Mitochondrial DNA and Exercise: Implications for Health and Injuries in Sports
Abstract
:1. Introduction
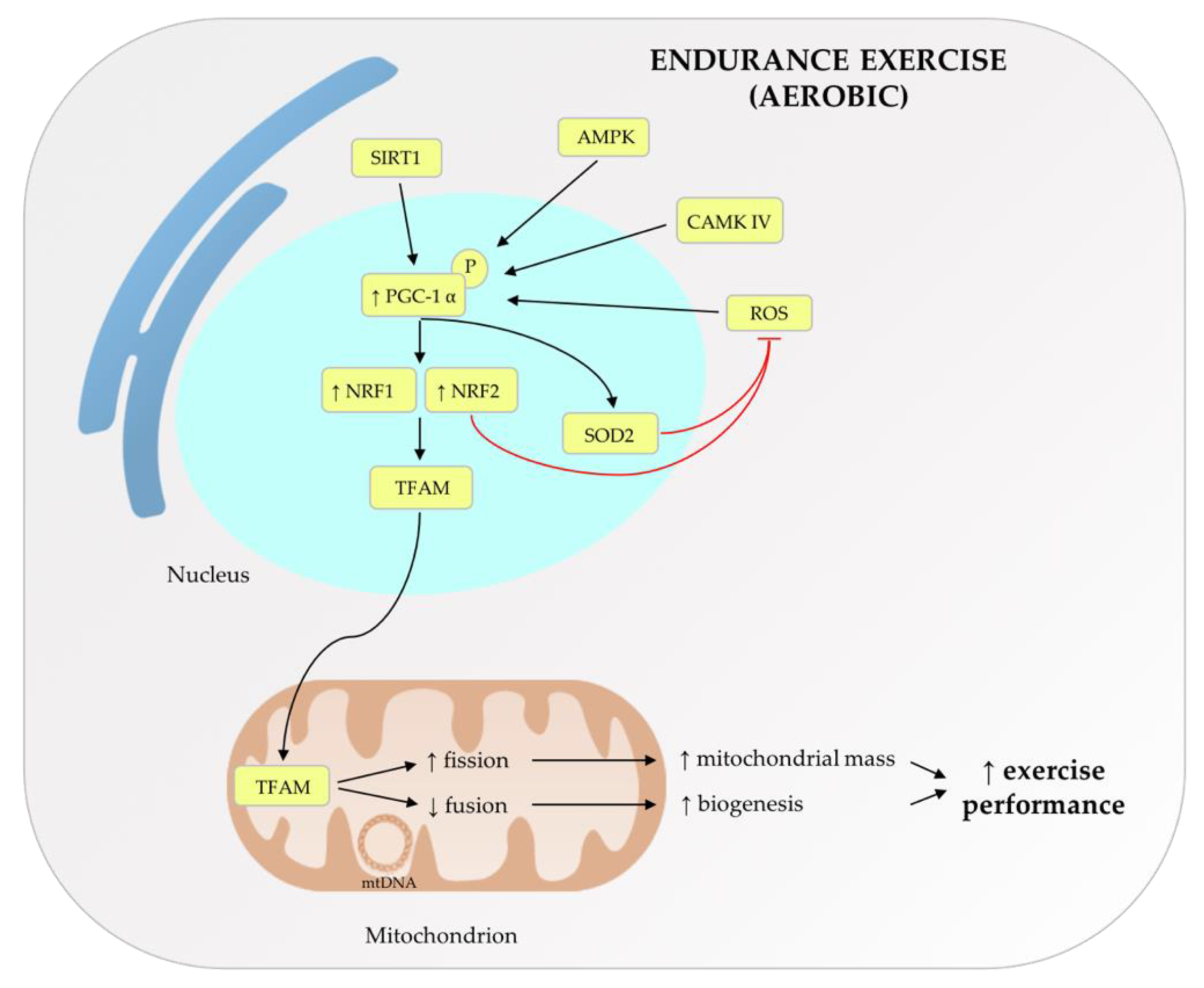
2. Role of Mitochondrial Metabolism and mtDNA Haplogroups on Exercise
3. Mitochondrial DNA as a Signaling and Pro-Inflammatory Molecule
3.1. Cellular Pathways Triggered by cf-mtDNA
3.2. Effects of cf-mtDNA on Central Nervous System
4. Release, Forms and Detection of cf-mtDNA
4.1. Mechanisms of Release and Removal of cf-mtDNA
4.2. Detection of cf-mtDNA in Plasma
5. The Effects of Trauma/Injuries on cf-mtDNA Release
5.1. The Effects of Traumatic Events on cf-mtDNA Release
5.2. Injuries/Trauma in Sports
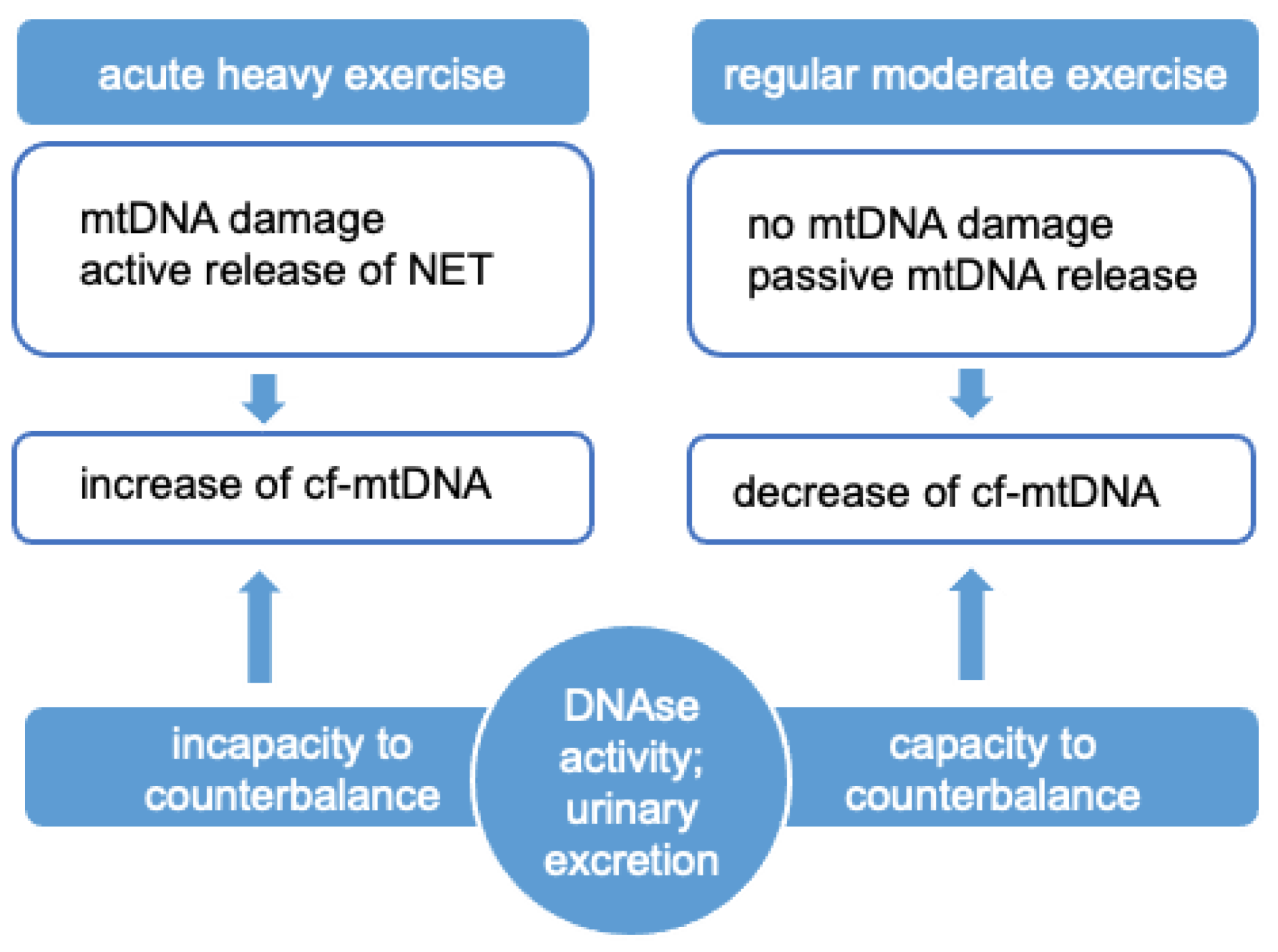
6. The Effects of Exercise on cf-mtDNA Release
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jee, H.; Park, J.; Oh, J.-G.; Lee, Y.-H.; Shin, K.-A.; Kim, Y.-J. Effect of a Prolonged Endurance Marathon on Vascular Endothelial and Inflammation Markers in Runners with Exercise-Induced Hypertension. Am. J. Phys. Med. Rehabil. 2013, 92, 513–522. [Google Scholar] [CrossRef]
- Kilbaugh, T.J.; Lvova, M.; Karlsson, M.; Zhang, Z.; Leipzig, J.; Wallace, D.C.; Margulies, S.S. Peripheral Blood Mitochondrial DNA as a Biomarker of Cerebral Mitochondrial Dysfunction following Traumatic Brain Injury in a Porcine Model. PLoS ONE 2015, 10, e0130927. [Google Scholar] [CrossRef] [Green Version]
- Beiter, T.; Fragasso, A.; Hudemann, J.; Nieß, A.M.; Simon, P. Short-Term Treadmill Running as a Model for Studying Cell-Free DNA Kinetics In Vivo. Clin. Chem. 2011, 57, 633–636. [Google Scholar] [CrossRef] [Green Version]
- Helmig, S.; Frühbeis, C.; Krämer-Albers, E.-M.; Simon, P.; Tug, S. Release of bulk cell free DNA during physical exercise occurs independent of extracellular vesicles. Graefe’s Arch. Eur. J. Appl. Physiol. 2015, 115, 2271–2280. [Google Scholar] [CrossRef]
- Shockett, P.E.; Khanal, J.; Sitaula, A.; Oglesby, C.; Meachum, W.A.; Castracane, V.D.; Kraemer, R.R. Plasma cell-free mitochondrial DNA declines in response to prolonged moderate aerobic exercise. Physiol. Rep. 2016, 4, e12672. [Google Scholar] [CrossRef] [PubMed]
- Stawski, R.; Walczak, K.; Kosielski, P.; Meissner, P.; Budlewski, T.; Padula, G.; Nowak, D. Repeated bouts of exhaustive exercise increase circulating cell free nuclear and mitochondrial DNA without development of tolerance in healthy men. PLoS ONE 2017, 12, e178216. [Google Scholar] [CrossRef] [PubMed]
- Blatteau, J.-E.; Gaillard, S.; De Maistre, S.; Richard, S.; Louges, P.; Gempp, E.; Druelles, A.; Lehot, H.; Morin, J.; Castagna, O.; et al. Reduction in the Level of Plasma Mitochondrial DNA in Human Diving, Followed by an Increase in the Event of an Accident. Front. Physiol. 2018, 9, 1695. [Google Scholar] [CrossRef]
- Hummel, E.M.; Hessas, E.; Müller, S.; Beiter, T.; Fisch, M.; Eibl, A.; Wolf, O.T.; Giebel, B.; Platen, P.; Kumsta, R.; et al. Cell-free DNA release under psychosocial and physical stress conditions. Transl. Psychiatry 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ohlsson, L.; Hall, A.; Lindahl, H.; Danielsson, R.; Gustafsson, A.; Lavant, E.; Ljunggren, L. Increased level of circulating cell-free mitochondrial DNA due to a single bout of strenuous physical exercise. Eur. J. Appl. Physiol. 2020, 120, 897–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, K.; Stawski, R.; Perdas, E.; Brzezinska, O.; Kosielski, P.; Galczynski, S.; Budlewski, T.; Padula, G.; Nowak, D. Circulating cell free DNA response to exhaustive exercise in average trained men with type I diabetes mellitus. Sci. Rep. 2021, 11, 4639. [Google Scholar] [CrossRef]
- Alexeyev, M.F. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009, 276, 5768–5787. [Google Scholar] [CrossRef] [Green Version]
- Perovic, A.; Unić, A.; Dumic, J. Recreational scuba diving: Negative or positive effects of oxidative and cardiovascular stress? Biochem. Med. 2014, 24, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Buzzacott, P.; Pollock, N.W.; Rosenberg, M. Exercise intensity inferred from air consumption during recreational scuba diving. Diving Hyperb. Med. J. 2014, 44, 74–78. [Google Scholar]
