Effect of Soybean and Maize Rotation on Soil Microbial Community Structure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Survey and Design
2.2. Samples Collection
2.3. Soil Physical and Chemical Properties
2.4. Determination of Soil Microbial Community Structure
2.5. Data Analysis
3. Results
3.1. Effects of Each Treatment on the Composition and Content of Phospholipid Fatty Acids
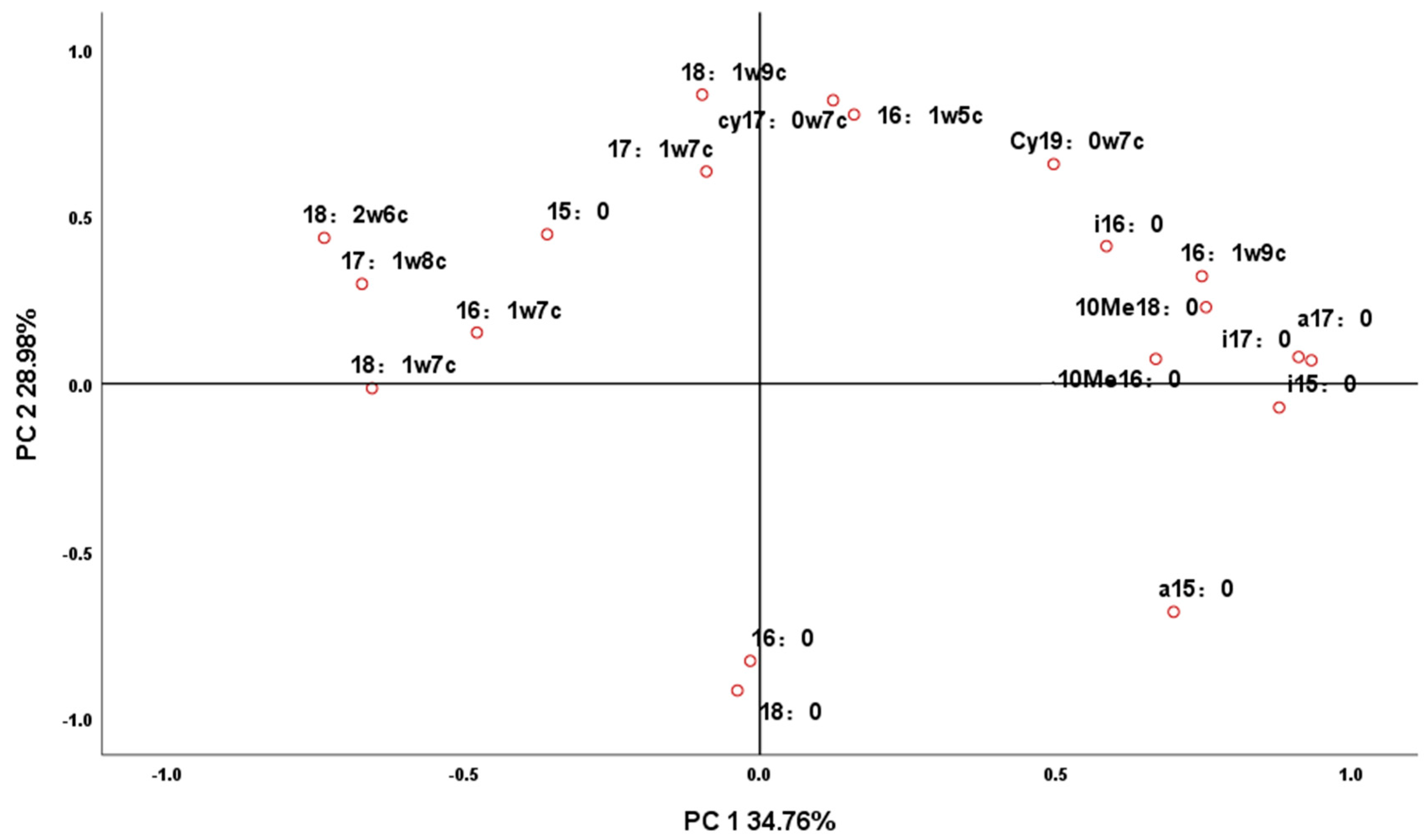
3.2. Principal Component Analysis of Microbial Fatty Acids in Soil by Each Treatment
3.3. Relationship between Soil Microbial Community Structure and Soil Properties
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Olsen, R.J.; Hensler, R.F.; Attoe, O.J.; Witzel, S.A.; Peterso, L.A. Fertilizer Nitrogen and Crop Rotation in relation to movement of Nitrate Nitrogen through Soil Profiles. Soil Sci. Soc. Am. J. 1970, 34, 448–452. [Google Scholar] [CrossRef]
- Jenkinson, D.S.; Ladd, J.N. Microbial Biomass in Soil: Measurement and Turnover. Soil Biochem. 1981, 5, 415–471. [Google Scholar]
- Wu, Q.; Lu, K.; Mao, X.; Qin, H.; Wang, H. Responses of Soil Nutrients and Microbial Biomass and Community Composition to Long-term Fertilization in Cultivated Land. Chin. Agric. Sci. Bull. 2015, 31, 150–156. [Google Scholar]
- Cavigelli, M.A.; Robertson, G.P. The functional significance of denitrifire community composition in a terrestrial ecosystem. Ecology 2000, 81, 1402–1414. [Google Scholar] [CrossRef]
- Zelles, L. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: A review. Biol. Fertil. Soils 1999, 29, 111–129. [Google Scholar] [CrossRef]
- Miller, M.; Dick, R.P. Dynamics of soil C and microbial biomass in whole soil and aggregates in two cropping systems. Appl. Soil Ecol. 1995, 2, 253–261. [Google Scholar] [CrossRef]
- Bucher, A.E.; Lanyon, L.E. Evaluating soil management with microbial community-level physiological profiles. Appl. Soil Ecol. 2005, 29, 59–71. [Google Scholar] [CrossRef]
- White, D.C.; Davis, W.M.; Nickels, J.S.; King, J.D.; Bobbie, R.J. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 1979, 40, 51–62. [Google Scholar] [CrossRef]
- Tunlid, A.H.; Baird, B.B.; Trexler, M.; Olsson, S.; Findlay, R.; Odham, G.C.; White, D. Determination of phospholipid ester-linked fatty acids and poly β-hydroxybutyrate for the estimation of bacterial biomass and activity in the rhizosphere of the rape plant Brassica napus (L.). Can. J. Microbiol. 1985, 31, 1113–1119. [Google Scholar] [CrossRef]
