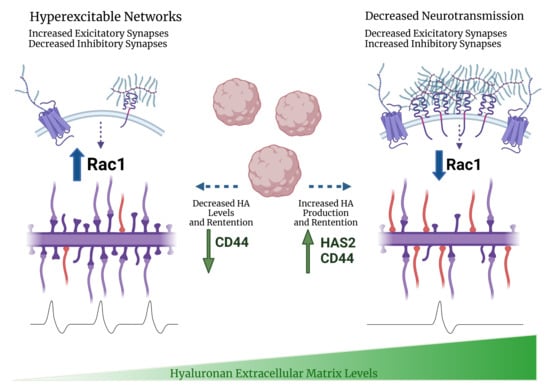
Synaptic Hyaluronan Synthesis and CD44-Mediated Signaling Coordinate Neural Circuit Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Lines
2.3. Three-Dimensional Cortical Spheroid Culture
2.4. HAS2 and CD44 Manipulation
2.5. Immunohistochemistry
2.6. Confocal Microscopy
2.7. Airyscan Microscopy
2.8. Three-Dimensional dSTORM Microscopy
2.9. Microelectrode Array Analysis
2.10. Western Blot Analysis
2.11. Quantitative Real Time PCR Analysis RT-PCR (qRT-PCR)
2.12. Statistical Analyses
3. Results
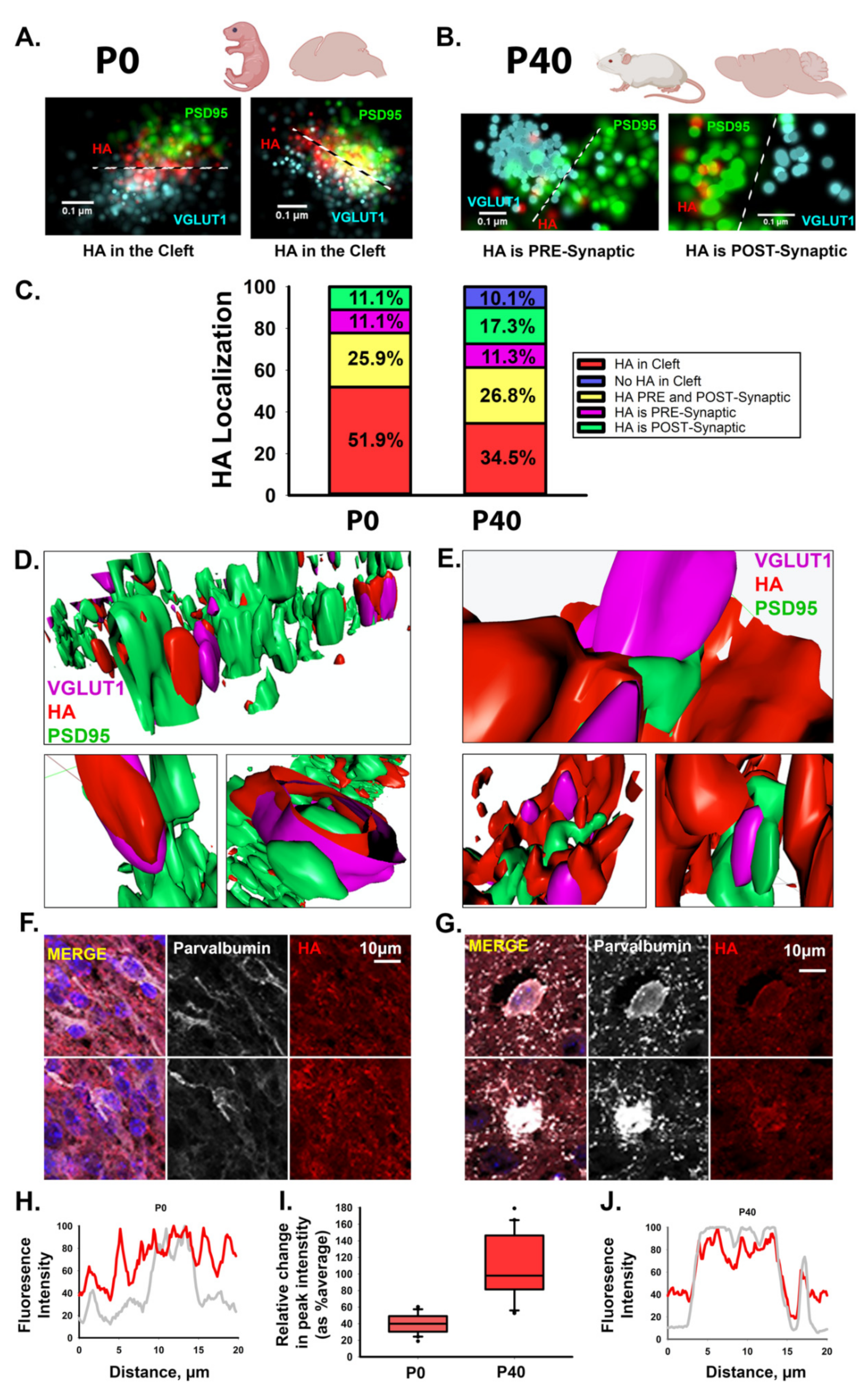
3.1. HA Localizes to the Synaptic Cleft of Excitatory Synapses during Cortical Development
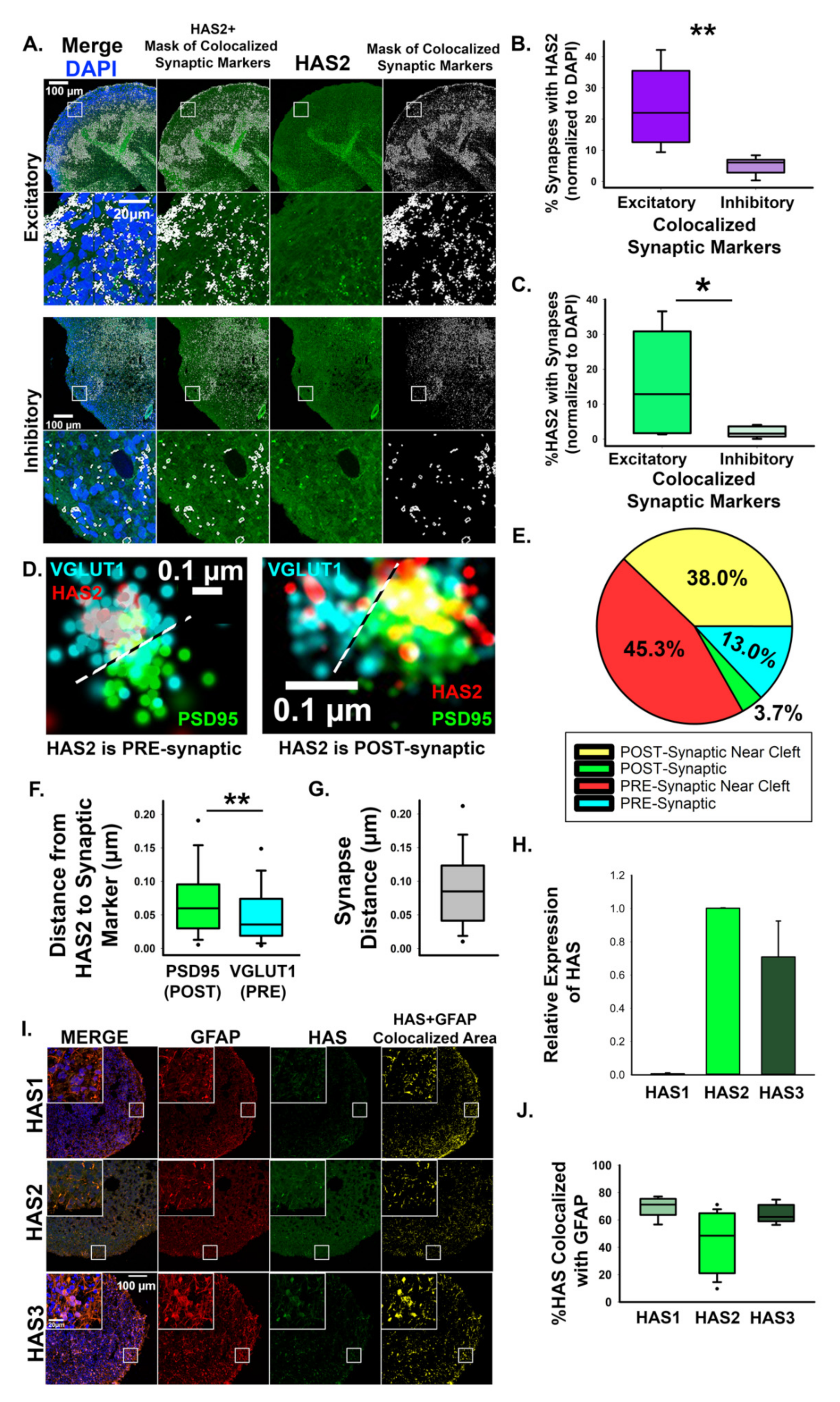
3.2. Manipulation of Hyaluronan Synthase 2 Alters HA Synthesis, Synapse Formation, and Excitability of Developing Neural Networks
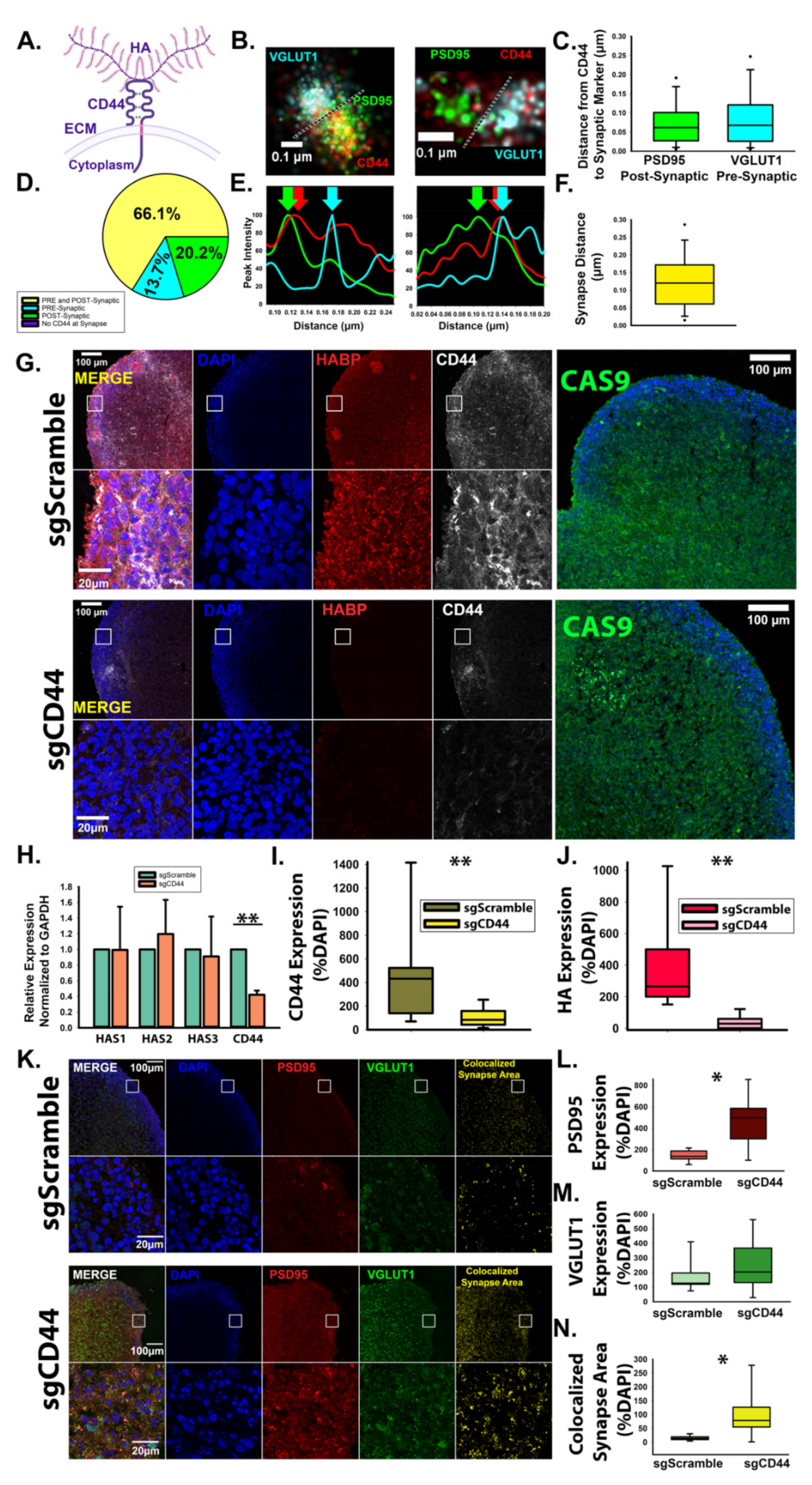
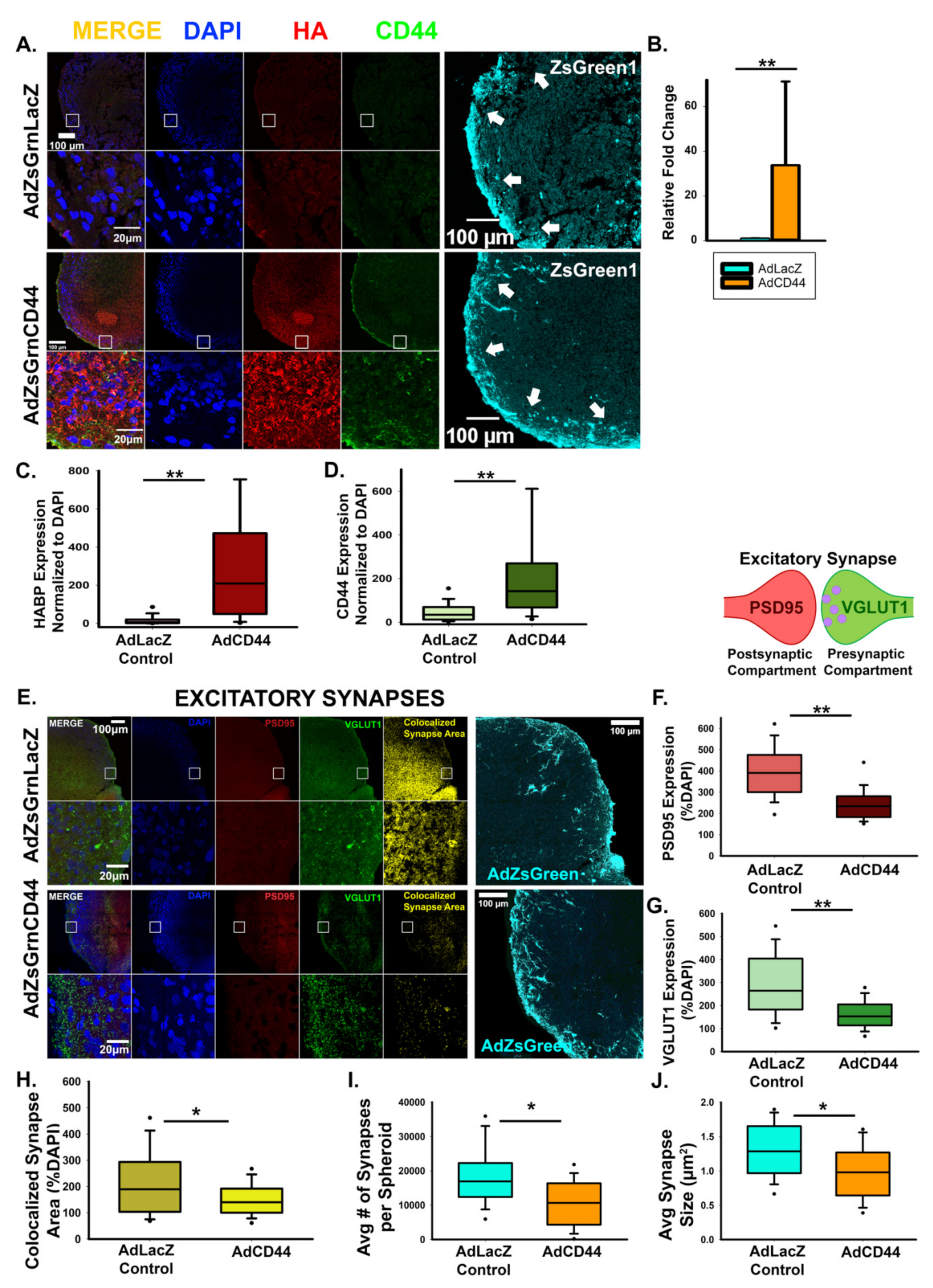
3.3. HA-CD44 Interactions at Developing Excitatory Synapses Attenuate Synaptogenesis through Regulation of RhoGTPase Signaling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Washbourne, P. Synapse Assembly and Neurodevelopmental Disorders. Neuropsychopharmacology 2014, 40, 4–15. [Google Scholar] [CrossRef] [Green Version]
- Zoghbi, H.Y.; Bear, M.F. Synaptic Dysfunction in Neurodevelopmental Disorders Associated with Autism and Intellectual Disabilities. Cold Spring Harb. Perspect. Biol. 2012, 4, a009886. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Penzes, P. Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism spectrum disorders. Curr. Mol. Med. 2015, 15, 146–167. [Google Scholar] [CrossRef]
- Gandal, M.J.; Haney, J.R.; Parikshak, N.N.; Leppa, V.; Ramaswami, G.; Hartl, C.; Schork, A.J.; Appadurai, V.; Buil, A.; Werge, T.M.; et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018, 359, 693–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frega, M.; Selten, M.; Mossink, B.; Keller, J.M.; Linda, K.; Moerschen, R.; Qu, J.; Koerner, P.; Jansen, S.; Oudakker, A.; et al. Distinct Pathogenic Genes Causing Intellectual Disability and Autism Exhibit a Common Neuronal Network Hyperactivity Phenotype. Cell Rep. 2020, 30, 173–186.e6. [Google Scholar] [CrossRef] [Green Version]
- Arranz, A.M.; Perkins, K.L.; Irie, F.; Lewis, D.P.; Hrabe, J.; Xiao, F.; Itano, N.; Kimata, K.; Hrabetova, S.; Yamaguchi, Y. Hyaluronan deficiency due to Has3 knock-out causes altered neuronal activity and seizures via reduction in brain extracellular space. J. Neurosci. 2014, 34, 6164–6176. [Google Scholar] [CrossRef] [Green Version]
