Comprehensive Analysis of ZmTBL Genes Reveals Their Roles in Maize Development and Abiotic Stress Responses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Identification and Evolutionary Analysis of ZmTBL Genes
2.3. Computational Prediction of ZmTBL Proteins’ Physicochemical Characteristics
2.4. Spatiotemporal Expression Analysis and Upstream Regulator Prediction of ZmTBLs
2.5. Expression Profiling and qPCR Analyze of the ZmTBLs Under Abiotic Stresses
2.6. Association Analysis Between ZmTBLs Variations and Maize Agronomic Traits
3. Results
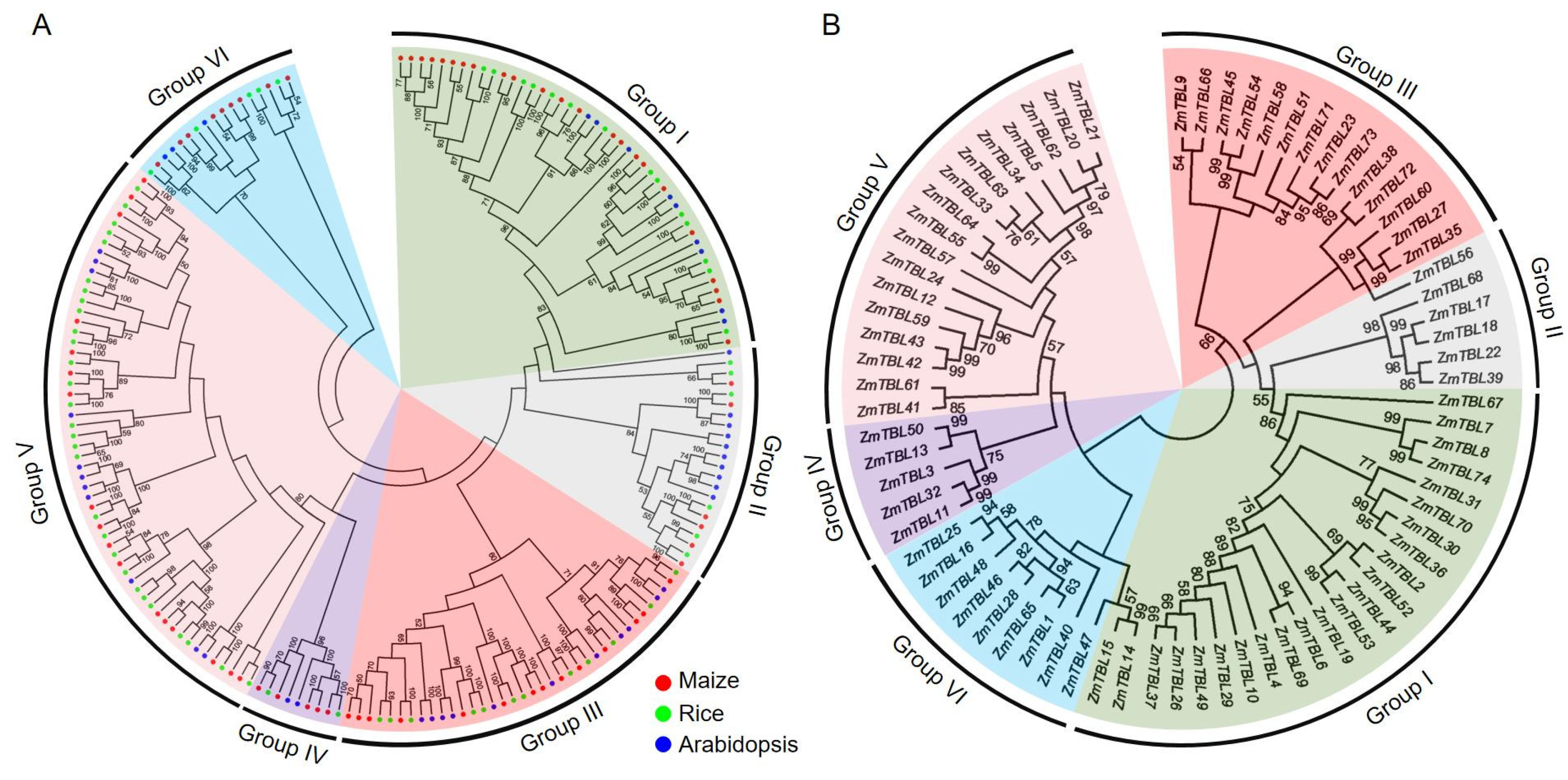
3.1. Identification and Phylogenetic Analysis of ZmTBLs
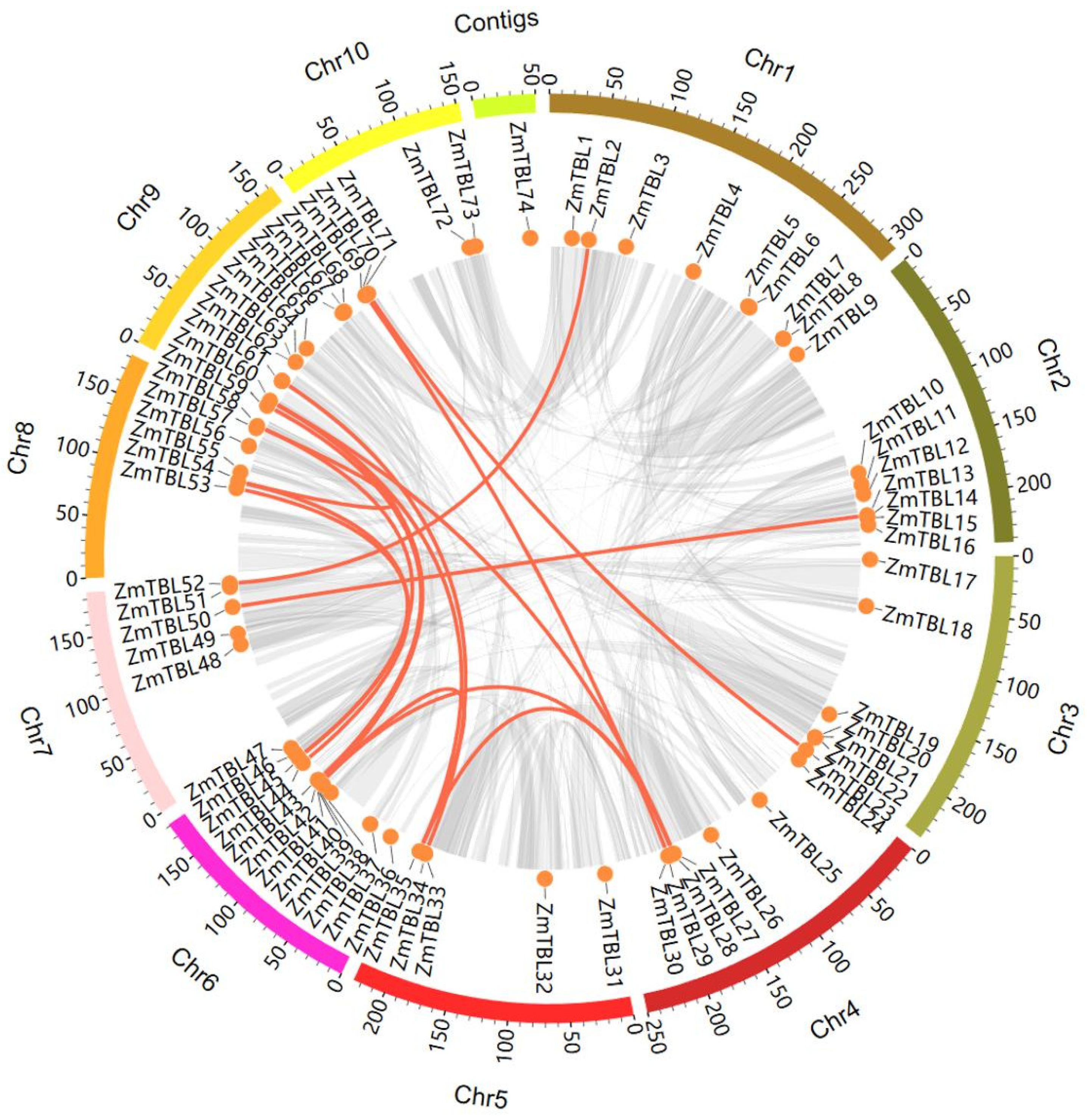
3.2. Chromosomal Localization and Syntenic Analysis of the ZmTBLs
3.3. Physicochemical Properties of ZmTBL Proteins
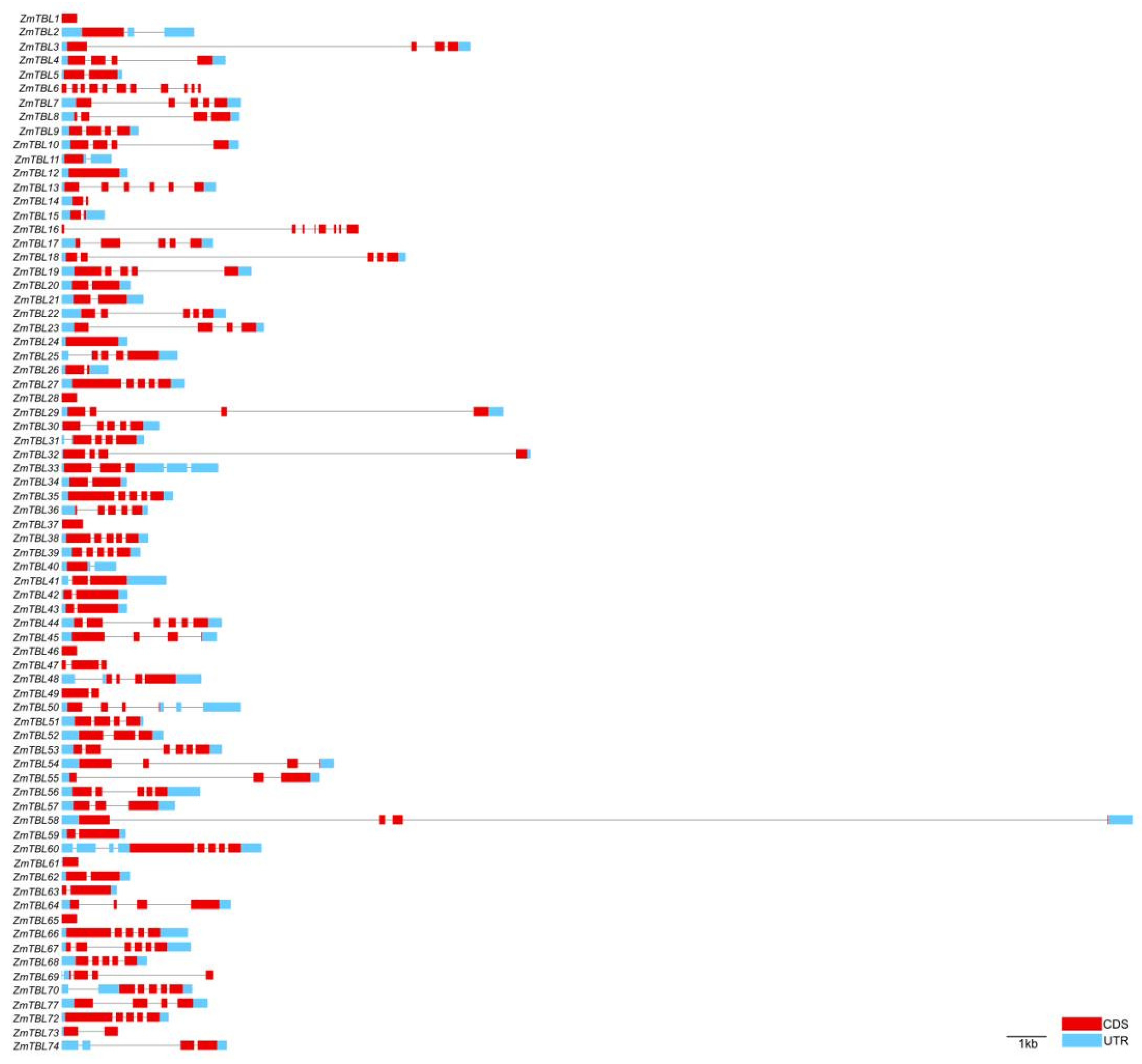
3.4. Gene Structure and Protein Conserved Motifs Analysis of ZmTBLs
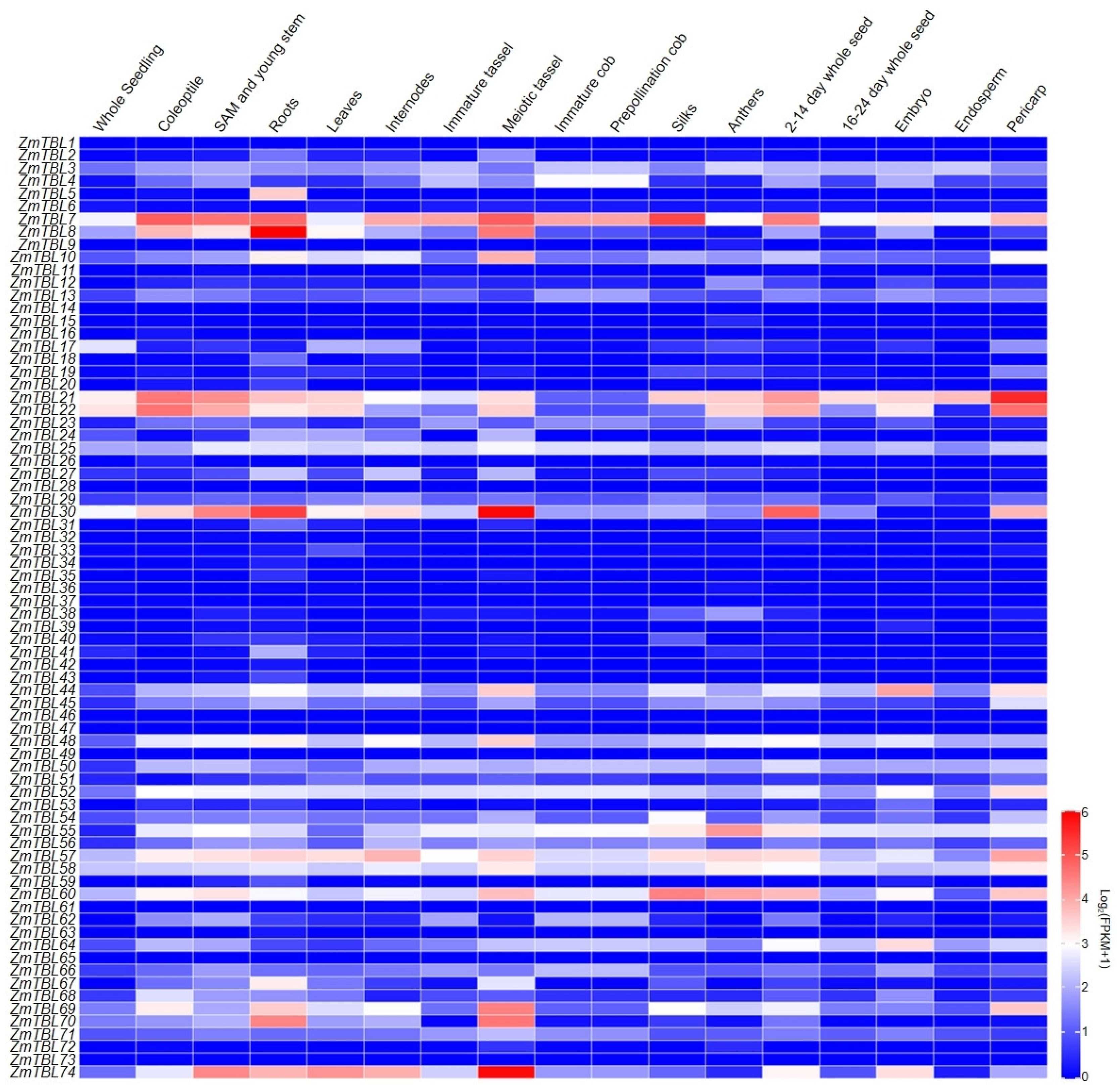
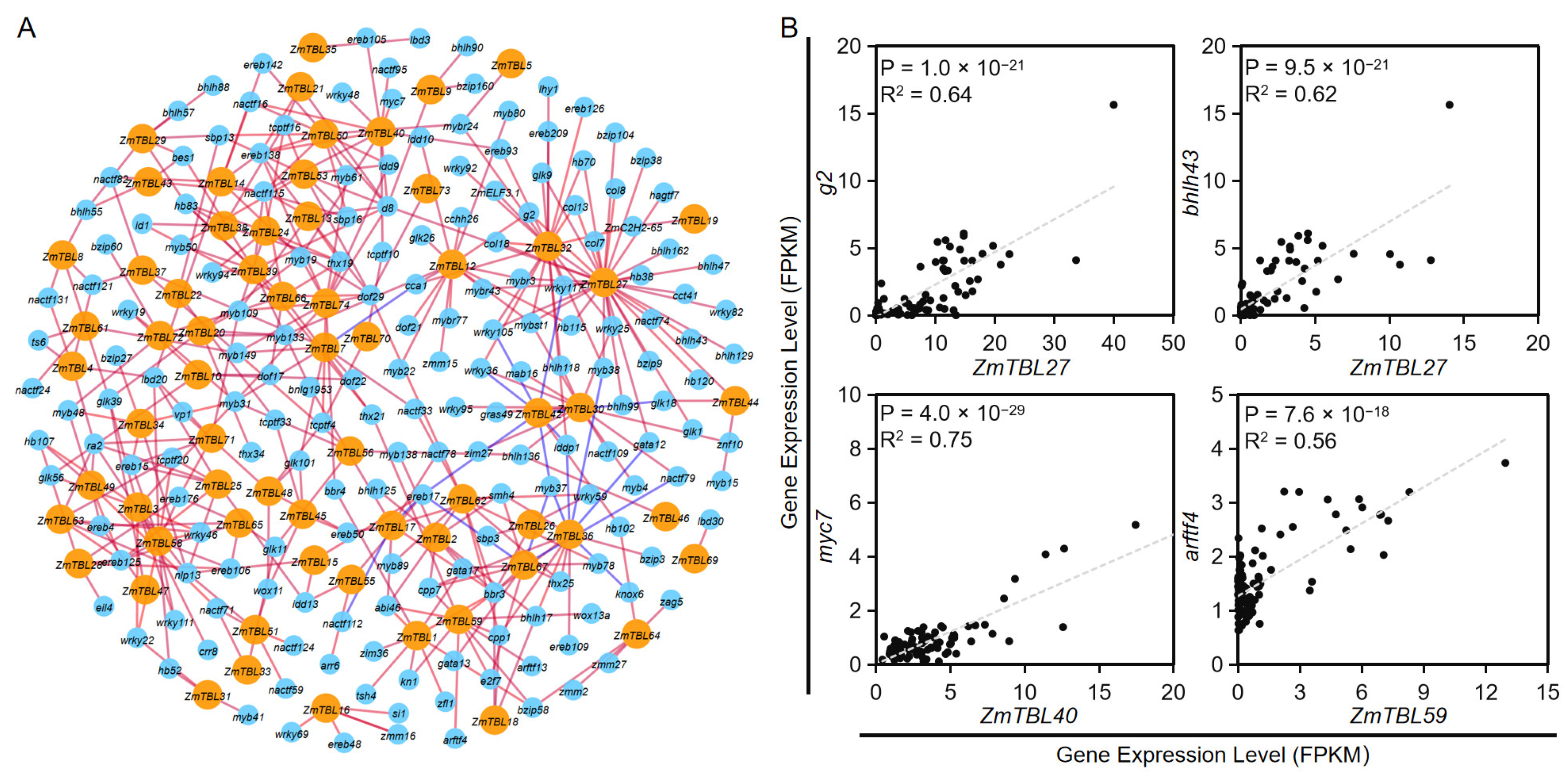
3.5. Tissue-Specific Expression and Upstream Regulator Analysis of ZmTBLs
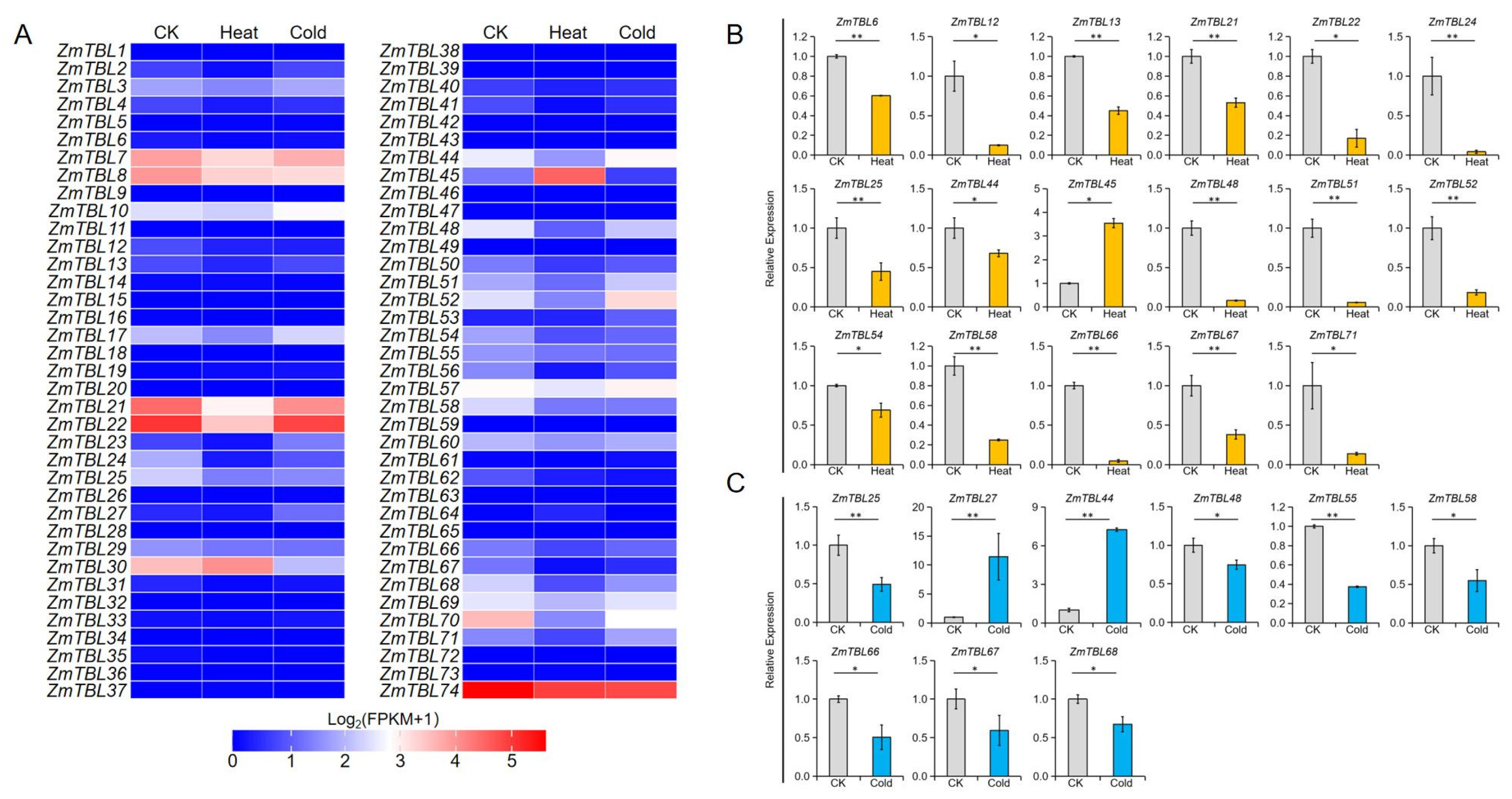
3.6. Potential Role of ZmTBLs in Regulating Maize Abiotic Stress Responses
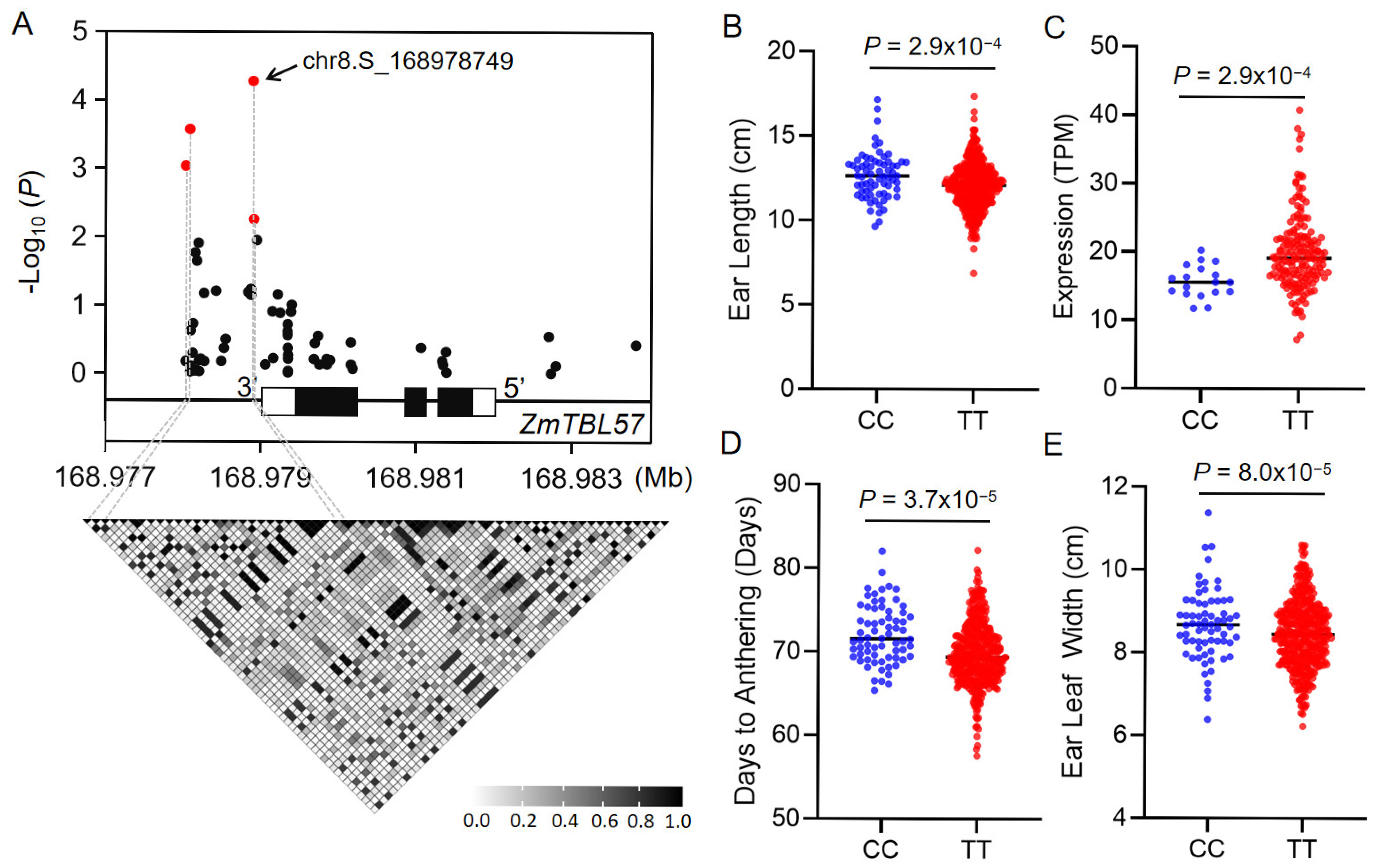
3.7. Variations in ZmTBLs Were Associated with Maize Agronomic Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pauly, M.; Ramírez, V. New Insights Into Wall Polysaccharide O-Acetylation. Front. Plant Sci. 2018, 9, 1210. [Google Scholar] [CrossRef]