- Roslev, P.; Iversen, N.; Henriksen, K. Direct fingerprinting of metabolically active bacteria in environmental samples by substrate specific radiolabelling and lipid analysis. J. Microbiol. Methods 1998, 31, 99–111. [Google Scholar] [CrossRef]
- ISSCAS. Physical and Chemical Analysis Methods of Soils; Institute of Soil Sciences, Chinese Academy of Sciences; Shanghai Science and Technology Press: Shanghai, China, 1978; pp. 7–59. (In Chinese) [Google Scholar]
- Hong, S.B.; Piao, S.L.; Chen, A.P.; Liu, Y.W.; Liu, L.L.; Peng, S.S.; Sardans, J.; Sun, Y.; Penuelas, J.; Zeng, H. Afforestation neutralizes soil pH. Nat. Commun. 2018, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Duan, Z.H. Changes in soil physical and chemical properties during reversal of desertification in Yanchi County of Ningxia Hui autonomous region, China. Enviorn. Geol. 2009, 57, 975–985. [Google Scholar] [CrossRef]
- Bossio, D.A.; Scow, K.M. Impacts of Carbon and Flooding on Soil Microbial Communities: Phospholipid Fatty Acid Profiles and Substrate Utilization Patterns. Microb. Ecol. 1998, 35, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, Y.; Wu, Y.; Xu, J. Effect of complex heavy metal pollution on biomass and community structure of soil microbes in paddy soil. Acta Pedol. Sin. 2008, 45, 1102–1109. [Google Scholar] [CrossRef]
- Frostegård, Å.; Bååth, E.; Tunlio, A. Shifts in the structure of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 1993, 25, 723–730. [Google Scholar] [CrossRef]
- Federle, T.W. Microbial distribution in the soil-new techniques. In Perspectives in Microbial Ecology, Proceedings of the International Symposium of Microbial Ecology IV, Ljubljana, Slovenia, January 1986; Megusar, F., Gantar, M., Eds.; Slovene Society for Microbiology: Ljubljana, Slovenia, 1986; pp. 493–498. [Google Scholar]
- Hamel, C.; Vujanovic, V.; Jeannotte, R.; Nakano-Hylander, A.; St-Arnaud, M. Negative feedback on a perennial crop: Fusarium crown and root rot of asparagus is related to changes in soil microbial community structure. Plant Soil 2005, 268, 75–87. [Google Scholar] [CrossRef]
- Tunlid, A.; Hoitink, J.H.A.; Low, C.; White, C.D. Characterization of bacteria that suppress rhizoctonia damping-off in bark compost media by analysis of Fatty Acid biomarkers. Appl. Environ. Microbiol. 1989, 55, 1368–1374. [Google Scholar]
- Zogg, G.P.; Zak, D.R.; Ringelberg, D.B.; White, D.C.; MacDonald, N.W.; Pregitzer, K.S. Compositional and Functional Shifts in Microbial Communities Due to Soil Warming. Soil Sci. Soc. Am. J. 1997, 61, 475. [Google Scholar] [CrossRef]
