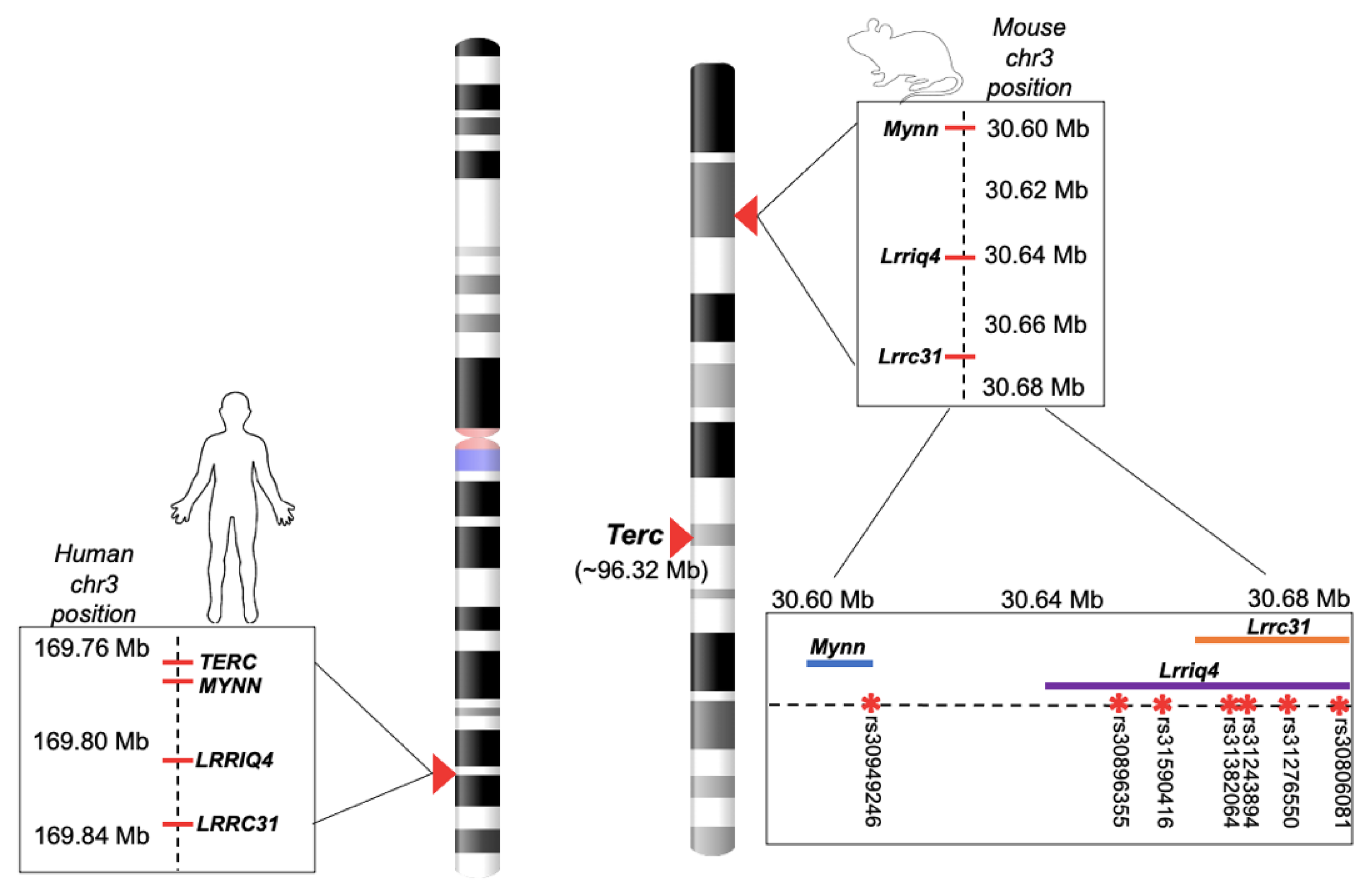
2.1.5. Experiment 1: SNP Query and Genotyping
2.1.6. Experiment 1: Statistical Analyses
2.2. Experiment 2
2.2.1. Experiment 2: Overview
2.2.2. Experiment 2: Strain Selection
2.2.3. Experiment 2: Subjects
2.2.4. Experiment 2: Liver Dissection and DNA Extraction
2.2.5. Experiment 2: Telomere Length Quantification
2.2.6. Experiment 2: Statistical Analyses
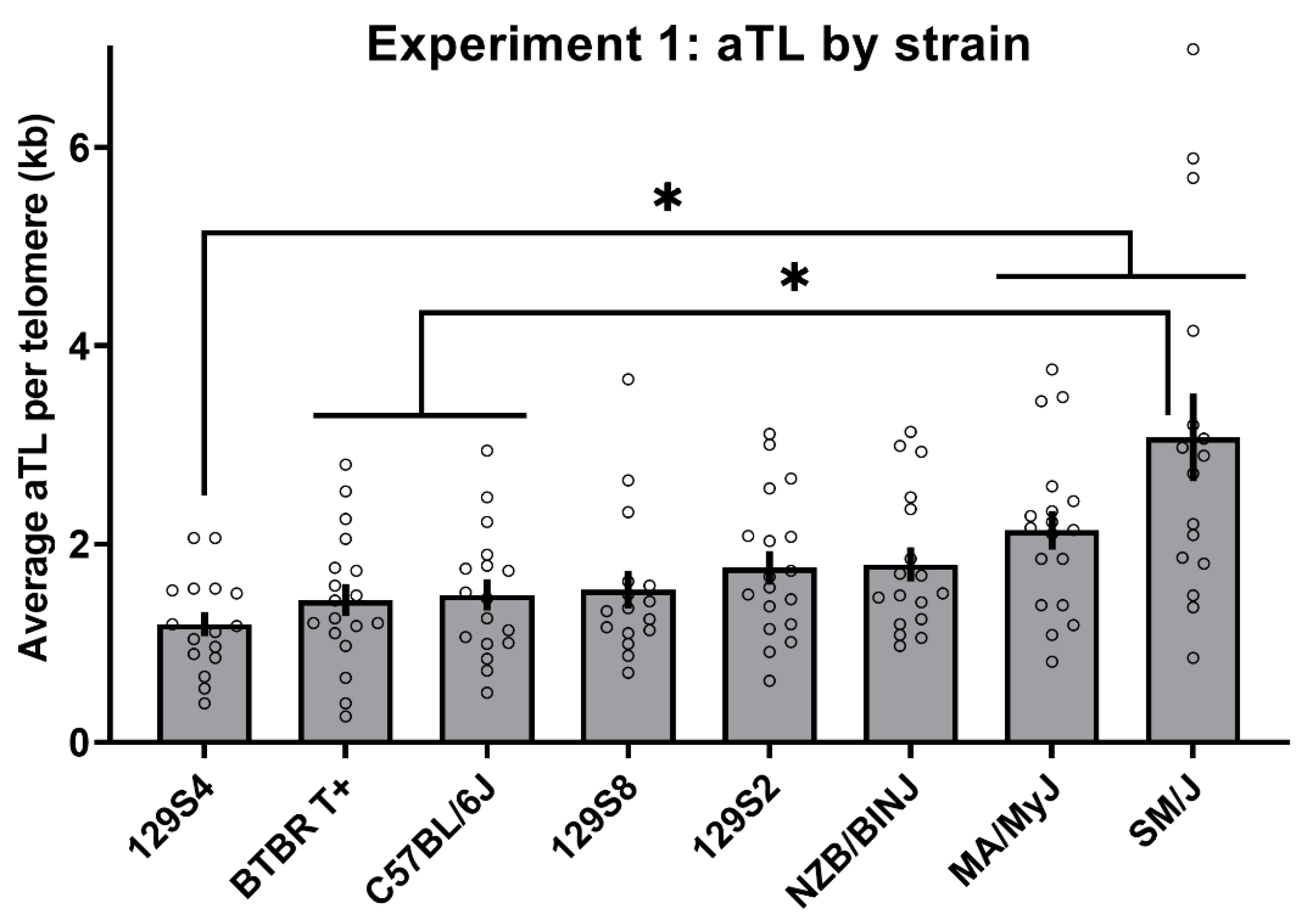
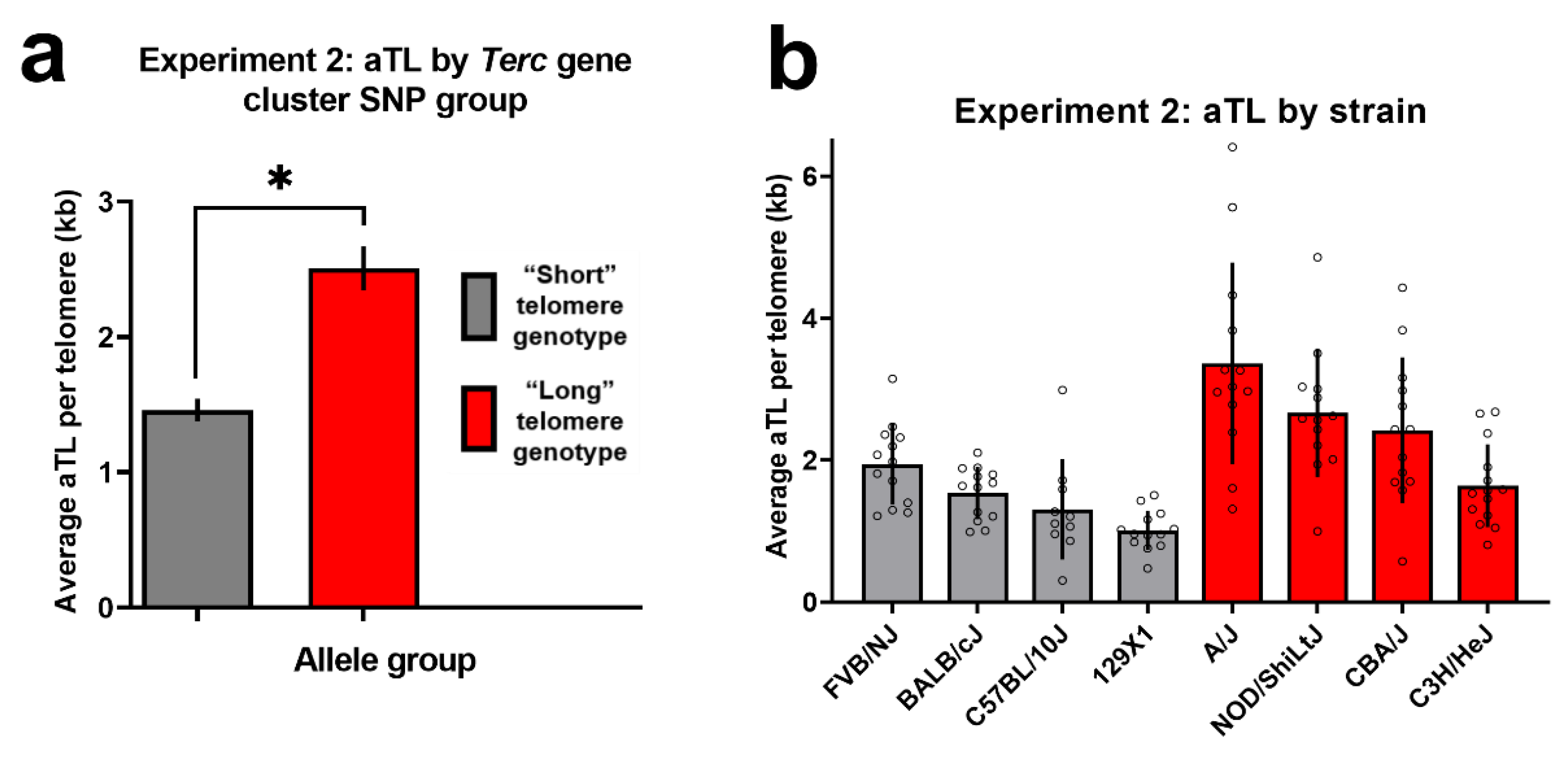
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Greider, C.W. Telomeres, telomerase and senescence. Bioessays 1990, 12, 363–369. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Telomeres, telomerase and cancer. Sci. Am. 1996, 274, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.Z.; Allsopp, R.C.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere end-replication problem and cell aging. J. Mol. Biol. 1992, 225, 951–960. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Greider, C.W.; Henderson, E.; Lee, M.S.; Shampay, J.; Shippen-Lentz, D. Recognition and elongation of telomeres by telomerase. Genome 1989, 31, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Prowse, K.R.; Greider, C.W. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. USA 1995, 92, 4818–4822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calado, R.T.; Dumitriu, B. Telomere dynamics in mice and humans. In Seminars in Hematology; WB Saunders: Philadelphia, PA, USA, 2013; Volume 50, pp. 165–174. [Google Scholar]
- Broer, L.; Codd, V.; Nyholt, D.R.; Deelen, J.; Mangino, M.; Willemsen, G.; Albrecht, E.; Amin, N.; Beekman, M.; De Geus, E.J.; et al. Meta-analysis of telomere length in 19 713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. 2013, 21, 1163–1168. [Google Scholar] [CrossRef] [Green Version]
