Abstract
Cysteine-rich polycomb-like protein (CPP) transcription factors (TFs) play critical roles in the process of plant growth and development, as well as stress responses. To date, no reports about CPP TFs have been published for lettuce (Lactuca sativa). In this study, six CPP TFs (LsCPP1-LsCPP6) were identified in lettuce. Phylogenetic analysis showed that LsCPP TFs were classified into two clades (Clade I and Clade II). Six LsCPP genes were distributed across four chromosomes. Cis-elements, which are involved in environmental stress, hormone response, and development processes, were identified in the promoters of LsCPP genes. LsCPP genes were induced by different tissues and the stem enlargement processes of stem lettuce. Plant hormones (SA, ABA) and abiotic stress (salt, drought) induced the expression of LsCPP genes. LsCPP4 was significantly induced after drought stress for 12 h. Notably, the expression level of LsCPP4 increased more than 10 times (12 h) and 150 times (24 h) after salt stress. ABA and SA significantly induced the expression profile of LsCPP6. This study not only provides the basis for future functional research of LsCPP genes, particularly their roles in lettuce stress resistance, but also provides a foundation for molecular breeding to enhance the agricultural traits in lettuce.
1. Introduction
Transcription factors (TFs), or trans-acting factors, specifically recognize and bind to cis-acting elements in the promoter regions of target genes [1]. TFs play pivotal roles in the processes of embryonic development, cell differentiation, and the transcriptional regulation of intracellular signals (such as hormones growth factors) and environmental cues (such as temperature, high salinity) [2,3,4]. Based on conserved DNA-binding domains, TFs are classified into numerous families including AP2/ERF, WRKY, MYB, bHLH, and Dof [5,6,7,8]. The model dicot plant Arabidopsis thaliana has 62 TF families (1533 genes), which accounts for 5.9% of its genome [8]. A total of 56 TF families (2408 genes), accounting for about 4% of its genome, are identified in the monocot crop Oryza sativa [9]. TFs in other important species have also been extensively identified, such as 63 families (over 2600 genes) in Zea mays, 58 families (over 3100 genes) in Triticum aestivum, and 55 families (over 1800 genes) in Solanum lycopersicum [10].
The cysteine-rich polycomb-like protein (CPP) TF family has been identified in many plants [11]. They contain one or two conserved cysteine-rich CXC motifs (CXCX4CX3YCXCX6CX3CXCX2C), and a variable-length connecting sequence is usually found between two CXC motifs [12,13]. Additionally, the CXC motifs usually contain a conserved R sequence (RNPXAFXPK) that mediates DNA binding and transcriptional regulation [14]. The first CPP gene TSO1, identified in Arabidopsis, plays a crucial role in regulating cell division and flowering [15,16]. Since then, CPP TFs families have been identified in many plant species, including 8 members in Arabidopsis [17], 5 members in cucumber [18], 6 members in tomato [19], 17 members in Moso bamboo [11], 10 members in apple [14], 10 members in quinoa [13], and 29 members in Populus trichocarpa [20].
CPP genes are involved in multiple biological processes, such as the regulation of growth and development. AtTSO1 encodes a floral-specific cell division protein; mutations in AtTSO1 disrupt cell division processes in floral meristem cells, which lead to impaired cell wall formation and increased DNA ploidy levels [15,16]. Additionally, AtTSO1 and MYB3R1 can coordinate cell proliferation and differentiation in shoot and root tissues by forming a regulatory module [21].
CPP genes also participate in the process of abiotic stress responses. The TaCPP5-1D gene of wheat (T. aestivum) has been significantly upregulated under drought stress [22]. When SlCPP3 is transiently downregulated in tomato, the plants show severe damage with more reactive oxygen species accumulation than the control plants under cold stress [19]. Additionally, MtCPP2 and MtCPP8 in barrel medic (Medicago truncatula) demonstrate a salt stress response [23]. Apart from plant development and abiotic stress, CPP TFs are involved in hormone responses. CsCPPs in the tea plant (Camellia sinensis) are significantly upregulated under abscisic acid (ABA) treatment [24]. In apple, MdCPPs show different expression responses to salicylic acid (SA), jasmonic acid (JA), and gibberellin (GA) [14].
Lettuce (Lactuca sativa), is extensively grown and consumed year-round across China. Lettuce is rich in vitamins, proteins, and phytochemicals (specifically flavonoids and terpenoids), which play critical roles for balancing human diets. However, lettuce can undergo adverse environmental conditions and pathogen attack during its growth and development processes. With the release of its genome data, multiple TF families including ABF [25], AP2/ERF [26], TCP [27], and YABBY [28] have been identified to provide key genetic resources for studying lettuce development and stress adaptation. To date, no studies about CPP TF families have been reported in lettuce. In this study, six LsCPP TFs are identified by genome-wide analysis in lettuce. Phylogenetic relationships, conserved domains, chromosomal distributions, and cis-regulatory elements of LsCPP TFs are systematically analyzed. The expression profiles of six LsCPPs under abiotic stresses, hormone treatments, as well as stem expansion stages of stem lettuce have been examined using quantitative reverse transcription polymerase chain reaction (qRT-PCR). These results not only elucidate the potential roles of CPP TFs in lettuce growth and stress adaptation, but also establish a molecular foundation for targeted breeding or genetic engineering (gene editing or overexpression) to enhance stress tolerance in lettuce.
2. Materials and Methods
2.1. Identification of Lettuce CPP TF Family Members
CPP TF family members in lettuce were identified by searching for the conserved CPP domains by using the Pfam database (http://pfam.sanger.ac.uk (accessed on 31 October 2024)) [29]. The coding sequences (CDS) and protein sequences of lettuce CPP TFs were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/ (accessed on 31 October 2024)). Domain validation was performed using HMMER (http://www.ebi.ac.uk/ (accessed on 5 November 2024)) [30]. The physicochemical properties of the proteins (isoelectric point, molecular weight and hydrophobicity) were analyzed via the ExPASy server (http://www.expasy.org/ (accessed on 10 November 2024)) [31].
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
The CPP sequences of A. thaliana, O. sativa, and S. lycopersicum were obtained from the TAIR database (http://www.arabidopsis.org/ (accessed on 6 March 2025)), the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/ (accessed on 6 March 2025)), and the Plant Transcription Factor Database (https://planttfdb.gao-lab.org/ (accessed on 6 March 2025)), respectively. Initial sequence alignment was performed using SnapGene 8.0 software. Multiple sequence alignment and the construction of a maximum likelihood (ML)-based phylogenetic tree with 1000 bootstrap replicates were conducted using MEGA 11.0.13 software [32]. The Interactive Tree of Life (iTOL) platform (https://itol.embl.de/ (accessed on 15 March 2025)) was used for topological adjustment branch coloring and functional annotation thereby visually elucidating the evolutionary relationships.
2.3. Conserved Domain, Gene Structure, Chromosomal Localization, and cis-Acting Elements Analysis
The MEME online tool (https://meme-suite.org/meme/tools/meme) and TBtools II software were used to analyze and identify the conserved motifs and exon–intron structures [33,34]. The number of motifs identified by the MEME tool was set to 15 and the optimal width of each motif was set to 6–50 residues. Chromosomal locations were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/gene (accessed on 17 April 2025)) and a chromosomal localization map was generated with MapChart. cis-acting elements in promoter regions (2000 bp upstream of the translation initiation site ATG) were scanned using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 10 May 2025)). These approaches systematically characterized the conserved domains, genomic organization, chromosomal distribution, and regulatory element composition of CPP.