- Zhong, R.; Adams, E.R.; Ye, Z.H. Ancient Origin of Acetyltransferases Catalyzing O-acetylation of Plant Cell Wall Polysaccharides. Plant Cell Physiol. 2024, 65, 1388–1398. [Google Scholar] [CrossRef]
- Gou, J.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef]
- Grantham, N.J.; Wurman-Rodrich, J.; Terrett, O.M.; Lyczakowski, J.J.; Stott, K.; Iuga, D.; Simmons, T.J.; Durand-Tardif, M.; Brown, S.P.; Dupree, R.; et al. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat. Plants 2017, 3, 859–865. [Google Scholar] [CrossRef]
- Lunin, V.V.; Wang, H.T.; Bharadwaj, V.S.; Alahuhta, M.; Peña, M.J.; Yang, J.Y.; Archer-Hartmann, S.A.; Azadi, P.; Himmel, M.E.; Moremen, K.W.; et al. Molecular Mechanism of Polysaccharide Acetylation by the Arabidopsis Xylan O-acetyltransferase XOAT1. Plant Cell 2020, 32, 2367–2382. [Google Scholar] [CrossRef] [PubMed]
- Schultink, A.; Naylor, D.; Dama, M.; Pauly, M. The role of the plant-specific ALTERED XYLOGLUCAN9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol. 2015, 167, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Yu, B.; Zhang, Q.; Peng, Y.; Yang, J.; Sun, Y.; Qin, P.; Jia, T.; Smeekens, S.; Teng, S. MYC2-Activated TRICHOME BIREFRINGENCE-LIKE37 Acetylates Cell Walls and Enhances Herbivore Resistance. Plant Physiol. 2020, 184, 1083–1096. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Teng, Q.; Zhong, R.; Ye, Z.H. Roles of Arabidopsis TBL34 and TBL35 in xylan acetylation and plant growth. Plant Sci. 2016, 243, 120–130. [Google Scholar] [CrossRef]