- Njajou, O.T.; Cawthon, R.M.; Damcott, C.M.; Wu, S.H.; Ott, S.; Garant, M.J.; Blackburn, E.H.; Mitchell, B.D.; Shuldiner, A.R.; Hsueh, W.C. Telomere length is paternally inherited and is associated with parental lifespan. Proc. Natl. Acad. Sci. USA 2007, 104, 12135–12139. [Google Scholar] [CrossRef] [Green Version]
- Al Khaldi, R.; Mojiminiyi, O.; AlMulla, F.; Abdella, N. Associations of TERC single nucleotide polymorphisms with human leukocyte telomere length and the risk of type 2 diabetes mellitus. PLoS ONE 2015, 10, e0145721. [Google Scholar] [CrossRef] [PubMed]
- Codd, V.; Mangino, M.; Van Der Harst, P.; Braund, P.S.; Kaiser, M.; Beveridge, A.J.; Rafelt, S.; Moore, J.; Nelson, C.; Soranzo, N.; et al. Common variants near TERC are associated with mean telomere length. Nat. Genet. 2010, 42, 197–199. [Google Scholar] [CrossRef] [Green Version]
- Jones, A.M.; Beggs, A.D.; Carvajal-Carmona, L.; Farrington, S.; Tenesa, A.; Walker, M.; Howarth, K.; Ballereau, S.; Hodgson, S.V.; Zauber, A.; et al. TERC polymorphisms are associated both with susceptibility to colorectal cancer and with longer telomeres. Gut 2012, 61, 248–254. [Google Scholar] [CrossRef] [Green Version]
- Kroupa, M.; Rachakonda, S.; Vymetalkova, V.; Tomasova, K.; Liska, V.; Vodenkova, S.; Cumova, A.; Rossnerova, A.; Vodickova, L.; Hemminki, K.; et al. Telomere length in peripheral blood lymphocytes related to genetic variation in telomerase, prognosis and clinicopathological features in breast cancer patients. Mutagenesis 2020, 35, 491–497. [Google Scholar] [CrossRef]