- Martin-Vilchez, S.; Whitmore, L.; Asmussen, H.; Zareno, J.; Horwitz, R.; Newell-Litwa, K. RhoGTPase Regulators Orchestrate Distinct Stages of Synaptic Development. PLoS ONE 2017, 12, e0170464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papariello, A.; Newell-Litwa, K. Human-Derived Brain Models: Windows into Neuropsychiatric Disorders and Drug Therapies. ASSAY Drug Dev. Technol. 2020, 18, 79–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papariello, A.; Taylor, D.; Soderstrom, K.; Litwa, K. CB1 antagonism increases excitatory synaptogenesis in a cortical spheroid model of fetal brain development. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wilson, E.; Rudisill, T.; Kirk, B.; Johnson, C.; Kemper, P.; Newell-Litwa, K. Cytoskeletal regulation of synaptogenesis in a model of human fetal brain development. J. Neurosci. Res. 2020, 98, 2148–2165. [Google Scholar] [CrossRef]
- Wilson, E.; Knudson, W.; Newell-Litwa, K. Hyaluronan regulates synapse formation and function in developing neural networks. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Wilson, E.S.; Newell-Litwa, K. Stem cell models of human synapse development and degeneration. Mol. Biol. Cell 2018, 29, 2913–2921. [Google Scholar] [CrossRef]
- Semple, B.D.; Blomgren, K.; Gimlina, K.; Ferriero, D.M.; Noble-Haeusslein, L.J. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013, 106–107, 1–16. [Google Scholar] [CrossRef]
- Frischknecht, R.; Seidenbecher, C.I. The crosstalk of hyaluronan-based extracellular matrix and synapses. Neuron Glia Biol. 2008, 4, 249–257. [Google Scholar] [CrossRef]
- Bondareff, W.; Sjöstrand, J. Cytochemistry of synaptosomes. Exp. Neurol. 1969, 24, 450–458. [Google Scholar] [CrossRef]
- Ferrer-Ferrer, M.; Dityatev, A. Shaping Synapses by the Neural Extracellular Matrix. Front. Neuroanat. 2018, 12, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochlamazashvili, G.; Henneberger, C.; Bukalo, O.; Dvoretskova, E.; Senkov, O.; Lievens, P.M.-J.; Westenbroek, R.; Engel, A.K.; Catterall, W.A.; Rusakov, D.A.; et al. The Extracellular Matrix Molecule Hyaluronic Acid Regulates Hippocampal Synaptic Plasticity by Modulating Postsynaptic L-Type Ca2+ Channels. Neuron 2010, 67, 116–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, S.; Rankin-Gee, E.; Risbud, R.M.; Porter, B.E.; Marsh, E.D. Normal Development of the Perineuronal Net in Humans; In Patients with and without Epilepsy. Neuroscience 2018, 384, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.W.; Oohashi, T.; Pizzorusso, T. The roles of perineuronal nets and the perinodal extracellular matrix in neuronal function. Nat. Rev. Neurosci. 2019, 20, 451–465. [Google Scholar] [CrossRef]