2.4. Interaction Network, Collinearity Analysis, and Secondary and Tertiary Structure Prediction
STRING (https://cn.string-db.org (accessed on 4 April 2025)) with the setting of a medium confidence score threshold (≥0.400) and medium stringency (5% FDR) was used to produce the interaction network of LsCPP TFs based on the orthologs in Arabidopsis. For the collinearity analysis of CPP genes, genomic sequences and annotation files of A. thaliana, O. sativa, and S. lycopersicum were downloaded from Ensembl Plants (http://plants.ensembl.org/index.html (accessed on 20 April 2025)). Interspecific synteny analysis was performed among L. sativa, A. thaliana, S. lycopersicum, and O. sativa. Additionally, SOPMA (https://npsa.lyon.inserm.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html (accessed on 21 April 2025)) was used to analyze the secondary structures of LsCPPs. Tertiary structures were modeled and analyzed via the SWISS-MODEL module on the Expasy platform (https://www.expasy.org (accessed on 30 November 2024)).
2.5. Experimental Materials and Treatments
The stem lettuce cultivar ‘Yonganhong’ saved in our laboratory was utilized in this study. Seeds were sown in plastic pots (15 cm diameter × 25 cm height) filled with a mixture of peat and vermiculite (1:1, v:v). Then they were put into a light culture chamber with a 12 h photoperiod and 20,000 µmol m2/s (lux) light intensity at 22 °C and 18 °C (day vs. night). Healthy and uniform seedlings, which grew ten true leaves, were selected for abiotic stress treatments (4 °C for low-temperature stress, 37 °C for high-temperature treatment, 200 mmol/L NaCl for salt stress, and 20% PEG6000 for drought stress); and hormone treatments (75 mol/L ABA, 0.5 mmol/L SA). The samples at different treatment times (0 h, 12 h, and 24 h) were collected and stored in a refrigerator at −80 °C for subsequent experiments. Some healthy seedlings were transplanted into the experimental field to ensure the normal growth of the stem lettuce. Then, the stem diameter of the stem lettuce was observed and measured. When the stem diameter reached 1 cm, 2 cm, 3 cm, and 4 cm, which were named stage S1, S2, S3, and S4, the stem samples were collected and stored at −80 °C for subsequent experiments. Different tissues (roots, stems, leaves, and flowers) were also collected at the flowering stage of the stem lettuce in order to detect the expression profiles of LsCPP genes. All experiments were performed with three independent biological replicates.
2.6. Heat Map and qRT-PCR Analysis
The RNA sequencing data of four stem enlargement stages (S1, S2, S3, and S4) from stem lettuce was obtained from our previous study [35]. The transcript abundance of six LsCPPs at four stem enlargement stages (S1, S2, S3, and S4) was obtained through RNA-Seq. The heat map of LsCPPs was generated using the FPKM by TBtools software. The total RNA of different samples including stem enlargement stages (S1, S2, S3, and S4), plant tissues (root, stem, leave, and flower), abiotic stress (4 °C, 37 °C, NaCl, PEG6000), and hormone treatment (SA, ABA) was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed using a commercial kit (Beijing Tsingke Biotech Co, Ltd., Beijing, China). qPrimerDB 2.0 database was used to design specific primers according to the mRNA sequences of LsCPP genes [36]. To ensure the specificity of the primers, the designed primers were subjected to blast in the lettuce genome, then, the gene amplification was carried out by PCR using the designed specific primers. For gene expression analysis, qRT-PCR was performed using SYBR Green Premix (Toyobo, Shanghai, China), and the relative expression levels were calculated using the 2−ΔΔCT method [37]. The expression levels of LsCPPs were normalized to the lettuce LsTIP41 gene, which was the most stable gene under drought, salt stress, as well as hormone treatment [38]. The primers used in this study can be found in Table S1.
3. Results
3.1. Identification and Analysis of CPP TFs in Lettuce
A total of six CPP family members (named LsCPP1-LsCPP6) were identified in the lettuce genome through a Pfam domain search. As shown in Table S2, the number of amino acids in the LsCPP TFs ranged from 274 (LsCPP3) to 722 (LsCPP1); the molecular weight of the LsCPP TFs was 30.80 kDa–78.70 kDa. The protein isoelectric point of the LsCPP TFs ranged from 5.33 (LsCPP4) to 8.03 (LsCPP2). The aliphatic index of the LsCPP TFs was between 55.80 (LsCPP3) and 61.69 (LsCPP1). In addition, the hydrophilicity range of the proteins in the LsCPP family TFs was from −0.823 (LsCPP5) to −0.606 (LsCPP3). All six LsCPP were hydrophilic nuclear proteins.
3.2. Phylogenetic Analysis of LsCPP TFs
The phylogenetic tree of the LsCPP TFs was produced. As shown in Figure 1, 29 CPP family members from lettuce, Arabidopsis, rice, and tomato were separated into three clades: I, II, and III. Clade I contained four OsCPPs, four AtCPPs, two LsCPPs (LsCPP5, LsCPP6), and two SlCPPs. Five OsCPPs, four AtCPPs, four LsCPPs (LsCPP1, LsCPP2, LsCPP3, LsCPP4), and two SlCPPs belonged to Clade II. Only two OsCPPs were classified into Clade III.
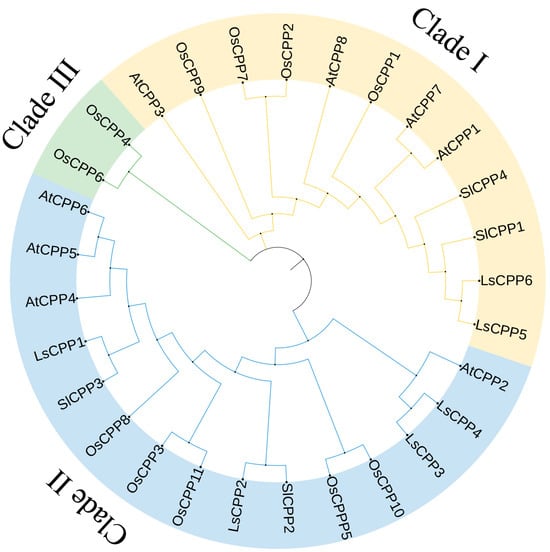
Figure 1.
Phylogenetic tree of CPP family members from lettuce (Ls), tomato (Sl), Arabidopsis (At), and rice (Os). Different branches are represented by different colors.
3.3. Analysis of Conserved Domains and Gene Structures
Protein sequence alignment showed that most LsCPPs had two CXC conserved domains and an R motif between the two conserved domains. As shown in Figure 2a, nine conserved cysteine (Cys) residues existed in CXC domains, and the R motif contained a relatively conserved RNPLAFXPK sequence. To investigate the structural variations in the LsCPP genes, this study conducted a comparative analysis of its full-length genome sequence and structural features. Significant differences in fragment length existed in the LsCPP genes. LsCPP3 contained the shortest genomic fragments, while the genomic fragment of LsCPP1 was the longest (Figure 2b).