- Nasi, M.; Patrizi, G.; Pizzi, C.; Landolfo, M.; Boriani, G.; Cas, A.D.; Cicero, A.F.; Fogacci, F.; Rapezzi, C.; Sisca, G.; et al. The role of physical activity in individuals with cardiovascular risk factors: An opinion paper from Italian Society of Cardiology-Emilia Romagna-Marche and SIC-Sport. J. Cardiovasc. Med. 2019, 20, 631–639. [Google Scholar] [CrossRef]
- Merry, T.L.; Chan, A.; Woodhead, J.S.T.; Reynolds, J.C.; Kumagai, H.; Kim, S.-J.; Lee, C. Mitochondrial-derived peptides in energy metabolism. Am. J. Physiol. Metab. 2020, 319, E659–E666. [Google Scholar] [CrossRef]
- Scott, R.A.; Fuku, N.; Onywera, V.O.; Boit, M.; Wilson, R.H.; Tanaka, M.; Goodwin, W.H.; Pitsiladis, Y.P. Mitochondrial Haplogroups Associated with Elite Kenyan Athlete Status. Med. Sci. Sports Exerc. 2009, 41, 123–128. [Google Scholar] [CrossRef]
- Picca, A.; Calvani, R.; Coelho-Junior, H.; Marzetti, E. Cell Death and Inflammation: The Role of Mitochondria in Health and Disease. Cells 2021, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Caielli, S.; Athale, S.; Domic, B.; Murat, E.; Chandra, M.; Banchereau, R.; Baisch, J.; Phelps, K.; Clayton, S.; Gong, M.; et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med. 2016, 213, 697–713. [Google Scholar] [CrossRef]
- Bae, J.H.; Jo, S.I.; Kim, S.J.; Lee, J.M.; Jeong, J.H.; Kang, J.S.; Cho, N.-J.; Kim, S.S.; Lee, E.Y.; Moon, J.-S. Circulating Cell-Free mtDNA Contributes to AIM2 Inflammasome-Mediated Chronic Inflammation in Patients with Type 2 Diabetes. Cells 2019, 8, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grazioli, S.; Pugin, J. Mitochondrial Damage-Associated Molecular Patterns: From Inflammatory Signaling to Human Diseases. Front. Immunol. 2018, 9, 832. [Google Scholar] [CrossRef]
- Zhong, F.; Liang, S.; Zhong, Z. Emerging Role of Mitochondrial DNA as a Major Driver of Inflammation and Disease Progression. Trends Immunol. 2019, 40, 1120–1133. [Google Scholar] [CrossRef]
- Koltai, E.; Hart, N.; Taylor, A.W.; Goto, S.; Ngo, J.K.; Davies, K.J.A.; Radak, Z. Age-associated declines in mitochondrial biogenesis and protein quality control factors are minimized by exercise training. Am. J. Physiol. Integr. Comp. Physiol. 2012, 303, R127–R134. [Google Scholar] [CrossRef] [Green Version]
- Erlich, A.T.; Tryon, L.D.; Crilly, M.J.; Memme, J.M.; Moosavi, Z.S.M.; Oliveira, A.N.; Beyfuss, K.; Hood, D.A. Function of specialized regulatory proteins and signaling pathways in exercise-induced muscle mitochondrial biogenesis. Integr. Med. Res. 2016, 5, 187–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefàno, E.; Marsigliante, S.; Vetrugno, C.; Muscella, A. Is mitochondrial DNA profiling predictive for athletic performance? Mitochondrion 2019, 47, 125–138. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta 2011, 1813, 1269–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leick, L.; Lyngby, S.S.; Wojtaszewski, J.; Pilegaard, H. PGC-1α is required for training-induced prevention of age-associated decline in mitochondrial enzymes in mouse skeletal muscle. Exp. Gerontol. 2010, 45, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Rygiel, K.A.; Picard, M.; Turnbull, D. The ageing neuromuscular system and sarcopenia: A mitochondrial perspective. J. Physiol. 2016, 594, 4499–4512. [Google Scholar] [CrossRef] [PubMed]
- Nogueiras, R.; Habegger, K.M.; Chaudhary, N.; Finan, B.; Banks, A.S.; Dietrich, M.O.; Horvath, T.L.; Sinclair, D.A.; Pfluger, P.T.; Tschöop, M.H. Sirtuin 1 and Sirtuin 3: Physiological Modulators of Metabolism. Physiol. Rev. 2012, 92, 1479–1514. [Google Scholar] [CrossRef] [Green Version]
- Newman, J.C.; He, W.; Verdin, E. Mitochondrial Protein Acylation and Intermediary Metabolism: Regulation by Sirtuins and Implications for Metabolic Disease. J. Biol. Chem. 2012, 287, 42436–42443. [Google Scholar] [CrossRef] [Green Version]