- Wen, T.H.; Binder, D.K.; Ethell, I.M.; Razak, K.A. The Perineuronal ‘Safety’ Net? Perineuronal Net Abnormalities in Neurological Disorders. Front. Mol. Neurosci. 2018, 11, 270. [Google Scholar] [CrossRef]
- Itano, N.; Kimata, K. Mammalian Hyaluronan Synthases. IUBMB Life 2002, 54, 195–199. [Google Scholar] [CrossRef]
- Itano, N.; Sawai, T.; Yoshida, M.; Lenas, P.; Yamada, Y.; Imagawa, M.; Shinomura, T.; Hamaguchi, M.; Yoshida, Y.; Ohnuki, Y.; et al. Three Isoforms of Mammalian Hyaluronan Synthases Have Distinct Enzymatic Properties. J. Biol. Chem. 1999, 274, 25085–25092. [Google Scholar] [CrossRef] [Green Version]
- Perkins, K.L.; Arranz, A.M.; Yamaguchi, Y.; Hrabetova, S. Brain extracellular space, hyaluronan, and the prevention of epileptic seizures. Rev. Neurosci. 2017, 28, 869–892. [Google Scholar] [CrossRef] [PubMed]
- Aruffo, A.; Stamenkovic, I.; Melnick, M. UCB and SB. CD44 is the Principal Cell Surface Receptor for Hyaluronate. Cell 1990, 61, 1303–1313. [Google Scholar] [CrossRef]
- Bourguignon, L.Y. Matrix Hyaluronan-Activated CD44 Signaling Promotes Keratinocyte Activities and Improves Abnormal Epidermal Functions. Am. J. Pathol. 2014, 184, 1912–1919. [Google Scholar] [CrossRef] [Green Version]
- Bourguignon, L.Y.W.; Singleton, P.A.; Zhu, H.; Diedrich, F. Hyaluronan-mediated CD44 Interaction with RhoGEF and Rho Kinase Promotes Grb2-associated Binder-1 Phosphorylation and Phosphatidylinositol 3-Kinase Signaling Leading to Cytokine (Macrophage-Colony Stimulating Factor) Production and Breast Tumor Progression. J. Biol. Chem. 2003, 278, 29420–29434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourguignon, L.Y.; Wong, G.; Xia, W.; Man, M.-Q.; Holleran, W.M.; Elias, P.M. Selective matrix (hyaluronan) interaction with CD44 and RhoGTPase signaling promotes keratinocyte functions and overcomes age-related epidermal dysfunction. J. Dermatol. Sci. 2013, 72, 32–44. [Google Scholar] [CrossRef] [Green Version]