- Zhong, R.; Zhou, D.; Chen, L.; Rose, J.P.; Wang, B.C.; Ye, Z.H. Plant Cell Wall Polysaccharide O-Acetyltransferases. Plants 2024, 13, 2304. [Google Scholar] [CrossRef]
- Chiniquy, D.; Underwood, W.; Corwin, J.; Ryan, A.; Szemenyei, H.; Lim, C.C.; Stonebloom, S.H.; Birdseye, D.S.; Vogel, J.; Kliebenstein, D.; et al. PMR5, an acetylation protein at the intersection of pectin biosynthesis and defense against fungal pathogens. Plant J. 2019, 100, 1022–1035. [Google Scholar] [CrossRef]
- Kabir, N.; Wang, X.; Lu, L.; Qanmber, G.; Liu, L.; Si, A.; Zhang, L.; Cao, W.; Yang, Z.; Yu, Y.; et al. Functional characterization of TBL genes revealed the role of GhTBL7 and GhTBL58 in cotton fiber elongation. Int. J. Biol. Macromol. 2023, 241, 124571. [Google Scholar] [CrossRef]
- Gao, Y.; He, C.; Zhang, D.; Liu, X.; Xu, Z.; Tian, Y.; Liu, X.H.; Zang, S.; Pauly, M.; Zhou, Y.; et al. Two Trichome Birefringence-Like Proteins Mediate Xylan Acetylation, Which Is Essential for Leaf Blight Resistance in Rice. Plant Physiol. 2017, 173, 470–481. [Google Scholar] [CrossRef]
- Zhong, K.; Zhang, P.; Wei, X.; Platre, M.P.; He, W.; Zhang, L.; Małolepszy, A.; Cao, M.; Hu, S.; Tang, S.; et al. Natural variation of TBR confers plant zinc toxicity tolerance through root cell wall pectin methylesterification. Nat. Commun. 2024, 15, 5823. [Google Scholar] [CrossRef]