- Hebert, A.S.; Dittenhafer-Reed, K.; Yu, W.; Bailey, D.J.; Selen, E.S.; Boersma, M.D.; Carson, J.J.; Tonelli, M.; Balloon, A.J.; Higbee, A.J.; et al. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Mol. Cell 2013, 49, 186–199. [Google Scholar] [CrossRef] [Green Version]
- Radak, Z.; Bori, Z.; Koltai, E.; Fatouros, I.G.; Jamurtas, A.Z.; Douroudos, I.I.; Terzis, G.; Nikolaidis, M.G.; Chatzinikolaou, A.; Sovatzidis, A.; et al. Age-dependent changes in 8-oxoguanine-DNA glycosylase activity are modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic. Biol. Med. 2011, 51, 417–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, S.L.; Rougé, P.M.-D.; Lanza, I.; McCrady-Spitzer, S.K.; Levine, J.A.; Middha, S.; Carter, R.E.; Klaus, K.A.; Therneau, T.M.; Highsmith, E.W.; et al. Mitochondrial Aging and Physical Decline: Insights from Three Generations of Women. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2015, 70, 1409–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, A.-M.; Adhihetty, P.J.; Buford, T.W.; Wohlgemuth, S.E.; Lees, H.A.; Nguyen, L.M.-D.; Aranda, J.M.; Sandesara, B.D.; Pahor, M.; Manini, T.M.; et al. The impact of aging on mitochondrial function and biogenesis pathways in skeletal muscle of sedentary high- and low-functioning elderly individuals. Aging Cell 2012, 11, 801–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radak, Z.; Torma, F.; Berkes, I.; Goto, S.; Mimura, T.; Posa, A.; Balogh, L.; Boldogh, I.; Suzuki, K.; Higuchi, M.; et al. Exercise effects on physiological function during aging. Free Radic. Biol. Med. 2019, 132, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Son, J.M.; Benayoun, B.A.; Lee, C. The Mitochondrial-Encoded Peptide MOTS-c Translocates to the Nucleus to Regulate Nuclear Gene Expression in Response to Metabolic Stress. Cell Metab. 2018, 28, 516–524.e7. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, J.C.; Lai, R.W.; Woodhead, J.S.T.; Joly, J.H.; Mitchell, C.J.; Cameron-Smith, D.; Lu, R.; Cohen, P.; Graham, N.A.; Benayoun, B.A.; et al. MOTS-c is an exercise-induced mitochondrial-encoded regulator of age-dependent physical decline and muscle homeostasis. Nat. Commun. 2021, 12, 470. [Google Scholar] [CrossRef]
- Woodhead, J.S.T.; D’Souza, R.F.; Hedges, C.P.; Wan, J.; Berridge, M.V.; Cameron-Smith, D.; Cohen, P.; Hickey, A.J.R.; Mitchell, C.J.; Merry, T.L. High-intensity interval exercise increases humanin, a mitochondrial encoded peptide, in the plasma and muscle of men. J. Appl. Physiol. 2020, 128, 1346–1354. [Google Scholar] [CrossRef]
- Jafari, A.; Hosseinpourfaizi, M.A.; Houshmand, M.; Ravasi, A.A. Effect of aerobic exercise training on mtDNA deletion in soleus muscle of trained and untrained Wistar rats. Br. J. Sports Med. 2005, 39, 517–520. [Google Scholar] [CrossRef]
- De Luca, A.; Nasi, M.; Di Giambenedetto, S.; Cozzi-Lepri, A.; Pinti, M.; Marzocchetti, A.; Mussini, C.; Fabbiani, M.; Bracciale, L.; Cauda, R.; et al. Mitochondrial DNA haplogroups and incidence of lipodystrophy in HIV-infected patients on long-term antiretroviral therapy. JAIDS J. Acquir. Immune Defic. Syndr. 2012, 59, 113–120. [Google Scholar] [CrossRef]
- Nasi, M.; Guaraldi, G.; Orlando, G.; Durante, C.; Pinti, M.; Nemes, E.; Nardini, G.; Passarino, G.; Cocchi, M.; Esposito, R.; et al. Mitochondrial DNA Haplogroups and Highly Active Antiretroviral Therapy—Related Lipodystrophy. Clin. Infect. Dis. 2008, 47, 962–968. [Google Scholar] [CrossRef] [Green Version]
- Niemi, A.-K.; Majamaa, K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur. J. Hum. Genet. 2005, 13, 965–969. [Google Scholar] [CrossRef]
- Castro, M.G.; Terrados, N.; Reguero, J.R.; Alvarez, V.; Coto, E. Mitochondrial haplogroup T is negatively associated with the status of elite endurance athlete. Mitochondrion 2007, 7, 354–357. [Google Scholar] [CrossRef]