- Bijata, M.; Labus, J.; Guseva, P.D.; Stawarski, M.; Butzlaff, M.; Dzwonek, J.; Schneeberg, J.; Böhm, K.; Michaluk, P.; Rusakov, D.; et al. Synaptic Remodeling Depends on Signaling between Serotonin Receptors and the Extracellular Matrix. Cell Rep. 2017, 19, 1767–1782. [Google Scholar] [CrossRef] [Green Version]
- Ponta, H.; Wainwright, D.; Herrlich, P. Molecules in focus The CD44 protein family. Int. J. Biochem. Cell Biol. 1998, 30, 299–305. [Google Scholar] [CrossRef]
- Konopka, A.; Zeug, A.; Skupień-Jaroszek, A.; Kaza, B.; Müller, F.; Chwedorowicz, A.; Ponimaskin, E.; Wilczynski, G.; Dzwonek, J. Cleavage of Hyaluronan and CD44 Adhesion Molecule Regulate Astrocyte Morphology via Rac1 Signalling. PLoS ONE 2016, 11, e0155053. [Google Scholar] [CrossRef] [Green Version]
- Roszkowska, M.; Skupień-Jaroszek, A.; Wójtowicz, T.; Konopka, A.; Gorlewicz, A.; Kisiel, M.; Bekisz, M.; Ruszczycki, B.; Doleżyczek, H.; Rejmak-Kozicka, E.; et al. CD44: A novel synaptic cell adhesion molecule regulating structural and functional plasticity of dendritic spines. Mol. Biol. Cell 2016, 27, 4055–4066. [Google Scholar] [CrossRef]
- Knudson, C.B.; Knudson, W. Hyaluronan and CD44: Modulators of chondrocyte metabolism. Clin. Orthop. Relat. Res. 2004, 427, S152–S162. [Google Scholar] [CrossRef]
- Yang, C.; Cao, M.; Liu, H.; He, Y.; Xu, J.; Du, Y.; Liu, Y.; Wang, W.; Cui, L.; Hu, J.; et al. The High and Low Molecular Weight Forms of Hyaluronan Have Distinct Effects on CD44 Clustering. J. Biol. Chem. 2012, 287, 43094–43107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorne, R.F.; Legg, J.W.; Isacke, C. The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events. J. Cell Sci. 2004, 117, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourguignon, L.Y.; Zhu, H.; Shao, L.; Chen, Y.W. Ankyrin–Tiam1 Interaction Promotes Rac1 Signaling and Metastatic Breast Tumor Cell Invasion and Migration. J. Cell Biol. 2000, 150, 177–192. [Google Scholar] [CrossRef] [Green Version]
- Sethi, M.K.; Zaia, J. Extracellular matrix proteomics in schizophrenia and Alzheimer’s disease. Anal. Bioanal. Chem. 2016, 409, 379–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, M.J.; Damodarasamy, M.; Pathan, J.L.; Chan, C.K.; Spiekerman, C.; Wight, T.N.; Banks, W.A.; Day, A.J.; Vernon, R.B.; Keene, C.D. Increased Hyaluronan and TSG-6 in Association with Neuropathologic Changes of Alzheimer’s Disease. J. Alzheimer’s Dis. 2019, 67, 91–102. [Google Scholar] [CrossRef]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Ishizuka, S.; Tsuchiya, S.; Ohashi, Y.; Terabe, K.; Askew, E.B.; Ishizuka, N.; Knudson, C.B.; Knudson, W. Hyaluronan synthase 2 (HAS2) overexpression diminishes the procatabolic activity of chondrocytes by a mechanism independent of extracellular hyaluronan. J. Biol. Chem. 2019, 294, 13562–13579. [Google Scholar] [CrossRef]