- Zhu, X.F.; Sun, Y.; Zhang, B.C.; Mansoori, N.; Wan, J.X.; Liu, Y.; Wang, Z.W.; Shi, Y.Z.; Zhou, Y.H.; Zheng, S.J. TRICHOME BIREFRINGENCE-LIKE27 affects aluminum sensitivity by modulating the O-acetylation of xyloglucan and aluminum-binding capacity in Arabidopsis. Plant Physiol. 2014, 166, 181–189. [Google Scholar] [CrossRef]
- Stranne, M.; Ren, Y.; Fimognari, L.; Birdseye, D.; Yan, J.; Bardor, M.; Mollet, J.C.; Komatsu, T.; Kikuchi, J.; Scheller, H.V.; et al. TBL10 is required for O-acetylation of pectic rhamnogalacturonan-I in Arabidopsis thaliana. Plant J. 2018, 96, 772–785. [Google Scholar] [CrossRef]
- Zhu, X.; Ma, X.; Hu, W.; Xing, Y.; Huang, S.; Chen, Z.; Fang, L. Genome-wide identification of TBL gene family and functional analysis of GhTBL84 under cold stress in cotton. Front. Plant Sci. 2024, 15, 1431835. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cao, Y.; Shi, Y.; Qin, F.; Jiang, C.; Yang, S. Genetic and molecular exploration of maize environmental stress resilience: Toward sustainable agriculture. Mol. Plant 2023, 16, 1496–1517. [Google Scholar] [CrossRef] [PubMed]
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic Stress in Crop Production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, P.; Jia, H.; Zhang, S.; Sun, X.; Nishawy, E.; Sun, X.; Dai, M. Regulatory mechanisms and breeding strategies for crop drought resistance. New Crops 2024, 1, 100029. [Google Scholar] [CrossRef]