- Blume, E.; Bischoff, M.; Reichert, J.M.; Moorman, T.; Konopka, A.; Turco, R.F. Surface and subsurface microbial biomass, community structure and metabolic activity as a function of soil depth and season. Appl. Soil Ecol. 2002, 20, 171–181. [Google Scholar] [CrossRef]
- Brant, J.B.; Myrold, D.D.; Sulzman, E.W. Root controls on soil microbial community structure in forest soils. Oecologia 2006, 148, 650–659. [Google Scholar] [CrossRef]
- Xu, Y.; Cai, X.; Zhu, X. Comparative analysis of microbial community structures in soils from rice-upland crop rotation fields by PLFA profile technique. Acta Agric. Zhejiangensis 2013, 25, 1056–1061. [Google Scholar] [CrossRef]
- Song, Y.S.P.; Deng, V. Acosta-Martínez, E. Katsalirou. Characterization of redox-related soil microbial communities along a river floodplain continuum by fatty acid methyl ester (fame) and 16s rrna genes. Appl. Soil Ecol. 2008, 40, 499–509. [Google Scholar] [CrossRef]
- Zhang, X.; Xin, X.; Zhu, A.; Yang, W.; Zhang, J.; Ding, S.; Mu, L.; Shao, L. Linking macroaggregation to soil microbial community and organic carbon accumulation under different tillage and residue managements. Soil Tillage Res. 2018, 178, 99–107. [Google Scholar] [CrossRef]
- Mccann, M.C.; Shi, J.; Roberts, K.N.; Carpita, C. Changes in pectin structure and localization during the growth of unadapted and nacl-adapted tobacco cells. Plant J. 2010, 5, 773–785. [Google Scholar] [CrossRef]
- Kiani, M.; Hernandez-Ramirez, G.; Quideau, S.; Smith, E.; Janzen, H.; Larney, F.J.; Puurveen, D. Quantifying sensitive soil quality indicators across contrasting long-term land management systems: Crop rotations and nutrient regimes. Agric. Ecosyst. Environ. 2017, 248, 123–135. [Google Scholar] [CrossRef]
- Chavarría, D.N.; Verdenelli, R.A.; Serri, D.L.; Restovich, S.B.; Andriulo, A.E.; Meriles, J.M.; Vargas-Gil, S. Effect of cover crops on microbial community structure and related enzyme activities and macronutrient availability. Eur. J. Soil Biol. 2016, 76, 74–82. [Google Scholar] [CrossRef]
- Zhang, B.; Li, Y.; Ren, T.; Tian, Z.; Wang, G.; He, X. Short-term effect of tillage and crop rotation on microbial community structure and enzyme activities of a clay loam soil. Biol. Fertil. Soils 2014, 50, 1077–1085. [Google Scholar] [CrossRef]
| Plants | Varieties | Sowing Rate | Plant Spacing | Row Spacing |
|---|---|---|---|---|
| Plant/ha | cm | cm | ||
| maize | Fengdan 189 | 67,500 | 33 | 45 |