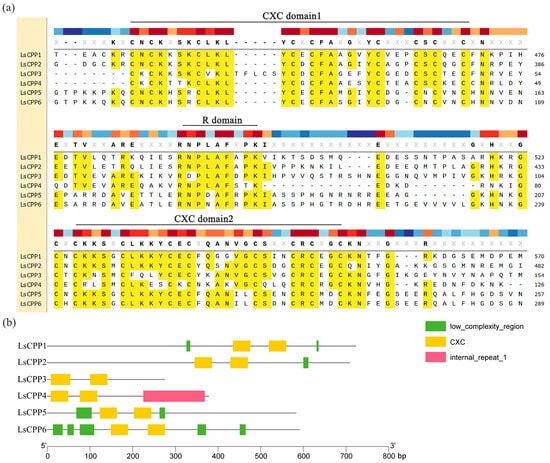
Figure 2.
Analysis of conserved domains and gene structures. (a) Multiple sequence alignment of the CXC domains in LsCPPs. (b) Gene structures of the LsCPP genes.
3.4. Analysis of Conserved Motifs of LsCPP TFs
The analysis of conserved motifs in lettuce LsCPP TFs was conducted by using MEME online tool. As shown in Figure 3a, 15 motifs were identified among six LsCPPs. Motif 1 was present in all six members. Motif 4 and Motif 5 were found in multiple LsCPPs, such as LsCPP1, LsCPP2, and LsCPP4. Except for LsCPP1 and LsCPP2, other family members (LsCPP3, LsCPP4, LsCPP5, and LsCPP6) lacked motif 10. LsCPP3 contained the lowest motifs. Although evolutionary divergence existed in the LsCPP TFs, certain motifs also were retained in specific subfamilies. For example, LsCPP5 and LsCPP6, which clustered in Clade I, shared same conserved motifs. LsCPP1 and LsCPP2 in Clade II contained the same motifs (motif 1, motif 2, motif 4, motif 5, motif 8 and motif 10).
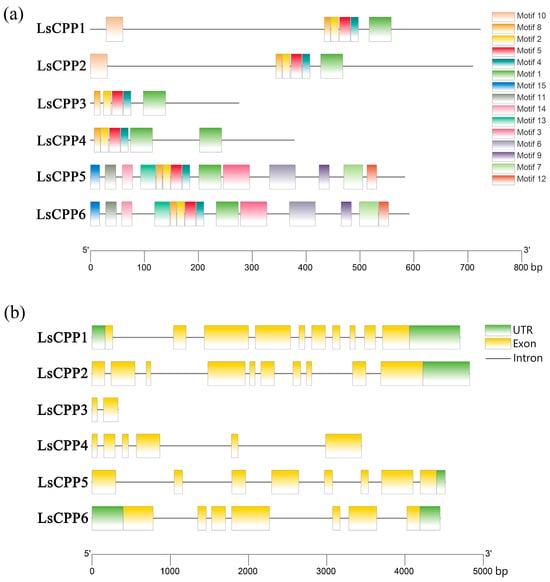
Figure 3.
Conserved motifs (a) and exon–intron structures (b) of LsCPP TFs.
Furthermore, the analysis of UTRs (untranslated regions) and exon/intron demonstrated that LsCPP3 contained only two exons, whereas LsCPP1 and LsCPP2 both harbored the highest number of exons (10) (Figure 3b). LsCPP1 and LsCPP6 had two UTRs, while LsCPP2 and LsCPP5 only contained one UTR.
3.5. Secondary and Tertiary Structure Prediction of Lettuce CPP
The secondary structures of six LsCPPs were identified. As shown in Figure 4a, the secondary structures of LsCPP1 and LsCPP2, both belonging to Clade II, were similar. A total of 606 random coils were identified in LsCPP2 and LsCPP1 contained 600 random coils. Six β-turns and seven β-turns were identified in LsCPP1 and LsCPP2, respectively. LsCPP5 and LsCPP6 contained 131 and 129 α-helices, respectively. A total of 72 α-helices, 14 extended chains, 12 β-turns, and 279 random coils were identified in LsCPP4. LsCPP3 ranked last in all metrics with the fewest α-helices (56), random coils (6), and β-turns (5). The LsCPP TFs had significant differences in secondary structure. Random coils and α-helices were predominant in most genes, while β-turns were usually fewer. This structural information is of great significance for understanding the functions of these genes and the protein folding mechanism.
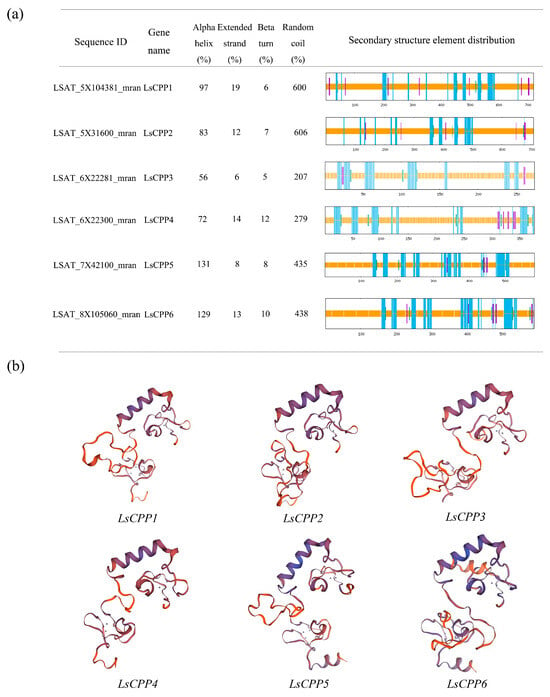
Figure 4.
Prediction of secondary and tertiary structures of LsCPPs. (a) Diagram of the secondary structures of LsCPPs. Color assignments for secondary structure elements: α-helix (blue), β-sheet (purple), β-turn (green), and random coil (yellow). (b) Tertiary structure models of LsCPP TFs.
The tertiary structures of LsCPP1-LsCPP6 proteins were predicted using the SwissModel online platform based on homology modeling (Figure 4b). The results showed that Clade II CPP TFs LsCPP1/LsCPP2 and Clade I CPP TFs LsCPP5/LsCPP6 shared similar and complex tertiary structures. These predictions provided a structural biological foundation for elucidating the functional mechanisms of CPPs in subsequent studies.
3.6. Cis-Element Analysis of LsCPP Genes
TFs can regulate gene transcription by binding to specific promoter sequences to respond to plant growth, development, and stress responses. As show in Figure 5, 27 classes of cis-acting elements, which were classified into four groups including developmental regulation (Box 4, GA-motif, MYB, MBSI, AE-box), hormone response (GARE-motif, P-box, TCA-element, TGACG-motif, ERE, MYC), environmental stress-related (TC-rich repes, ARE, LTR, ABRE), and light response (MRE, Sp1, TCCC-motif, ACE, GT1-motif) were identified. Notably, MYC (a MeJA response element) was present in all LsCPP genes; seven and eight MYC cis-elements were found in LsCPP2 and LsCPP6, respectively. LsCPP5 contained the fewest kinds of cis-acting elements. GA-motif was uniquely detected in the promoter of LsCPP2. Plant hormone-responsive elements, including ABA-responsive element ABRE and SA-related TCA-element, were also detected in LsCPP genes. Multiple hormone-responsive elements were found in the promoter region of LsCPP3, LsCPP4, and LsCPP6. The promoter of LsCPP5 contained many development-related cis-element. The prevalence of light-responsive elements (e.g., Sp1, ACE) further implies cross-talk between photomorphogenesis and stress adaptation mechanisms in lettuce.