- Yang, Z.; Qin, F. The battle of crops against drought: Genetic dissection and improvement. J. Integr. Plant Biol. 2023, 65, 496–525. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Song, H.; Qi, M.; Wang, M.; Bai, Y.; Sun, Y.; Yu, H. Impact of High-Temperature Stress on Maize Seed Setting: Cellular and Molecular Insights of Thermotolerance. Int. J. Mol. Sci. 2025, 26, 1283. [Google Scholar] [CrossRef] [PubMed]
- Si, C.; He, C.; Teixeira da Silva, J.A.; Yu, Z.; Duan, J. Metabolic accumulation and related synthetic genes of O-acetyl groups in mannan polysaccharides of Dendrobium officinale. Protoplasma 2022, 259, 641–657. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, S.; Liu, X.; Zhang, Z. Global Investigation of TBL Gene Family in Rose (Rosa chinensis) Unveils RcTBL16 Is a Susceptibility Gene in Gray Mold Resistance. Front. Plant Sci. 2021, 12, 738880. [Google Scholar] [CrossRef]
- Zhong, R.; Cui, D.; Ye, Z.H. A group of Populus trichocarpa DUF231 proteins exhibit differential O-acetyltransferase activities toward xylan. PLoS ONE 2018, 13, e0194532. [Google Scholar] [CrossRef]
- He, Z.; Zhang, J.; Jia, H.; Zhang, S.; Sun, X.; Nishawy, E.; Zhang, H.; Dai, M. Genome-wide identification and analyses of ZmAPY genes reveal their roles involved in maize development and abiotic stress responses. Mol. Breed. 2024, 44, 37. [Google Scholar] [CrossRef]