- Miyamoto-Mikami, E.; Fuku, N.; Takahashi, H.; Ohiwa, N.; Scott, R.; Pitsiladis, Y.P.; Higuchi, M.; Kawahara, T.; Tanaka, M. Mitochondrial haplogroups associated with elite Japanese athlete status. Br. J. Sports Med. 2011, 45, 1179–1183. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef] [Green Version]
- West, A.P.; Shadel, G.S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 2017, 17, 363–375. [Google Scholar] [CrossRef] [PubMed]
- White, M.J.; McArthur, K.; Metcalf, D.; Lane, R.M.; Cambier, J.; Herold, M.; van Delft, M.; Bedoui, S.; Lessene, G.; Ritchie, M.; et al. Apoptotic Caspases Suppress mtDNA-Induced STING-Mediated Type I IFN Production. Cell 2014, 159, 1549–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, A.P.; Khoury-Hanold, W.; Staron, M.; Tal, M.; Pineda, C.M.; Lang, S.M.; Bestwick, M.; Duguay, B.A.; Raimundo, N.; MacDuff, D.A.; et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 2015, 520, 553–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Chiu, Y.-H.; Chen, Z.J. The cGAS-cGAMP-STING Pathway of Cytosolic DNA Sensing and Signaling. Mol. Cell 2014, 54, 289–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, G.N. STING: Infection, inflammation and cancer. Nat. Rev. Immunol. 2015, 15, 760–770. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Sun, L.; Chen, Z. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016, 17, 1142–1149. [Google Scholar] [CrossRef]
- Imajo, M.; Tsuchiya, Y.; Nishida, E. Regulatory mechanisms and functions of MAP kinase signaling pathways. IUBMB Life 2006, 58, 312–317. [Google Scholar] [CrossRef]
- Marongiu, L.; Gornati, L.; Artuso, I.; Zanoni, I.; Granucci, F. Below the surface: The inner lives of TLR4 and TLR9. J. Leukoc. Biol. 2019, 106, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Martinez, I.; Santoro, N.; Chen, Y.; Hoque, R.; Ouyang, X.; Caprio, S.; Shlomchik, M.J.; Coffman, R.L.; Candia, A.; Mehal, W.Z. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J. Clin. Investig. 2016, 126, 859–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, C.G.; Webb, R.C. The toll of the gridiron: Damage-associated molecular patterns and hypertension in American football. FASEB J. 2016, 30, 34–40. [Google Scholar] [CrossRef] [Green Version]
- Nakahira, K.; Haspel, J.A.; Rathinam, V.A.; Lee, S.-J.; Dolinay, T.; Lam, H.C.; Englert, J.A.; Rabinovitch, M.; Cernadas, M.; Kim, H.P.; et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011, 12, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Latz, E.; Xiao, T.; Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013, 13, 397–411. [Google Scholar] [CrossRef]
- Wen, H.; Miao, E.A.; Ting, J.P.-Y. Mechanisms of NOD-like Receptor-Associated Inflammasome. Act. Immun. 2013, 39, 432–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dib, B.; Lin, H.; Maidana, D.E.; Tian, B.; Miller, J.B.; Bouzika, P.; Miller, J.W.; Vavvas, D.G. Mitochondrial DNA has a pro-inflammatory role in AMD. Biochim. Biophys. Acta (BBA)—Bioenerg. 2015, 1853, 2897–2906. [Google Scholar] [CrossRef] [Green Version]
- Barnett, K.; Coronas-Serna, J.; Zhou, W.; Ernandes, M.J.; Cao, A.; Kranzusch, P.J.; Kagan, J.C. Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction between Self- and Viral DNA. Cell 2019, 176, 1432–1446.e11. [Google Scholar] [CrossRef] [Green Version]
- Gentili, M.; Lahaye, X.; Nadalin, F.; Nader, G.P.; Lombardi, E.P.; Hervé, S.; De Silva, N.S.; Rookhuizen, D.C.; Zueva, E.; Goudot, C.; et al. The N-Terminal Domain of cGAS Determines Preferential Association with Centromeric DNA and Innate Immune Activation in the Nucleus. Cell Rep. 2019, 26, 2377–2393.e13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wu, J.; Du, F.; Xu, H.; Sun, L.; Chen, Z.; Brautigam, C.; Zhang, X.; Chen, Z.J. The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop. Cell Rep. 2014, 6, 421–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diner, E.J.; Burdette, D.L.; Wilson, S.C.; Monroe, K.M.; Kellenberger, C.A.; Hyodo, M.; Hayakawa, Y.; Hammond, M.C.; Vance, R.E. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep. 2013, 3, 1355–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, J.S.; Quarato, G.; Cloix, C.; Lopez, J.; O’Prey, J.; Pearson, M.; Chapman, J.; Sesaki, H.; Carlin, L.M.; Passos, J.F.; et al. Mitochondrial inner membrane permeabilisation enables mt DNA release during apoptosis. EMBO J. 2018, 37, e99238. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging: An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef]