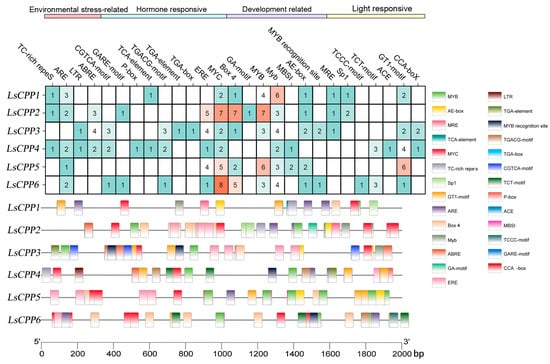
Figure 5.
The cis-acting elements in the promoter region of LsCPP genes. The upper part shows the number and classification of various cis-acting elements in each LsCPP gene promoter region. The lower part uses various colored squares to represent different cis-acting elements and their positions within the promoter region.
3.7. Chromosomal Location of LsCPP Genes
The chromosomal positions of six LsCPPs were mapped. As shown in Figure 6, the six LsCPPs were distributed on four chromosomes (Chr 5, Chr 6, Chr 7, and Chr 8). Two LsCPPs (LsCPP3 and LsCPP4) were located on Chr 6, and LsCPP1 and LsCPP2 were localized to Chr 5. Chromosome 7 and Chromosome 8 harbored one gene, LsCPP5 and LsCPP6, respectively.
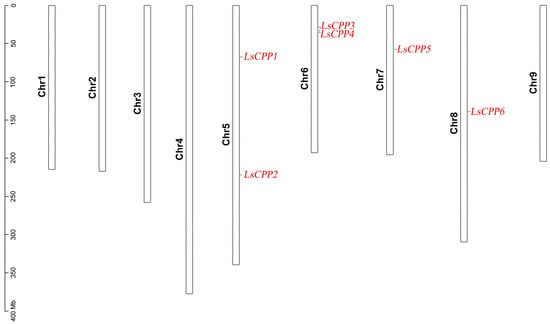
Figure 6.
The distribution of LsCPP genes on the lettuce chromosome.
3.8. Heatmap Analysis of LsCPP Gene Family During Stem Enlargement Stage
To investigate the expression patterns of the LsCPP gene family, transcriptome data from lettuce stems at different swelling stages were integrated. As shown in Figure 7, four LsCPP genes (LsCPP1, LsCPP2, LsCPP5, and LsCPP6) showed different expression patterns during the process of stem enlargement. LsCPP1 maintained high expression throughout the four stem enlargement stages (S1, S2, S3, and S4). No obvious changes were found in the expression levels of LsCPP3 and LsCPP4. Compared with stage S1, the expression pattern of LsCPP6 decreased at stages S2, S3, and S4.
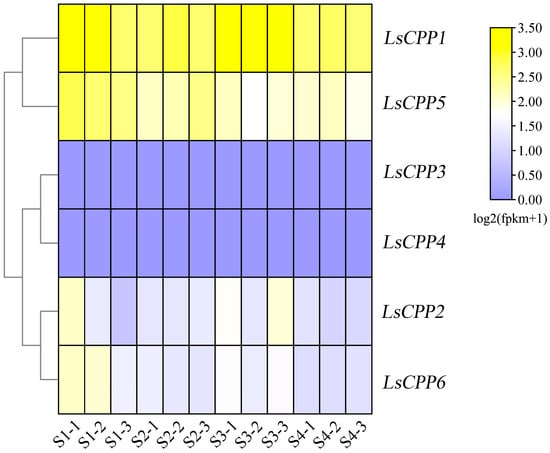
Figure 7.
Expression analysis of LsCPP genes in different tissues based on RNA-seq. S1, S2, S3, and S4 represent stem diameters of stem lettuce of 1 cm, 2 cm, 3 cm and 4 cm, respectively, with three biological replicates performed for each stage.
3.9. Expression Profiles Analysis of LsCPP Genes by qRT-PCR
3.9.1. Expression Levels of LsCPP Genes at Different Stem Expansion Stages and Different Tissues in Stem Lettuce
The LsCPP genes in lettuce showed significant expression differences across stem enlargement processes (S1, S2, S3, and S4). According to Figure 8a, LsCPP2 and LsCPP5 exhibited slightly increased expression at stage S4 compared with stage S1, while LsCPP6 showed moderate increases at stage S4. The expression level of LsCPP4 also showed an increase at stage S3 compared with stage S1. Overall, these results suggested that the LsCPP genes might primarily regulate later stages of lettuce stem development. The expression patterns of six LsCPP genes in different tissues (root, stem, leaf, and flower) were also measured (Figure 8b). Compared with the roots, LsCPP1 showed low expression levels in stems, leaves, and flowers, whereas LsCPP2 was highly expressed in stems, leaves, and flower compared with roots. Additionally, LsCPP5 and LsCPP6 showed elevated expression in leaf tissues compared with other tissues. Notably, LsCPP4 exhibited significantly lower expression in stem and leaves compared with the roots.
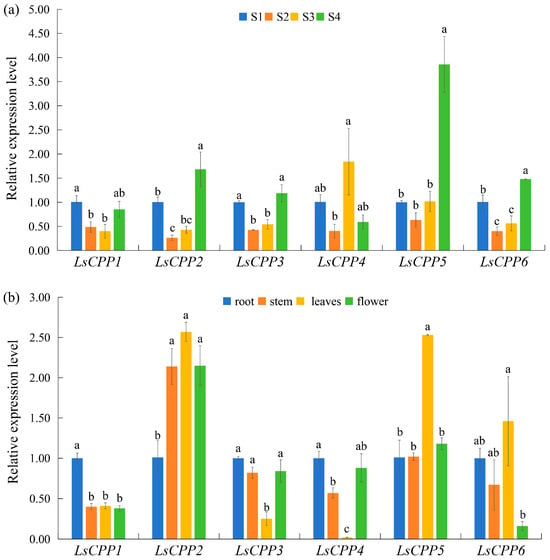
Figure 8.
Expression analysis of lettuce LsCPP genes in different stem enlargement stages (a) and different tissues (b) based on qRT-PCR. (a) S1, S2, S3, and S4 represent stem diameters of stem lettuce of 1 cm, 2 cm, 3 cm and 4 cm, respectively. Different lowercase letters indicate statistically significant differences (p ≤ 0.05) as determined by one-way ANOVA.
3.9.2. Expression Levels of LsCPP Genes Under Abiotic Stresses
The dynamic expression patterns of the LsCPP genes in stem lettuce under abiotic stress (4 °C, 37 °C, 200 mmol/L NaCl, and 20% PEG6000) were evaluated by qRT-PCR (Figure 9). After 4 °C for 12 h, most LsCPP genes (LsCPP1, LsCPP2, LsCPP3, and LsCPP5) showed a significant downregulation trend; however, compared with CK, LsCPP2 and LsCPP6 showed an upward trend in expression levels after 24 h of 4 °C. For 37 °C treatment, the expression level of LsCPP5 exhibited a downward trend after 12 h. No significant difference in expression levels was detected among all six LsCPP genes after 37 °C treatment for 24 h.