- Bischoff, V.; Nita, S.; Neumetzler, L.; Schindelasch, D.; Urbain, A.; Eshed, R.; Persson, S.; Delmer, D.; Scheible, W.R. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 2010, 153, 590–602. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhong, W.; Qian, J.; Jin, M.; Tian, P.; Zhu, W.; Zhang, H.; Sun, Y.; Feng, J.W.; Liu, X.; et al. A multi-omics integrative network map of maize. Nat. Genet. 2023, 55, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Tu, X.; Mejía-Guerra, M.K.; Valdes Franco, J.A.; Tzeng, D.; Chu, P.Y.; Shen, W.; Wei, Y.; Dai, X.; Li, P.; Buckler, E.S.; et al. Author Correction: Reconstructing the maize leaf regulatory network using ChIP-seq data of 104 transcription factors. Nat. Commun. 2023, 14, 1586. [Google Scholar] [CrossRef]
- Woodhouse, M.R.; Sen, S.; Schott, D.; Portwood, J.L.; Freeling, M.; Walley, J.W.; Andorf, C.M.; Schnable, J.C. qTeller: A tool for comparative multi-genomic gene expression analysis. Bioinformatics 2021, 38, 236–242. [Google Scholar] [CrossRef]
- Zhang, P.; Fan, Y.; Sun, X.; Chen, L.; Terzaghi, W.; Bucher, E.; Li, L.; Dai, M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019, 98, 697–713. [Google Scholar] [CrossRef]
- Liu, H.; Luo, X.; Niu, L.; Xiao, Y.; Chen, L.; Liu, J.; Wang, X.; Jin, M.; Li, W.; Zhang, Q.; et al. Distant eQTLs and Non-coding Sequences Play Critical Roles in Regulating Gene Expression and Quantitative Trait Variation in Maize. Mol. Plant 2017, 10, 414–426. [Google Scholar] [CrossRef]