- Sethi, I.; Sharma, V.; Sharma, I.; Singh, G.; Bhat, G.R.; Bhanwer, A.S.; Sharma, S.; Rai, E. Telomere maintenance genes are associated with type 2 diabetes susceptibility in northwest Indian population group. Sci. Rep. 2020, 10, 6444. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.; Zhang, Z.; Yu, L.; Cao, L.; Zhou, D.; Kan, M.; Li, B.; Zhang, D.; He, L.; Liu, Y. Common variants near TERC are associated with leukocyte telomere length in the Chinese Han population. Eur. J. Hum. Genet. 2011, 19, 721–723. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Sun, F.; Cui, X.; Janz, S. Genetic predisposition to multiple myeloma. Blood Genom. 2020, 4, 9–18. [Google Scholar] [CrossRef]
- Polat, F.; Yilmaz, M.; Diler, S.B. The association of MYNN and TERC gene polymorphisms and bladder cancer in a Turkish population. Urol. J. 2019, 16, 50–55. [Google Scholar]
- Went, M. Deciphering Genetic Susceptibility to Multiple Myeloma. Ph.D. Thesis, University of London, London, UK, 2020. [Google Scholar]
- Kolishovski, G.; Lamoureux, A.; Hale, P.; Richardson, J.E.; Recla, J.M.; Adesanya, O.; Simons, A.; Kunde-Ramamoorthy, G.; Bult, C.J. The JAX Synteny Browser for mouse-human comparative genomics. Mamm. Genome 2019, 30, 353–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Hong, W.Y.; Cho, M.; Sim, M.; Lee, D.; Ko, Y.; Kim, J. Synteny Portal: A web-based application portal for synteny block analysis. Nucleic Acids Res. 2016, 44, W35–W40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, W.E.; Shay, J.W. Telomere dynamics in cancer progression and prevention: Fundamental differences in human and mouse telomere biology. Nat. Med. 2000, 6, 849–851. [Google Scholar] [CrossRef] [PubMed]