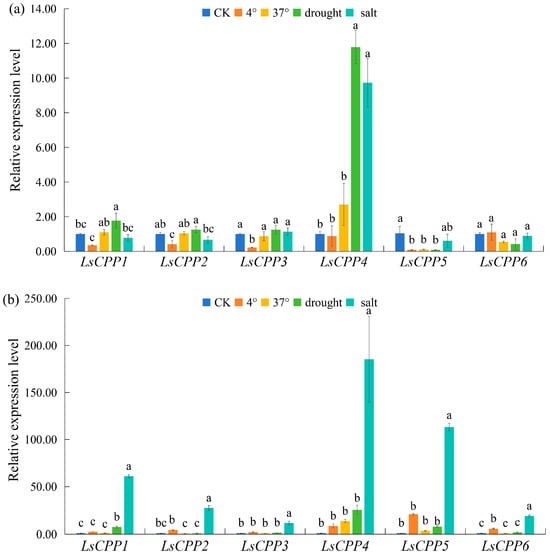
Figure 9.
Expression analysis of lettuce LsCPP genes under abiotic stress based on qRT-PCR. (a): 12 h after different abiotic stress, (b): 24 h after different abiotic stress. Different lowercase letters denote statistically significant differences (p ≤ 0.05) as determined by one-way ANOVA.
Under drought conditions, LsCPP1 and LsCPP4 showed an upward trend after 12 h. Compared with CK, a roughly 11 times increase in expression level was identified in LsCPP4. After 24 h of drought treatment, the expression level of LsCPP1 showed a slight increase compared with CK. LsCPP4 also exhibited the most significant increase (about 10 times) after NaCl treatment for 12 h. Notably, all six LsCPP genes showed varying degrees of upregulation after 24 h of salt stress. The expression levels of LsCPP4 and LsCPP5 increased about 170-fold and 100-fold compared with CK, respectively.
3.9.3. Expression Levels of LsCPP Genes Under Plant Hormone Treatment
As shown in Figure 10, plant hormone treatment (ABA and SA) significantly affected the expression dynamics of the LsCPP genes in stem lettuce. After ABA treatment for 12 h, the expression pattern of LsCPP6 showed the most pronounced induction effect with a 20-fold increase. However, after ABA treatment for 24 h, the expression level of LsCPP6 decreased compared with the control group. The expression level of LsCPP5 showed the opposite expression pattern with LsCPP6: the expression increased about three-fold after ABA treatment for 24 h. Similar expression patterns in response to ABA treatment were found in LsCPP1, which increased about six-fold at 24 h. For SA treatment, the expression levels of LsCPP5 and LsCPP6 showed obvious increases of 15 times and 35 times at 12 h. LsCPP1, LsCPP3, and LsCPP4 showed no difference in expression at 12 h. After SA treatment for 24 h, three LsCPP genes (LsCPP1, LsCPP4, and LsCPP5) significantly increased, while the expression level of LsCPP6 decreased. No difference in expression existed in LsCPP2 and LsCPP3 after SA treatment for 24 h.
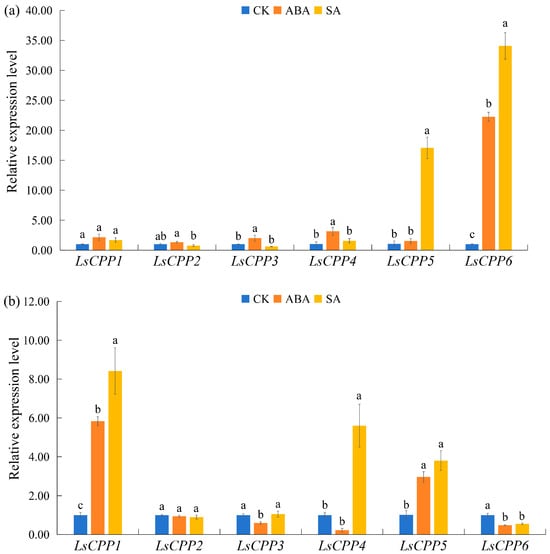
Figure 10.
Expression analysis of lettuce LsCPP genes under hormone treatment based on qRT-PCR. (a): 12 h after hormone treatment, (b): 24 h after hormone treatment. Different lowercase letters indicate statistically significant differences (p ≤ 0.05) as determined by one-way ANOVA.
3.10. Synteny Analysis of LsCPP Genes
Synteny refers to the conservation of gene arrangement order across different species or genomic regions. By analyzing the syntenic blocks of LsCPP genes, it is possible to infer whether they have undergone evolutionary events such as gene duplication (e.g., whole-genome duplication or segmental duplication) or rearrangement, and to identify functionally conserved key regions. As shown in Figure 11a, the collinear relationship between LsCPP5 and LsCPP6 indicated that the LsCPP gene family might have expanded within the species through duplication events, with the potential functional divergence or division of labor among its members.
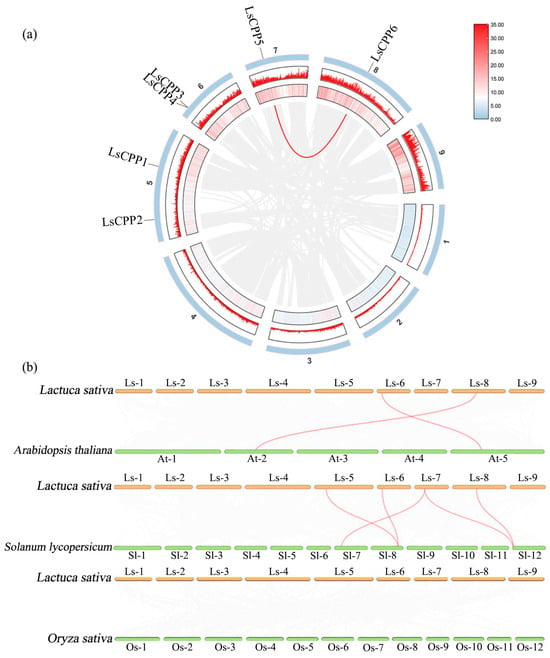
Figure 11.
Synteny analysis of the CPP genes. (a) Synteny analysis of the CPP genes in lettuce. Red lines indicate duplication events in LsCPPs. (b) Synteny analysis of the CPP genes among lettuce, Arabidopsis, tomato, and rice.
To further determine the evolutionary relationship, CPP genes of Arabidopsis, tomato, and rice were chosen for synteny analysis with lettuce. Two pairs of orthologous CPP genes were identified between Arabidopsis and lettuce. The collinear pairs between Arabidopsis and lettuce were clustered on the same branch, such as LsCPP6 and AT2G20110.2 (Figure 11b, Table S3). Five pairs of orthologous CPP genes were identified between tomato and lettuce. Similarly, collinear pairs between tomato and lettuce were clustered on the same branch, such as LsCPP2 and SlCPP2, LsCPP5 and SlCPP4, and LsCPP6 and SlCPP4. However, no collinear pairs of CPP genes were identified between rice and lettuce, indicating that the genetic relationship between lettuce, Arabidopsis, and tomato was more advanced than lettuce and rice.