- Olivier, N.; Keller, D.; Rajan, V.S.; Gönczy, P.; Manley, S. Simple buffers for 3D STORM microscopy. Biomed. Opt. Express 2013, 4, 885–899. [Google Scholar] [CrossRef] [Green Version]
- Dempsey, G.T.; Vaughan, J.C.; Chen, K.H.; Bates, M.; Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 2011, 8, 1027–1036. [Google Scholar] [CrossRef]
- Jourquin, J.; Tremblay, E.; Décanis, N.; Charton, G.; Hanessian, S.; Chollet, A.-M.; Le Diguardher, T.; Khrestchatisky, M.; Rivera, S. Neuronal activity-dependent increase of net matrix metalloproteinase activity is associated with MMP-9 neurotoxicity after kainate. Eur. J. Neurosci. 2003, 18, 1507–1517. [Google Scholar] [CrossRef]
- Bloom, F.E.; Aghajanian, G.K. Fine structural and cytochemical analysis of the staining of synaptic junctions with phosphotungstic acid. J. Ultrastruct. Res. 1968, 22, 361–375. [Google Scholar] [CrossRef]
- Cohen, M.; Klein, E.; Geiger, B.; Addadi, L. Organization and Adhesive Properties of the Hyaluronan Pericellular Coat of Chondrocytes and Epithelial Cells. Biophys. J. 2003, 85, 1996–2005. [Google Scholar] [CrossRef] [Green Version]
- Fowke, T.M.; Karunasinghe, R.N.; Bai, J.-Z.; Jordan, S.; Gunn, A.; Dean, J.M. Hyaluronan synthesis by developing cortical neurons in vitro. Sci. Rep. 2017, 7, 44135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, R.; Gachechiladze, M.A.; Ludwig, C.H.; Laurie, M.T.; Hong, J.Y.; Nathaniel, D.; Prabhu, A.V.; Fernandopulle, M.S.; Patel, R.; Abshari, M.; et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 2020, 104, 239–255. [Google Scholar] [CrossRef]
- Deans, P.J.M.; Brennand, K.J. Applying stem cells and CRISPR engineering to uncover the etiology of schizophrenia. Curr. Opin. Neurobiol. 2021, 69, 193–201. [Google Scholar] [CrossRef]
- Su, W.; Foster, S.C.; Xing, R.; Feistel, K.; Olsen, R.H.J.; Acevedo, S.F.; Raber, J.; Sherman, L.S. CD44 Transmembrane Receptor and Hyaluronan Regulate Adult Hippocampal Neural Stem Cell Quiescence and Differentiation. J. Biol. Chem. 2017, 292, 4434–4445. [Google Scholar] [CrossRef] [Green Version]
- Kazanis, I.; Ffrench-Constant, C. Extracellular matrix and the neural stem cell niche. Dev. Neurobiol. 2011, 71, 1006–1017. [Google Scholar] [CrossRef] [Green Version]