- Manning, E.L.; Crossland, J.; Dewey, M.J.; Van Zant, G. Influences of inbreeding and genetics on telomere length in mice. Mamm. Genome 2002, 13, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Hemann, M.T.; Strong, M.A.; Hao, L.Y.; Greider, C.W. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 2001, 107, 67–77. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Blasco, M.A.; Gottlieb, G.J.; Horner, J.W.; Greider, C.W.; DePinho, R.A. Essential role of mouse telomerase in highly proliferative organs. Nature 1998, 392, 569–574. [Google Scholar] [CrossRef]
- Muñoz-Lorente, M.A.; Cano-Martin, A.C.; Blasco, M.A. Mice with hyper-long telomeres show less metabolic aging and longer lifespans. Nat. Commun. 2019, 10, 4723. [Google Scholar] [CrossRef] [Green Version]
- Bednarek, A.; Budunova, I.; Slaga, T.J.; Aldaz, C.M. Increased telomerase activity in mouse skin premalignant progression. Cancer Res. 1995, 55, 4566–4569. [Google Scholar]
- Broccoli, D.; Godley, L.A.; Donehower, L.A.; Varmus, H.E.; De Lange, T. Telomerase activation in mouse mammary tumors: Lack of detectable telomere shortening and evidence for regulation of telomerase RNA with cell proliferation. Mol. Cell. Biol. 1996, 16, 3765–3772. [Google Scholar] [CrossRef] [Green Version]
- O’Callaghan, N.J.; Fenech, M. A quantitative PCR method for measuring absolute telomere length. Biol. Proced. Online 2011, 13, 3. [Google Scholar] [CrossRef] [Green Version]
- Cawthon, R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002, 30, e47. [Google Scholar] [CrossRef]