| soybean | Red bean 3 | 210,000 | 12 | 40 |
| Microbial Type | Phospholipid Fatty Acid Labelled |
|---|---|
| Bacteria in general (B) | i14:0, i15:1, i15:0, a15:0, i16:0, i17:0, a17:0, 16:1 ω 7cis, 16:1 ω 9cis, 17:1 ω 7cis, 17:1 ω 8cis, 18:1 ω 7cis, 18:1 ω 5cis, cy17:0, cy19:0, 16:12 OH, 16:0, 18:0 |
| Gram-positive bacteria (G+) | i14:0, i15:1, i15:0, a15:0, i16:0, 16:0, i17:0, a17:0 |
| Gram-negative bacteria (G-) | 16:1 ω 7cis, 16:1 ω 9cis, 17:1 ω 7cis, 17:1 ω 8cis, 1:1 ω 7cis, 18:1 ω 7cis, 18:1 ω 5cis, cy17:0, cy19:0, 16:12 OH |
| Actinomycetes (Act) | 10Me16:0, 10Me17:0, 10Me18:0 |
| Fungi (Fug) | 16:1 ω 5cis, 18:1 ω 9cis, 18:2 ω 6cis, 18:2 ω 9cis, 18:3 ω 6cis |
| Treatment | G+ | G- | AMF | Act | OB | ||
|---|---|---|---|---|---|---|---|
| a15:0 | 16:0 | i16:0 | 17:1 ω 7c | 16:1 ω 5c | 10Me18:0 | 18:0 | |
| CM + OF | † 4.4 ± 0.2 bc | † 11.7 ± 1.0 bc | † 3.9 ± 0.3 a | † 2.1 ± 0.3 ab | † 3.9 ± 0.1 a | † 1.9 ± 0.2 b | † 2.1 ± 0.2 c |
| CM + NP | 4.4 ± 0.1 c | 11.6 ± 0.3 c | 3.6 ± 0.1 c | 2.0 ± 0.3 ab | 3.9 ± 0.1 a | 1.8 ± 0.1 b | 2.1 ± 0.1 c |
| CM + 0 | 4.4 ± 0.0 abc | 11.8 ± 0.5 abc | 3.7 ± 0.0 abc | 2.2 ± 0.4 a | 3.9 ± 0.2 a | 1.9 ± 0.1 b | 2.2 ± 0.2 bc |
| SM + OF | 4.7 ± 0.1 abc | 11.6 ± 0.4 c | 3.8 ± 0.1 abc | 1.9 ± 0.2 ab | 3.9 ± 0.1 a | 2.2 ± 0.2 a | 2.3 ± 0.2 bc |
| SM + NP | 4.6 ± 0.3 abc | 11.7 ± 0.2 bc | 3.9 ± 0.1 ab | 1.9 ± 0.1 ab | 3.7 ± 0.1 a | 1.9 ± 0.1 b | 2.3 ± 0.1 ab |
| SM + 0 | 4.7 ± 0.3 ab | 12.0 ± 0.4 abc | 3.8 ± 0.1 abc | 1.9 ± 0.1 ab | 3.7 ± 0.2 a | 2.1 ± 0.4 ab | 2.2 ± 0.2 bc |
| CS + OF | 4.6 ± 0.2 abc | 12.7 ± 0.6 a | 3.6 ± 0.1 bc | 1.8 ± 0.1 b | 3.2 ± 0.2 b | 1.8 ± 0.1 ab | 2.5 ± 0.1 a |
| CS + NP | 4.8 ± 0.1 a | 12.7 ± 0.1 ab | 3.8 ± 0.1 abc | 2.0 ± 0.1 ab | 3.1 ± 0.2 b | 1.8 ± 0.1 b | 2.5 ± 0.1 a |
| CS + 0 | 4.6 ± 0.1 abc | 12.1 ± 0.4 abc | 3.8 ± 0.1 abc | 1.8 ± 0.2 b | 3.1 ± 0.1 b | 1.8 ± 0.1 b | 2.5 ± 0.1 a |
| Cropping system | ** | ** | — | * | ** | * | ** |
| Fertilizer | — | — | — | — | — | — | — |
| Interaction effect | — | — | * | — | — | — | — |
| Treatment | Gram-Positive Bacteria | Gram-Negative Bacteria | Fungi | Actinomycetes | Other Bacteria | Fungi/Bacteria |
|---|---|---|---|---|---|---|
| G+ | G- | |||||
| CM + OF | † 37.3 ± 0.5 bc | † 33.2 ± 0.3 ab | † 3.4 ± 0.3 ab | † 17.5 ± 0.8 a | † 8.3 ± 1.3 ab | † 0.049 a |
| CM + NP | 36.2 ± 0.6 c | 33.6 ± 0.4 ab | 3.4 ± 0.5 a | 17.0 ± 1.5 a | 9.7 ± 0.4 a | 0.050 a |
| CM + 0 | 37.0 ± 0.4 bc | 32.1 ± 0.6 b | 3.0 ± 0.5 abc | 18.1 ± 0.3 a | 9.9 ± 0.5 a | 0.043 ab |