- Casini, P.; Nardi, I.; Ori, M. Hyaluronan is required for cranial neural crest cells migration and craniofacial development. Dev. Dyn. 2012, 241, 294–302. [Google Scholar] [CrossRef]
- Preston, M.; Sherman, L.S. Neural Stem Cell Niches: Roles for the Hyaluronan-Based Extracellular Matrix. Front. Biosci. 2011, 3, 1165–1179. [Google Scholar] [CrossRef] [Green Version]
- Farhy-Tselnicker, I.; Allen, N.J. Astrocytes, neurons, synapses: A tripartite view on cortical circuit development. Neural Dev. 2018, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.-S.; Allen, N.J.; Eroglu, C. Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb. Perspect. Biol. 2015, 7, a020370. [Google Scholar] [CrossRef] [Green Version]
- Distler, C.; Dreher, Z.; Stone, J. Contact spacing among astrocytes in the central nervous system: An hypothesis of their structural role. Glia 1991, 4, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Halassa, M.M.; Fellin, T.; Takano, H.; Dong, J.-H.; Haydon, P.G. Synaptic Islands Defined by the Territory of a Single Astrocyte. J. Neurosci. 2007, 27, 6473–6477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroon, T.; van Hugte, E.; Van Linge, L.; Mansvelder, H.; Meredith, R. Early postnatal development of pyramidal neurons across layers of the mouse medial prefrontal cortex. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, C.M.; Swanson, R.A. Astrocyte Glutamate Transport: Review of Properties, Regulation, and Physiological Functions. Glia 2000, 32, 1–14. [Google Scholar] [CrossRef]
- Mahmoud, S.; Gharagozloo, M.; Simard, C.; Gris, D. Astrocytes Maintain Glutamate Homeostasis in the CNS by Controlling the Balance between Glutamate Uptake and Release. Cells 2019, 8, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.K.; Nishioka, T.; Shimizu, H.; Takahashi, K.; Kakegawa, W.; Mikami, T.; Hirayama, Y.; Koizumi, S.; Yoshida, S.; Yuzaki, M.; et al. Hyaluronan synthesis supports glutamate transporter activity. J. Neurochem. 2019, 150, 249–263. [Google Scholar] [CrossRef]
- Carulli, D.; Rhodes, K.E.; Brown, D.J.; Bonnert, T.P.; Pollack, S.J.; Oliver, K.; Strata, P.; Fawcett, J. Composition of perineuronal nets in the adult rat cerebellum and the cellular origin of their components. J. Comp. Neurol. 2006, 494, 559–577. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.W.; Cua, R.; Keough, M.B.; Haylock-Jacobs, S.; Yong, V.W. Pathophysiology of the brain extracellular matrix: A new target for remyelination. Nat. Rev. Neurosci. 