- Yang, N.; Lu, Y.; Yang, X.; Huang, J.; Zhou, Y.; Ali, F.; Wen, W.; Liu, J.; Li, J.; Yan, J. Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet. 2014, 10, e1004573. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhao, Y.; Shen, R.; Wang, B.; Xie, Y.; Ma, X.; Zheng, Z.; Wang, H. Characterization of Maize Phytochrome-Interacting Factors in Light Signaling and Photomorphogenesis. Plant Physiol. 2019, 181, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, R.; Sun, Y.; Zhang, M.; Li, S.; Xu, Y.; Song, J.; Li, J.; Qi, J.; Wang, L.; et al. ZmMYC2s play important roles in maize responses to simulated herbivory and jasmonate. J. Integr. Plant Biol. 2023, 65, 1041–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Wei, S.; Gao, Y.; Pei, H.; Geng, R.; Lu, Z.; Wang, P.; Zhou, W. Maize GOLDEN2-LIKE proteins enhance drought tolerance in rice by promoting stomatal closure. Plant Physiol. 2024, 194, 774–786. [Google Scholar] [CrossRef]
- Li, J.; Wu, F.; He, Y.; He, B.; Gong, Y.; Yahaya, B.S.; Xie, Y.; Xie, W.; Xu, J.; Wang, Q.; et al. Maize Transcription Factor ZmARF4 Confers Phosphorus Tolerance by Promoting Root Morphological Development. Int. J. Mol. Sci. 2022, 23, 2361. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Gao, J.; Lin, H.X.; Lin, Y. The molecular basis of heat stress responses in plants. Mol. Plant 2023, 16, 1612–1634. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, Y.; Zhu, J.K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Makarevitch, I.; Waters, A.J.; West, P.T.; Stitzer, M.; Hirsch, C.N.; Ross-Ibarra, J.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-wide Association Studies in Maize: Praise and Stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.; Ye, W.; Zhang, J.; Mu, Y.; Teng, F.; Zhang, S.; He, Z.; Jia, H.; Sun, X. Comprehensive Analysis of ZmTBL Genes Reveals Their Roles in Maize Development and Abiotic Stress Responses. Agronomy 2025, 15, 2121. https://doi.org/10.3390/agronomy15092121
Yu S, Ye W, Zhang J, Mu Y, Teng F, Zhang S, He Z, Jia H, Sun X. Comprehensive Analysis of ZmTBL Genes Reveals Their Roles in Maize Development and Abiotic Stress Responses. Agronomy. 2025; 15(9):2121. https://doi.org/10.3390/agronomy15092121
Chicago/Turabian StyleYu, Sijia, Wenju Ye, Jie Zhang, Yang Mu, Feng Teng, Shilong Zhang, Zhenghua He, Haitao Jia, and Xiaopeng Sun. 2025. "Comprehensive Analysis of ZmTBL Genes Reveals Their Roles in Maize Development and Abiotic Stress Responses" Agronomy 15, no. 9: 2121. https://doi.org/10.3390/agronomy15092121
APA StyleYu, S., Ye, W., Zhang, J., Mu, Y., Teng, F., Zhang, S., He, Z., Jia, H., & Sun, X. (2025). Comprehensive Analysis of ZmTBL Genes Reveals Their Roles in Maize Development and Abiotic Stress Responses. Agronomy, 15(9), 2121. https://doi.org/10.3390/agronomy15092121