- Nasi, M.; De Biasi, S.; Gibellini, L.; Bianchini, E.; Pecorini, S.; Bacca, V.; Guaraldi, G.; Mussini, C.; Pinti, M.; Cossarizza, A. Ageing and inflammation in patients with HIV infection. Clin. Exp. Immunol. 2017, 187, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Pinti, M.; Cevenini, E.; Nasi, M.; De Biasi, S.; Salvioli, S.; Monti, D.; Benatti, S.; Gibellini, L.; Cotichini, R.; Stazi, M.A.; et al. Circulating mitochondrial DNA increases with age and is a familiar trait: Implications for “inflamm-aging”. Eur. J. Immunol. 2014, 44, 1552–1562. [Google Scholar] [CrossRef]
- Cossarizza, A.; Pinti, M.; Nasi, M.; Gibellini, L.; Manzini, S.; Roat, E.; De Biasi, S.; Bertoncelli, L.; Montagna, J.P.; Bisi, L.; et al. Increased plasma levels of extracellular mitochondrial DNA during HIV infection: A new role for mitochondrial damage-associated molecular patterns during inflammation. Mitochondrion 2011, 11, 750–755. [Google Scholar] [CrossRef]
- Nasi, M.; Cristani, A.; Pinti, M.; Lamberti, I.; Gibellini, L.; De Biasi, S.; Guazzaloca, A.; Trenti, T.; Cossarizza, A. Decreased Circulating mtDNA Levels in Professional Male Volleyball Players. Int. J. Sports Physiol. Perform. 2016, 11, 116–121. [Google Scholar] [CrossRef]
- Pinti, M.; Ferraro, D.; Nasi, M. Microglia activation: A role for mitochondrial DNA? Neural Regen. Res. 2021, 16, 2393–2394. [Google Scholar] [CrossRef]
- Nasi, M.; De Gaetano, A.; Bianchini, E.; De Biasi, S.; Gibellini, L.; Neroni, A.; Mattioli, M.; Pinti, M.; Tartaro, D.L.; Borella, R.; et al. Mitochondrial damage-associated molecular patterns stimulate reactive oxygen species production in human microglia. Mol. Cell. Neurosci. 2020, 108, 103538. [Google Scholar] [CrossRef]
- Simpson, D.S.A.; Oliver, P.L. ROS Generation in Microglia: Understanding Oxidative Stress and Inflammation in Neurodegenerative Disease. Antioxidants 2020, 9, 743. [Google Scholar] [CrossRef]
- Gambardella, S.; Limanaqi, F.; Ferese, R.; Biagioni, F.; Campopiano, R.; Centonze, D.; Fornai, F. ccf-mtDNA as a Potential Link Between the Brain and Immune System in Neuro-Immunological Disorders. Front. Immunol. 2019, 10, 1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podlesniy, P.; Figueiro-Silva, J.; Llado, A.; Antonell, A.; Sanchez-Valle, R.; Alcolea, D.; Lleo, A.; Molinuevo, J.L.; Serra, N.; Trullas, R. Low cerebrospinal fluid concentration of mitochondrial DNA in preclinical Alzheimer disease. Ann. Neurol. 2013, 74, 655–668. [Google Scholar] [CrossRef] [PubMed]
- Tapiola, T.; Alafuzoff, I.; Herukka, S.-K.; Parkkinen, L.; Hartikainen, P.; Soininen, H.; Pirttilä, T. Cerebrospinal Fluid β-Amyloid 42 and Tau Proteins as Biomarkers of Alzheimer-Type Pathologic Changes in the Brain. Arch. Neurol. 2009, 66, 382–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasi, M.; Bianchini, E.; De Biasi, S.; Gibellini, L.; Neroni, A.; Mattioli, M.; Pinti, M.; Iannone, A.; Mattioli, A.V.; Simone, A.M.; et al. Increased plasma levels of mitochondrial DNA and pro-inflammatory cytokines in patients with progressive multiple sclerosis. J. Neuroimmunol. 2020, 338, 577107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trumpff, C.; Michelson, J.; Lagranha, C.J.; Taleon, V.; Karan, K.R.; Sturm, G.; Lindqvist, D.; Fernström, J.; Moser, D.; Kaufman, B.A.; et al. Stress and circulating cell-free mitochondrial DNA: A systematic review of human studies, physiological considerations, and technical recommendations. Mitochondrion 2021, 59, 225–245. [Google Scholar] [CrossRef]