2013, 14, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Takegahara, N.; Walsh, M.C.; Choi, Y. CD44 Can Compensate for IgSF11 Deficiency by Associating with the Scaffold Protein PSD-95 during Osteoclast Differentiation. Int. J. Mol. Sci. 2020, 21, 2646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wlodarczyk, J.; Mukhina, I.; Kaczmarek, L.; Dityatev, A. Extracellular matrix molecules, their receptors, and secreted proteases in synaptic plasticity. Dev. Neurobiol. 2011, 71, 1040–1053. [Google Scholar] [CrossRef]
- Pletikos, M.; Sousa, A.M.; Sedmak, G.; Meyer, K.A.; Zhu, Y.; Cheng, F.; Li, M.; Kawasawa, Y.I.; Šestan, N. Temporal Specification and Bilaterality of Human Neocortical Topographic Gene Expression. Neuron 2014, 81, 321–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nat. Cell Biol. 2011, 478, 483–489. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.B.; Kawasawa, Y.I.; Mason, C.E.; Krsnik, Z.; Coppola, G.; Bogdanović, D.; Geschwind, D.H.; Mane, S.M.; State, M.W.; Šestan, N. Functional and Evolutionary Insights into Human Brain Development through Global Transcriptome Analysis. Neuron 2009, 62, 494–509. [Google Scholar] [CrossRef] [Green Version]
- Yuste, R.; Bonhoeffer, T. Genesis of dendritic spines: Insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 2004, 5, 24–34. [Google Scholar] [CrossRef]
- Martín-Villar, E.; Fernández-Muñoz, B.; Parsons, M.; Yurrita, M.M.; Megías, D.; Pérez-Gómez, E.; Jones, G.E.; Quintanilla, M. Podoplanin Associates with CD44 to Promote Directional Cell Migration. Mol. Biol. Cell 2010, 21, 4387–4399. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Askew, E.B.; Knudson, C.B.; Knudson, W. CRISPR/Cas9 knockout of HAS2 in rat chondrosarcoma chondrocytes demonstrates the requirement of hyaluronan for aggrecan retention. Matrix Biol. 2016, 56, 74–94. [Google Scholar] [CrossRef] [Green Version]
- Newell-Litwa, K.A.; Badoual, M.; Asmussen, H.; Patel, H.; Whitmore, L.; Horwitz, A.R. ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J. Cell Biol. 2015, 210, 225–242. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Yang, X.; Shi, L. Rho GTPase Regulators and Effectors in Autism Spectrum Disorders: Animal Models and Insights for Therapeutics. Cells 2020, 9, 835. [Google Scholar] [CrossRef] [Green Version]