3.11. Protein–Protein Interaction (PPI) Network Analysis of LsCPP TFs
Protein interaction network prediction based on known interaction groups is an effective method for studying unknown protein networks in plants. To investigate the regulatory mechanisms of LsCPPs, an interaction network was produced using STRING online tool. As shown in Figure 12, complex interactions existed between LsCPP TFs and other factors such as MYB3R3, DPB, and SWEET6. All six LsCPP TFs were shown to interact with MYB3R3 [39], while LsCPP3, LsCPP4, and LsCPP5 interacted with the Q8L637_ARATH and F5D21.3 proteins, which are involved in energy metabolism and cellular signaling. LsCPP5 (TCX5) showed a complex interaction with multiple proteins such as the ALY protein (ALY1 and ALY2), SWEET, MYB3R1, and other LsCPPs (LsCPP1, LsCPP2, LsCPP3, and LsCPP4). The co-expression relationships between LsCPPs and other factors indicate potential regulatory mechanisms among these proteins.
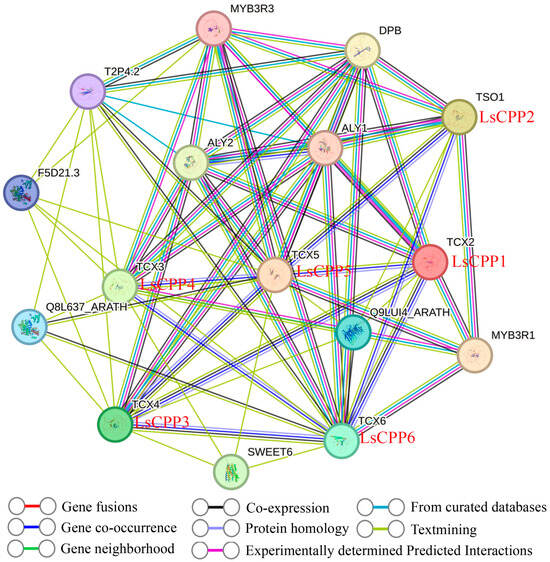
Figure 12.
Protein–protein interaction analysis. Different colors of the connecting lines represent distinct sources of interaction evidence.
4. Discussion
The CPP TF family contains two conserved CXC domains and a conserved R motif between these domains [40]. The CXC domains, widely present in both animals and plants, contained nine conserved cysteine residues, with the C1 domain exhibiting higher conservation than C2. This feature is consistently observed in CPP TF families across species such as rice, Arabidopsis, and cucumber [17,40]. As a transcription factor, CPP regulates the expression of downstream target genes through protein–DNA interactions [19,41]. Soybean CPP1 has been identified to interact with the promoter of Gmlbc3; the DNA-binding domain of CPP1 was the CXC domain, which contained two similar Cys-rich domains with 9 and 10 Cys, respectively [41].
In this study, six LsCPP TFs (LsCPP1-LsCPP6) were identified in the lettuce genome. Their physicochemical properties and subcellular localization align with typical TF characteristics (Table S2). Notably, LsCPP members display substantial variation in amino acid numbers (274–722) and molecular weights (30.80 kDa–78.70 kDa); similar phenomenon has also been reported in CPP families of other plants. For example, among eight AtCPP members in Arabidopsis, the shortest (AtCPP3) contained only 89 amino acids, while AtCPP1 comprised 728 amino acids [17]. Similarly, the molecular weight range (10.50 kDa–75.20 kDa) of six CPP family members in tomato was comparable to that of lettuce [19]. Despite these variations, all LsCPPs harbored a conserved CXC domain and were localized to the nucleus, indicating their core function in DNA binding-mediated transcriptional regulation. Six LsCPP TFs in lettuce and eight AtCPP TFs in Arabidopsis were classified into two clades (I and II), while eleven CPP TFs in rice were distributed into three clades (I, II, and III); the different numbers and clades of CPP TFs suggest that divergence existed between lettuce, Arabidopsis, and other species during the evolutionary process [42]. Gene duplication events, which play a vital role during the process of gene family expansion, have been identified to enhance plant’s adaptability [43]. In this study, only one orthologous gene pair was identified in lettuce, while three orthologous gene pairs existed in maize plants. The lower gene replication in lettuce may be related to the small number of CPP family members in lettuce [42]. Two, five, and zero orthologous gene pairs were identified in lettuce and Arabidopsis, tomato, rice, respectively, which also showed gene expansion events that existed in plant species. Similar results were identified in the gene families of other species [44,45].
MYB3R3 has been shown to be associated with cold stress, salt stress, and drought stress responses [39]. AtTSOI, a kind of CPP TF, was confirmed to interact with the MYB3R1 protein, which could form a regulatory module to coordinate cell proliferation and differentiation in buds and roots [21]. An interaction network analysis showed that most LsCPP TFs (LsCPP1, LsCPP2, LsCPP4, LsCPP5, and LsCPP6) could interact with MYB3R1 and MYB3R3, which indicated that these LsCPP TFs might play a similar function in response to abiotic stress and plant growth processes. The interaction relationship between LsCPP TFs with the TSO1 protein needs further verification.
The promoters of CPP genes in different plants have been confirmed to contain many cis-elements related to plant growth and development as well as abiotic stress [13,23]. An analysis of the cis-acting elements in the promoter regions of LsCPP genes revealed an abundance of hormone-responsive elements, development-responsive elements, and environmental stress-related elements (Figure 5). Studies have indicated that the expression levels of the CPP gene family vary across different plant tissues [40]. Most of them have high expression levels in young tissues. Some CPP members could be specifically expressed during the pollen development stage, the young leaf stage, and the actively growing terminal bud stage [16,46]. In wheat, TaCPP1-1A and TaCPP13-4D were highly regulated in leaves, while their expression decreased markedly in mature spikes [22]. In contrast, members of the AtCPP family in Arabidopsis (such as AtCPP5-AtCPP7) are mainly involved in the regulation of floral organ development and cell division, while the OsCPPs in rice are related to seed germination and hormone signal transduction [17]. PeCPP05 is highly expressed only in bamboo flowers and is basically not expressed in roots [11]. In lettuce, three LsCPP genes (LsCPP2, LsCPP5, and LsCPP6) showed higher expression levels in the leaf than in the root, stems and flower. These differences existed in the expression profiles of different tissues and indicated that the functions of CPP TFs may have diverged due to the ecological adaptation requirements of different species. Two LsCPP (LsCPP2 and LsCPP5) were identified to play potential roles during the process of stem enlargement in stem lettuce (Figure 8). The heat map and qRT-PCR analysis both showed that LsCPP genes might participated in the process of stem enlargement in stem lettuce. The expression levels of LsCPP1 in stage S2 and stage S4 were decreased compared with stage S1. LsCPP6 also showed decreased expression in stages S2 and S3 than in stage S1. Some deviations existed in the expression profile of LsCPP2 by RNA sequence and qRT-PCR, which might be related to some deviations in RNA sequencing and the complexity of transcriptional regulation.