- Cooper, J.C.; Dealtry, G.B.; Ahmed, M.A.; Arck, P.C.; Klapp, B.F.; Blois, S.M.; Fernández, N. An impaired breeding phenotype in mice with a genetic deletion of beta-2 microglobulin and diminished MHC class I expression: Role in reproductive fitness. Biol. Reprod. 2007, 77, 274–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakao, R.; Okauchi, H.; Hashimoto, C.; Wada, N.; Oishi, K. Determination of reference genes that are independent of feeding rhythms for circadian studies of mouse metabolic tissues. Mol. Genet. Metab. 2017, 121, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Grubb, S.C.; Bult, C.J.; Bogue, M.A. Mouse phenome database. Nucleic Acids Res. 2014, 42, D825–D834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, J.A.; Lloyd, S.; Hafezparast, M.; Lennon-Pierce, M.; Eppig, J.T.; Festing, M.F.; Fisher, E.M. Genealogies of mouse inbred strains. Nat. Genet. 2000, 24, 23–25. [Google Scholar] [CrossRef] [PubMed]
- White, H. A heteroskedasticity-consistent covariance matrix estimator and a direct test for heteroskedasticity. Econom. J. Econom. Soc. 1980, 48, 817–838. [Google Scholar] [CrossRef]
- Harrell, F.E. Ordinal logistic regression. In Regression Modeling Strategies; Springer: Cham, Switzerland, 2015; pp. 311–325. [Google Scholar]
- Kipling, D.; Cooke, H.J. Hypervariable ultra-long telomeres in mice. Nature 1990, 347, 400–402. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Gu, Y.; Xu, W.; Nie, D.; Zhang, D.; Dai, J.; Zhao, X.; Zhang, M.; Wang, Z.; Chen, Z.; Qiao, Z. Nicotine induces Nme2-mediated apoptosis in mouse testes. Biochem. Biophys. Res. Commun. 2016, 472, 573–579. [Google Scholar] [CrossRef]
- Vera, E.; de Jesus, B.B.; Foronda, M.; Flores, J.M.; Blasco, M.A. The rate of increase of short telomeres predicts longevity in mammals. Cell Rep. 2012, 2, 732–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, E.L.; Richardson, D.S. Sex differences in telomeres and lifespan. Aging Cell 2011, 10, 913–921. [Google Scholar] [CrossRef]