- Tejada-Simon, M.V. Modulation of actin dynamics by Rac1 to target cognitive function. J. Neurochem. 2015, 133, 767–779. [Google Scholar] [CrossRef]
- Bonneh-Barkay, D.; Wiley, C.A. Brain Extracellular Matrix in Neurodegeneration. Brain Pathol. 2009, 19, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.J.; Vernon, R.B.; Damodarasamy, M.; Chan, C.K.; Wight, T.N.; Bentov, I.; Banks, W.A. Microvasculature of the Mouse Cerebral Cortex Exhibits Increased Accumulation and Synthesis of Hyaluronan With Aging. J. Gerontol. Ser. A Boil. Sci. Med. Sci. 2016, 72, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Koffie, R.M.; Hyman, B.T.; Spires-Jones, T.L. Alzheimer’s disease: Synapses gone cold. Mol. Neurodegener. 2011, 6, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashyap, G.; Bapat, D.; Das, D.; Gowaikar, R.; Amritkar, R.E.; Rangarajan, G.; Ravindranath, V.; Ambika, G. Synapse loss and progress of Alzheimer’s disease -A network model. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yoshiyama, Y.; Higuchi, M.; Zhang, B.; Huang, S.-M.; Iwata, N.; Saido, T.C.; Maeda, J.; Suhara, T.; Trojanowski, J.Q.; Lee, V.M.-Y. Synapse Loss and Microglial Activation Precede Tangles in a P301S Tauopathy Mouse Model. Neuron 2007, 53, 337–351. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, J.; Savage, J.C.; Tremblay, M.È. Synaptic Loss in Alzheimer’s Disease: Mechanistic Insights Provided by Two-Photon in vivo Imaging of Transgenic Mouse Models. Front. Cell. Neurosci. 2020, 14, 445. [Google Scholar] [CrossRef]
- DeKosky, S.T.; Scheff, S.W. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: Correlation with cognitive severity. Ann. Neurol. 1990, 27, 457–464. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilson, E.S.; Litwa, K. Synaptic Hyaluronan Synthesis and CD44-Mediated Signaling Coordinate Neural Circuit Development. Cells 2021, 10, 2574. https://doi.org/10.3390/cells10102574
Wilson ES, Litwa K. Synaptic Hyaluronan Synthesis and CD44-Mediated Signaling Coordinate Neural Circuit Development. Cells. 2021; 10(10):2574. https://doi.org/10.3390/cells10102574
Chicago/Turabian StyleWilson, Emily S., and Karen Litwa. 2021. "Synaptic Hyaluronan Synthesis and CD44-Mediated Signaling Coordinate Neural Circuit Development" Cells 10, no. 10: 2574. https://doi.org/10.3390/cells10102574
APA StyleWilson, E. S., & Litwa, K. (2021). Synaptic Hyaluronan Synthesis and CD44-Mediated Signaling Coordinate Neural Circuit Development. Cells, 10(10), 2574. https://doi.org/10.3390/cells10102574