- Rostami, A.; Lambie, M.; Yu, C.W.; Stambolic, V.; Waldron, J.N.; Bratman, S.V. Senescence, Necrosis, and Apoptosis Govern Circulating Cell-free DNA Release Kinetics. Cell Rep. 2020, 31, 107830. [Google Scholar] [CrossRef]
- Suzuki, N.; Kamataki, A.; Yamaki, J.; Homma, Y. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta 2008, 387, 55–58. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Safdar, A.; Tarnopolsky, M.A. Exosomes as Mediators of the Systemic Adaptations to Endurance Exercise. Cold Spring Harb. Perspect. Med. 2018, 8, a29827. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Shi, H.; Zeng, T.; Liu, H.; Su, Y.; Cheng, X.; Ye, J.; Yin, Y.; Liu, M.; Zheng, H.; et al. Increased neutrophil extracellular traps activate NLRP3 and inflammatory macrophages in adult-onset Still’s disease. Arthritis Res. 2019, 21, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousefi, S.; Mihalache, C.C.O.; Kozlowski, E.; Schmid, I.; Simon, H.-U. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 2009, 16, 1438–1444. [Google Scholar] [CrossRef]
- Itagaki, K.; Kaczmarek, E.; Lee, Y.T.; Tang, I.T.; Isal, B.; Adibnia, Y.; Sandler, N.; Grimm, M.J.; Segal, B.H.; Otterbein, L.E.; et al. Mitochondrial DNA Released by Trauma Induces Neutrophil Extracellular Traps. PLoS ONE 2015, 10, e120549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Itagaki, K.; Hauser, C.J. Mitochondrial dna is released by shock and activates neutrophils via P38 map kinase. Shock 2010, 34, 55–59. [Google Scholar] [CrossRef]
- Loffek, S.; Schilling, O.; Franzke, C.-W. Series “matrix metalloproteinases in lung health and disease”: Biological role of matrix metalloproteinases: A critical balance. Eur. Respir. J. 2011, 38, 191–208. [Google Scholar] [CrossRef] [Green Version]
- Keyel, P.A. Dnases in health and disease. Dev. Biol. 2017, 429, 1–11. [Google Scholar] [CrossRef]
- Velders, M.; Treff, G.; Machus, K.; Bosnyák, E.; Steinacker, J.M.; Schumann, U. Exercise is a potent stimulus for enhancing circulating DNase activity. Clin. Biochem. 2014, 47, 471–474. [Google Scholar] [CrossRef] [Green Version]
- Strasser, A.; Jost, P.J.; Nagata, S. The Many Roles of FAS Receptor Signaling in the Immune System. Immunity 2009, 30, 180–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botezatu, I.; Serdyuk, O.; Potapova, G.; Shelepov, V.; Alechina, R.; Molyaka, Y.; Anan’Ev, V.; Bazin, I.; Garin, A.; Narimanov, M.; et al. Genetic Analysis of DNA Excreted in Urine: A New Approach for Detecting Specific Genomic DNA Sequences from Cells Dying in an Organism. Clin. Chem. 2000, 46, 1078–1084. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Ren, J.; Ren, H.; Wu, J.; Wu, X.; Liu, S.; Wang, G.; Gu, G.; Guo, K.; Li, J. Urinary Mitochondrial DNA Identifies Renal Dysfunction and Mitochondrial Damage in Sepsis-Induced Acute Kidney Injury. Oxidative Med. Cell. Longev. 2018, 2018, 8074936. [Google Scholar] [CrossRef]
- Nasi, M.; Bianchini, E.; Tartaro, D.L.; De Biasi, S.; Mattioli, M.; Paolini, A.; Gibellini, L.; Pinti, M.; De Gaetano, A.; D’Alisera, R.; et al. Effects of whole-body cryotherapy on the innate and adaptive immune response in cyclists and runners. Immunol. Res. 2020, 68, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Busani, S.; De Biasi, S.; Nasi, M.; Paolini, A.; Venturelli, S.; Tosi, M.; Girardis, M.; Cossarizza, A. Increased Plasma Levels of Mitochondrial DNA and Normal Inflammasome Gene Expression in Monocytes Characterize Patients With Septic Shock Due to Multidrug Resistant Bacteria. Front. Immunol. 2020, 11, 768. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; He, J.; Shen, L.; Fang, H.; Nie, H.; Jin, T.; Wei, X.; Xin, Y.; Jiang, Y.; Li, H.; et al. The mitochondrial DNA 4,977-bp deletion and its implication in copy number alteration in colorectal cancer. BMC Med. Genet. 2011, 12, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Qu, Y.; Gao, K.; Yang, Q.; Shi, B.; Hou, P.; Ji, M. High copy number of mitochondrial DNA (mtDNA) predicts good prognosis in glioma patients. Am. J. Cancer Res. 2015, 5, 1207–1216. [Google Scholar]