- Watson, R.L.; Bird, E.J.; Underwood, S.; Wilbourn, R.V.; Fairlie, J.; Watt, K.; Salvo-Chirnside, E.; Pilkington, J.G.; Pemberton, J.M.; McNeilly, T.N.; et al. Sex differences in leucocyte telomere length in a free-living mammal. Mol. Ecol. 2017, 26, 3230–3240. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.A.; James, J.R.; Siegel, S.J.; Gould, T.J. Withdrawal from chronic nicotine administration impairs contextual fear conditioning in C57BL/6 mice. J. Neurosci. 2005, 25, 8708–8713. [Google Scholar] [CrossRef] [Green Version]
- Portugal, G.S.; Wilkinson, D.S.; Kenney, J.W.; Sullivan, C.; Gould, T.J. Strain-dependent effects of acute, chronic, and withdrawal from chronic nicotine on fear conditioning. Behav. Genet. 2012, 42, 133–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeid, D.; Mooney-Leber, S.; Seemiller, L.R.; Goldberg, L.R.; Gould, T.J. Terc Gene Cluster Variants Predict Liver Telomere Length in Mice. Cells 2021, 10, 2623. https://doi.org/10.3390/cells10102623
Zeid D, Mooney-Leber S, Seemiller LR, Goldberg LR, Gould TJ. Terc Gene Cluster Variants Predict Liver Telomere Length in Mice. Cells. 2021; 10(10):2623. https://doi.org/10.3390/cells10102623
Chicago/Turabian StyleZeid, Dana, Sean Mooney-Leber, Laurel R. Seemiller, Lisa R. Goldberg, and Thomas J. Gould. 2021. "Terc Gene Cluster Variants Predict Liver Telomere Length in Mice" Cells 10, no. 10: 2623. https://doi.org/10.3390/cells10102623
APA StyleZeid, D., Mooney-Leber, S., Seemiller, L. R., Goldberg, L. R., & Gould, T. J. (2021). Terc Gene Cluster Variants Predict Liver Telomere Length in Mice. Cells, 10(10), 2623. https://doi.org/10.3390/cells10102623