CPP genes can be involved in the process of abiotic/biotic stress as well as plant hormones and plant growth and development [19,20,24,47,48,49]. In this study, qRT-PCR showed that LsCPP genes participated in the response to different abiotic stress. The expression levels of LsCPP4 increased significantly after NaCl and drought treatment (Figure 9). Similar results are reported in CPP TFs from other plants. ZmCPP2 could be phosphorylated by ZmSK1, which reduced drought tolerance in maize [49]. Rice’s OsCPP5 gene decreased the tolerance to salt stress by interacting with other factors such as TCP TFs [50]. Potato StCPP3 decreased the resistance to Ralstonia solanacearum by interacting with HrpB7 proteins [48]. In this study, the expression of LsCPP6 in lettuce increased significantly at 12 h under salt stress, suggesting functional conservation in stress resistance mechanisms.
5. Conclusions
The CPP TF family plays important roles in multiple plant species, especially in growth, development, and stress responses. In this study, CPP family members have been identified in lettuce. Bioinformatics approaches including phylogenetic analysis, chromosomal localization, and cis-element analyses have been conducted in order to conduct a functional analysis of LsCPP TFs. The results indicated that the LsCPP gene family in lettuce might regulate leaf development, stress tolerance, and the stem swelling process of lettuce. Subsequent functional verification studies, such as gene knockout or overexpression experiments, will help to further clarify the specific action mechanisms of the LsCPP gene family in lettuce.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15092120/s1, Table S1: Primer sequences used in the text; Table S2: Characteristic features of LsCPP TFs; Table S3: The paralogs and orthologs gene of CPP TFs between lettuce, Arabidopsis, tomato, and rice. Table S4: CPP family members of lettuce (Ls), tomato (Sl), Arabidopsis (At), and rice (Os).
Author Contributions
Methodology, Y.H. and M.L.; Software, M.Z., L.J., Z.C., C.W., and Q.Z.; Visualization, M.Z., L.J., Z.C., and C.W.; Investigation, M.Z., L.J., Z.C., and C.W.; Writing—original draft, M.Z.; Writing—review and editing, P.X., Q.Z., M.L., and Y.H.; Resources, Y.H. and M.L.; Funding acquisition, P.X., Q.Z., M.L., and Y.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32102405, 32002027, and 32202455); the Natural Science Foundation of Shandong Province (ZR2024MC152 and ZR2022QC016); and the Innovation team of youth technology project of high school in Shandong Province (2021KJ055).
Data Availability Statement
The original contributions presented in the study are publicly available. The data can be found here: NCBI, PRJNA844256.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| ABA | Abscisic acid |
| CDS | Coding sequence |
| CPP | Cysteine-rich polycomb-like protein |
| GA | Gibberellin |
| JA | Jasmonic acid |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| SA | Salicylic acid |
| TFs | Transcription factors |
| UTR | Untranslated region |
References
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, 23–24. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Q.; Qin, Z.; Huang, J. Transcription factor OsNAC055 regulates GA-mediated lignin biosynthesis in rice straw. Plant Sci. 2022, 325, 111455. [Google Scholar] [CrossRef]
- Li, J.; Li, B.; Zhou, X.; Ma, X.; Xie, R.; Sun, Y.; Su, S. An ALOG transcription factor targets a TALE homeobox gene during corolla abscission in Torenia fournieri. New Phytol. 2025, 247, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Shi, Y.; Guo, L.; Fu, D.; Li, M.; Zhang, X.; Li, Z.; Zhuang, J.; Yang, X.; Zuo, J.; et al. A natural variant of COOL1 gene enhances cold tolerance for high-latitude adaptation in maize. Cell 2025, 188, 1315–1329. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Gao, F.; Dubos, C. The Arabidopsis bHLH transcription factor family. Trends Plant Sci. 2024, 29, 668–680. [Google Scholar] [CrossRef]
- Xu, P.; Yang, Y.; Zhao, Z.; Hu, J.; Xie, J.; Wang, L.; Zheng, H.; Cai, W. The transcription factor Dof3.6/OBP3 regulates iron homeostasis in Arabidopsis. EMBO J. 2025, 44, 251–268. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Zhang, F.; Fang, C.; Liang, W. Molecular mechanisms regulating lamina joint development in rice. Agronomy 2024, 14, 1562. [Google Scholar] [CrossRef]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Xuan, X.; Su, S.; Jiao, Y.; Guo, H.; Zhang, Z. Comprehensive analysis of the CPP gene family in Moso bamboo: Insights into their role in rapid shoot growth. BMC Genom. 2024, 25, 1173. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, F.; Ma, J.; Liu, H.; Ye, H.; Zhao, P.; Wang, J. Comprehensive evolutionary analysis of CPP genes in Brassica napus L. and its two diploid progenitors revealing the potential molecular basis of allopolyploid adaptive advantage under salt stress. Front. Plant Sci. 2022, 13, 873071. [Google Scholar] [CrossRef] [PubMed]
- Bakhtari, B.; Zamani, E. Genome-wide identification and characterization of Cysteine-Rich Polycomb-like Protein and E2F/DP gene families in quinoa (Chenopodium quinoa). Genet. Resour. Crop Evol. 2025, 72, 6387–6405. [Google Scholar] [CrossRef]
- Jiang, L.; Zhu, M.; Huang, Y.; Zhang, Q. Genome-wide characterization and expression analysis of the cysteine-rich polycomb-like protein gene family in response to hormone signaling in apple (Malus domestica). Int. J. Mol. Sci. 2025, 26, 5528. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Running, M.P.; Meyerowitz, E.M. TSO1 functions in cell division during Arabidopsis flower development. Development 1997, 124, 665–672. [Google Scholar] [CrossRef]
- Sijacic, P.; Wang, W.; Liu, Z. Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PLoS Genet. 2011, 7, 11. [Google Scholar] [CrossRef]
- Yang, Z.; Gu, S.; Wang, X.; Li, W.; Tang, Z.; Xu, C. Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and rice. J. Mol. Evol. 2008, 67, 266–277. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Ye, S.; Jiang, L.; Liu, S. Genome-wide identification and characterization of cysteine-rich polycomb-like protein CPP family genes in cucumber (Cucumis sativus) and their roles in stress responses. Biologia 2018, 73, 425–435. [Google Scholar] [CrossRef]
- Sun, Y.; Jia, X.; Chen, D.; Fu, Q.; Chen, J.; Yang, W.; Yang, H.; Xu, X. Genome-wide identification and expression analysis of cysteine-rich polycomb-like protein (CPP) gene family in tomato. Int. J. Mol. Sci. 2023, 24, 5762. [Google Scholar] [CrossRef]