- Tsai, N.-W.; Lin, T.-K.; Chen, S.-D.; Chang, W.-N.; Wang, H.-C.; Yang, T.-M.; Lin, Y.-J.; Jan, C.-R.; Huang, C.-R.; Liou, C.-W.; et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin. Chim. Acta 2011, 412, 476–479. [Google Scholar] [CrossRef]
- McGill, M.R.; Staggs, V.S.; Sharpe, M.R.; Lee, W.M.; Jaeschke, H. Acute Liver Failure Study Group Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology 2014, 60, 1336–1345. [Google Scholar] [CrossRef] [Green Version]
- Pyle, A.; Burn, D.J.; Gordon, C.; Swan, C.; Chinnery, P.F.; Baudouin, S.V. Fall in circulating mononuclear cell mitochondrial DNA content in human sepsis. Intensive Care Med. 2010, 36, 956–962. [Google Scholar] [CrossRef] [Green Version]
- Becker, Y.; Loignon, R.-C.; Julien, A.-S.; Marcoux, G.; Allaeys, I.; Lévesque, T.; Rollet-Labelle, E.; Benk-Fortin, H.; Cloutier, N.; Melki, I.; et al. Anti-mitochondrial autoantibodies in systemic lupus erythematosus and their association with disease manifestations. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Lood, C.; Blanco, L.P.; Purmalek, M.; Carmona-Rivera, C.; De Ravin, S.S.; Smith, C.K.; Malech, H.L.; Ledbetter, J.A.; Elkon, K.B.; Kaplan, M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016, 22, 146–153. [Google Scholar] [CrossRef] [Green Version]
- Matyszewski, M.; Morrone, S.R.; Sohn, J. Digital signaling network drives the assembly of the AIM2-ASC inflammasome. Proc. Natl. Acad. Sci. USA 2018, 115, E1963–E1972. [Google Scholar] [CrossRef] [Green Version]
- Pinti, M.; Mussini, C.; Cossarizza, A. Mitochondrial DNA: A proinflammatory ‘enemy from within’ during HIV infection? Cell Death Dis. 2012, 3, e307. [Google Scholar] [CrossRef]
- Oka, T.; Hikoso, S.; Yamaguchi, O.; Taneike, M.; Takeda, T.; Tamai, T.; Oyabu, J.; Murakawa, T.; Nakayama, H.; Nishida, K.; et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nat. Cell Biol. 2012, 485, 251–255. [Google Scholar] [CrossRef]
- Terluk, M.R.; Kapphahn, R.J.; Soukup, L.M.; Gong, H.; Gallardo, C.; Montezuma, S.R.; Ferrington, D.A. Investigating Mitochondria as a Target for Treating Age-Related Macular Degeneration. J. Neurosci. 2015, 35, 7304–7311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Ren, J.; Wu, J.; Li, G.; Wu, X.; Liu, S.; Wang, G.; Gu, G.; Ren, H.; Hong, Z.; et al. Urinary Mitochondrial DNA Levels Identify Acute Kidney Injury in Surgical Critical Illness Patients. Shock 2017, 48, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, D.; Capili, A.; Choi, M.E. Mitochondrial dysfunction in kidney injury, inflammation, and disease: Potential therapeutic approaches. Kidney Res. Clin. Pract. 2020, 39, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Emma, F.; Montini, G.; Parikh, S.M.; Salviati, L. Mitochondrial dysfunction in inherited renal disease and acute kidney injury. Nat. Rev. Nephrol. 2016, 12, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Krysko, D.; Agostinis, P.; Krysko, O.; Garg, A.; Bachert, C.; Lambrecht, B.N.; Vandenabeele, P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011, 32, 157–164. [Google Scholar] [CrossRef]
- Simmons, J.D.; Lee, Y.-L.; Mulekar, S.; Kuck, J.L.; Brevard, S.B.; Gonzalez, R.P.; Gillespie, M.N.; Richards, W.O. Elevated Levels of Plasma Mitochondrial DNA DAMPs Are Linked to Clinical Outcome in Severely Injured Human Subjects. Ann. Surg. 2013, 258, 591–598. [Google Scholar] [CrossRef] [Green Version]
- Lam, N.Y.; Rainer, T.H.; Chiu, R.W.; Joynt, G.M.; Lo, Y.D. Plasma Mitochondrial DNA Concentrations after Trauma. Clin. Chem. 2004, 50, 213–216. [Google Scholar] [CrossRef] [Green Version]
- Filho, E.M.R.; Simon, D.; Ikuta, N.; Klovan, C.; Dannebrock, F.A.; de Oliveira, C.O.; Regner, A. Elevated Cell-Free Plasma DNA Level as an Independent Predictor of Mortality in Patients with Severe Traumatic Brain Injury. J. Neurotrauma 2014, 31, 1639–1646. [Google Scholar] [CrossRef] [Green Version]