| SM + OF | 38.1 ± 0.8 ab | 32.4 ± 0.8 b | 2.6 ± 0.1 c | 17.7 ± 0.5 a | 9.2 ± 0.6 ab | 0.036 b |
| SM + NP | 37.5 ± 0.8 ab | 32.7 ± 0.5 b | 2.9 ± 0.3 abc | 17.1 ± 0.6 a | 9.7 ± 1.0 a | 0.041 ab |
| SM + 0 | 38.0 ± 0.9 ab | 32.3 ± 0.8 b | 2.6 ± 0.3 bc | 18.1 ± 0.8 a | 9.0 ± 0.6 ab | 0.037 b |
| CS + OF | 37.0 ± 0.7 bc | 34.4 ± 0.3 a | 3.4 ± 0.8 ab | 16.9 ± 0.8 a | 8.3 ± 0.6 b | 0.048 ab |
| CS + NP | 38.5 ± 0.7 a | 32.7 ± 1.6 b | 2.6 ± 0.2 bc | 18.1 ± 0.5 a | 8.1 ± 0.4 b | 0.037 b |
| CS + 0 | 37.8 ± 0.7 ab | 33.6 ± 1.5 ab | 2.8 ± 0.5 abc | 17.3 ± 1.0 a | 8.5 ± 0.4 ab | 0.039 ab |
| Cropping system | ** | * | * | — | ** | * |
| Fertilizer | — | — | — | — | — | — |
| Interaction effect | * | — | — | — | — | — |
| Treatment | Gram-Negative Bacteria | Treatment | Gram-Positive Bacteria |
|---|---|---|---|
| G- | G+ | ||
| CM + OF | † 33.2 ± 0.3 aA | SM + OF | † 38.1 ± 0.8 aA |
| CM + NP | 33.6 ± 0.4 aA | SM + NP | 37.5 ± 0.8 bB |
| CM + 0 | 32.1 ± 0.6 bB | SM + 0 | 38.0 ± 0.9 aA |
| Phospholipid Fatty Acid | Component | |
|---|---|---|
| 1 | 2 | |
| a17:0 | 0.932 | 0.054 |
| i17:0 | 0.911 | 0.064 |
| i15:0 | 0.876 | −0.086 |
| 10Me18:0 | 0.757 | 0.216 |
| 16:1 ω9c | 0.752 | 0.309 |
| 18:2 ω6c | −0.728 | 0.448 |
| 10Me16:0 | 0.670 | 0.062 |
| 17:1 ω8c | −0.667 | 0.310 |
| 18:1 ω7c | −0.655 | −0.003 |
| i16:0 | 0.592 | 0.401 |
| 16:1 ω7c | −0.475 | 0.161 |
| 18:0 | −0.053 | −0.917 |
| 18:1 ω9c | −0.083 | 0.866 |
| cy17:0 ω7c | 0.138 | 0.846 |
| 16:0 | −0.030 | −0.829 |
| 16:1 ω5c | 0.173 | 0.802 |
| a15:0 | 0.687 | −0.695 |
| cy19:0 ω7c | 0.507 | 0.648 |
| 17:1 ω7c | −0.080 | 0.636 |
| 15:0 | −0.352 | 0.453 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Sun, J.; Li, L.; Wang, X.; Li, X.; Qu, J. Effect of Soybean and Maize Rotation on Soil Microbial Community Structure. Agronomy 2019, 9, 42. https://doi.org/10.3390/agronomy9020042
Zhang P, Sun J, Li L, Wang X, Li X, Qu J. Effect of Soybean and Maize Rotation on Soil Microbial Community Structure. Agronomy. 2019; 9(2):42. https://doi.org/10.3390/agronomy9020042
Chicago/Turabian StyleZhang, Peng, Jiying Sun, Lijun Li, Xinxin Wang, Xiaoting Li, and Jiahui Qu. 2019. "Effect of Soybean and Maize Rotation on Soil Microbial Community Structure" Agronomy 9, no. 2: 42. https://doi.org/10.3390/agronomy9020042
APA StyleZhang, P., Sun, J., Li, L., Wang, X., Li, X., & Qu, J. (2019). Effect of Soybean and Maize Rotation on Soil Microbial Community Structure. Agronomy, 9(2), 42. https://doi.org/10.3390/agronomy9020042