- Dzinyela, R.; Manda, T.; Hwarari, D.; Yang, L.M.; Movahedi, A. Genomic characterization and expression analysis of the cysteine-rich polycomb-like protein gene family of Populus trichocarpa. S. Afr. J. Bot. 2025, 181, 67–82. [Google Scholar] [CrossRef]
- Wang, W.; Sijacic, P.; Xu, P.; Lian, H.; Liu, Z. Arabidopsis TSO1 and MYB3R1 form a regulatory module to coordinate cell proliferation with differentiation in shoot and root. Proc. Natl. Acad. Sci. USA 2018, 115, 13. [Google Scholar] [CrossRef]
- Ullah, U.; Buttar, Z.A.; Shalmani, A.; Muhammad, I.; Ud-Din, A.; Ali, H. Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions. Open Life Sci. 2022, 171, 544–562. [Google Scholar] [CrossRef]
- Tian, J.Y.; Wang, Q.X.; Zheng, S.W. Genome-wide identification and expression profile analysis of the CPP gene family in Medicago truncatula. Acta Prataculturae Sin. 2022, 317, 111–121. [Google Scholar]
- Nan, H.; Lin, Y.L.; Wang, X.H.; Gao, L.Z. Comprehensive genomic analysis and expression profiling of cysteine-rich polycomb-like transcription factor gene family in tea tree. Hortic. Plant J. 2021, 7, 469–478. [Google Scholar] [CrossRef]
- Li, Z.F.; Yang, X.Y.; Li, Z.X.; Zou, X.L.; Jiang, C.Y.; Zhu, J.X.; Zhang, Y.J.; Han, Y.Y.; Liu, C.J.; Hao, J.J. Genome-wide analysis of ABF gene family in lettuce (Lactuca sativa L.) reveals the negative roles of LsABF1 in thermally induced bolting. J. Hortic. Sci. Bi. 2025, 100, 53–67. [Google Scholar] [CrossRef]
- Park, S.; Shi, A.; Meinhardt, L.W.; Mou, B. Genome-wide characterization and evolutionary analysis of the AP2/ERF gene family in lettuce Lactuca sativa. Sci. Rep. 2023, 131, 21990. [Google Scholar] [CrossRef]
- Yun, Y.J.; Kim, S.S.; Lee, J.H.; Kim, Y.C. Overexpression of lettuce TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factor genes (LsTCP13 and LsTCP17) promotes flowering time through upregulation of AtFT and AtAP1 in Arabidopsis. Plant Biotechnol. Rep. 2023, 17, 509–517. [Google Scholar] [CrossRef]
- Luo, K.S.; Zhang, D.C.; Zhai, Z.D.; Liu, X.; Zhou, J.; Zhang, B.; Li, D.Y. Genome-wide analysis of YABBY gene family in lettuce (Lactuca sativa) and functional characterization of LsaFILd. J. Plant Growth Regul. 2023, 42, 2124–2135. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Ceric, G.; Forslund, K.; Eddy, S.R.; Sonnhammer, E.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, 281–288. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the swiss bioinformatics resource portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11 molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Nystrom, S.L.; McKay, D.J. Memes: A motif analysis environment in R using tools from the MEME Suite. PLoS Comput. Biol. 2021, 17, e1008991. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Huang, Y.; Li, Y.; Liu, Z.; Chen, W.; Wang, Y.; Wang, X.; Liu, Y.; Zheng, Y. Combined analysis of the transcriptome and metabolome provides insights into the fleshy stem expansion mechanism in stem lettuce. Front. Plant Sci. 2022, 13, 1101199. [Google Scholar] [CrossRef]
- Li, X.D.; Meng, B.Y.; Zhang, Z.; Wei, L.J.; Chang, W.; Wang, Y.H.; Zhang, K.; Li, T.; Lu, K. qPrimerDB 2.0: An updated comprehensive gene-specific qPCR primer database for 1172 organisms. Nucleic Acids Res. 2024, 53, D205–D210. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Borowski, J.M.; Galli, V.; Messias Rda, S.; Perin, E.C.; Buss, J.H.; Silva, S.D.d.A.e.; Rombaldi, C.V. Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta 2014, 239, 1187–1200. [Google Scholar] [CrossRef]
- Zheng, Q.; Takei-Hoshi, R.; Okumura, H.; Ito, M.; Kawaguchi, K.; Otagaki, S.; Matsumot, S.; Luo, Z.; Zhang, Q.; Shiratake, K. Genome editing of SlMYB3R3, a cell cycle transcription factor gene of tomato, induces elongated fruit shape. J. Exp. Bot. 2022, 73, 7312–7325. [Google Scholar] [CrossRef]
- Lu, T.; Dou, Y.; Zhang, C. Fuzzy clustering of CPP family in plants with evolution and interaction analyses. BMC Bioinform. 2013, 14, S10. [Google Scholar] [CrossRef]
- Cvitanich, C.; Pallisgaard, N.; Nielsen, K.A.; Hansen, A.C.; Larsen, K.; Pihakaski-Maunsbach, K.; Marcker, K.A.; Jensen, E.O. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc. Natl. Acad. Sci. USA 2000, 97, 8163–8168. [Google Scholar] [CrossRef]
- Gu, L.; Kang, T.Y.; Zeng, T.; Wang, H.C.; Zhu, B.; Du, X.Y.; Liu, Y.L. Comprehensive identification and expression analysis of the CPP gene family in maize (Zea mays L.). BMC Plant Biol. 2025, 25, 731. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, X.; Bao, J.; Shan, Y.; Zhang, M.; Shen, Y.; Abubakar, Y.S.; Lu, G.; Wang, Z.; Wang, A. Genome-wide identification and expression analysis of SNARE genes in Brassica napus. Plants 2022, 11, 711. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, Z.P.; Cheng, Y.; Ruan, M.Y.; Ye, Q.J.; Wang, R.Q.; Zhou, G.Z.; Liu, J.; Liu, C.C.; Wan, H.J. Divergent retention of sucrose metabolism genes after whole genome triplication in the tomato (Solanum lycopersicum). Plants 2023, 12, 4145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Yuan, W.C.; Zhao, Y.J.; Ren, Y.; Zhao, X.Q.; Yuan, Z.H. Genome-wide identification and evolutionary analysis of AOMT gene family in pomegranate (Punica granatum). Agronomy 2021, 11, 318. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, H.K.; Wang, Y.M.; Yuan, C.P.; Zhang, Y.Y.; Li, H.Y.; Yan, X.F.; Li, Q.Y.; Dong, Y.S. Genome-wide identification and expression analysis of the CPP-like gene family in soybean. Genet. Mol. Res. 2015, 14, 1260–1268. [Google Scholar] [CrossRef]
- Huang, J.; Xu, W.; Zhai, J.; Hu, Y.; Guo, J.; Zhang, C.; Zhao, Y.; Zhang, L.; Martine, C.; Ma, H.; et al. Nuclear phylogeny and insights into whole-genome duplications and reproductive development of Solanaceae plants. Plant Commun. 2023, 4, 100595. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Cheng, L.X.; Guan, X.Y.; Liang, Y.; Xue, Y.J.; Zhao, W.Y.; Zhang, Z.Y.; Chang, X.Y.; Liang, L.Q.; Gao, G. StCPP3 interacts with type III secretion protein HrpB7 and negatively regulates plant resistance against Ralstonia solanacearum. Biochem. Biophys. Res. Commun. 2025, 742, 151105. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Liu, W.J.; Niu, Y.X.; Li, Q.; Zhao, C.Y.; Pan, Y.T.; Li, G.D.; Bian, X.L.; Miao, Y.D.; Zhang, A.Y. The maize GSK3-like kinase ZmSK1 negatively regulates drought tolerance by phosphorylating the transcription factor ZmCPP2. Plant Cell 2025, 37, koaf032. [Google Scholar] [CrossRef]
- Almeida, D.M.; Gregorio, G.B.; Oliveira, M.M.; Saibo, N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 2017, 93, 61–77. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).