- Prins, M.; Hales, A.; Reger, M.; Giza, C.; Hovda, D. Repeat Traumatic Brain Injury in the Juvenile Rat Is Associated with Increased Axonal Injury and Cognitive Impairments. Dev. Neurosci. 2010, 32, 510–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barkhoudarian, G.; Hovda, D.A.; Giza, C.C. The Molecular Pathophysiology of Concussive Brain Injury—An Update. Phys. Med. Rehabil. Clin. N. Am. 2016, 27, 373–393. [Google Scholar] [CrossRef]
- Faust, H.E.; Reilly, J.P.; Anderson, B.J.; Ittner, C.A.; Forker, C.M.; Zhang, P.; Weaver, B.A.; Holena, D.N.; Lanken, P.N.; Christie, J.D.; et al. Plasma Mitochondrial DNA Levels Are Associated with ARDS in Trauma and Sepsis Patients. Chest 2020, 157, 67–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulndreaj, A.; Tzekou, A.; Mothe, A.J.; Siddiqui, A.M.; Dragas, R.; Tator, C.H.; Torlakovic, E.E.; Fehlings, M.G. Characterization of the Antibody Response after Cervical Spinal Cord Injury. J. Neurotrauma 2017, 34, 1209–1226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.-C.; Lin, Y.-T.; Hsu, S.-Y.; Tsai, N.-W.; Lai, Y.-R.; Su, B.Y.-J.; Kung, C.-T.; Lu, C.-H. Serial plasma DNA levels as predictors of outcome in patients with acute traumatic cervical spinal cord injury. J. Transl. Med. 2019, 17, 329. [Google Scholar] [CrossRef] [Green Version]

| Pathway | Effects | Cell Type | References |
|---|---|---|---|
| cGAS-STING signaling | IFN-stimulated genes induction | Neutrophils Macrophages | [5,34,35,36] |
| TLR-9 | Proinflammatory cytokines and chemokines production IFN-I production | Hepatocytes Cardiomyocytes Leukocytes Neutrophils Endothelial cells | [18,37,38,39,40] |
| NLRP3 inflammasome | IL-1β and IL-18 activation | Macrophages RPE cells | [41,42,43,44] |
| References | Subjects | Age * (Years) | Type of Exercise | Effects on cf-mtDNA |
|---|---|---|---|---|
| [105] | n = 9 well-trained men | 29.3 ± 8.5 | Incremental exhaustive treadmill | =before, immediately after and after 30 min of rest |
| [106] | n = 5 healthy, physically active men | 26.8 ± 2.2 | Incremental exhaustive treadmill | =before, immediately after, 10, 30 and 90 min of rest |
| [51] | n = 12 male volleyball players; n = 20 non-athletes | 27.5 ± 3.9; 29.5 ± 4.5 | Volleyball | ↓ in the first in-season training period and = in the second in-season training period. |
| [107] | n = 7 healthy moderately trained men | 22.4 ± 1.2 | Ninety min treadmill | ↓ during exercise in comparison to control group (same subjects in seated position) |
| [108] | n = 12 average-trained men | 34.0 ± 5.2 | Exhaustive treadmill | ↑ post-exercise; ↓ pre-exercise over the study period |
| [109] | n = 8 free divers; n = 22 non-divers | 36.9 ± 9.6; 52.3 ± 14.5 | Diving | divers have less mtDNA than non-divers |
| [110] | n = 20 healthy males | 23.3 ± 3.8 | Incremental exhaustive treadmill | ↑ post-exercise; ↓ after 15 and 30 min of rest |
| [111] | n = 8 healthy volunteers (4 women and 4 men) | 38.6 ± 14.4 | Incremental exhaustive ergometer cycle | ↑ during exercise, compared to baseline values and after 30 and 90 min of rest |
| [112] | n = 14 men with T1DM **; n = 11 healthy controls | 29.3 ± 5.3; 34.0 ± 5.2 | Exhaustive treadmill run | =before and after exercise in both groups |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanini, G.; De Gaetano, A.; Selleri, V.; Savino, G.; Cossarizza, A.; Pinti, M.; Mattioli, A.V.; Nasi, M. Mitochondrial DNA and Exercise: Implications for Health and Injuries in Sports. Cells 2021, 10, 2575. https://doi.org/10.3390/cells10102575
Zanini G, De Gaetano A, Selleri V, Savino G, Cossarizza A, Pinti M, Mattioli AV, Nasi M. Mitochondrial DNA and Exercise: Implications for Health and Injuries in Sports. Cells. 2021; 10(10):2575. https://doi.org/10.3390/cells10102575
Chicago/Turabian StyleZanini, Giada, Anna De Gaetano, Valentina Selleri, Gustavo Savino, Andrea Cossarizza, Marcello Pinti, Anna Vittoria Mattioli, and Milena Nasi. 2021. "Mitochondrial DNA and Exercise: Implications for Health and Injuries in Sports" Cells 10, no. 10: 2575. https://doi.org/10.3390/cells10102575






