Genome-Wide Association Study of Soybean Mosaic Virus Resistance with a GFP-Based Rapid Evaluation System
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
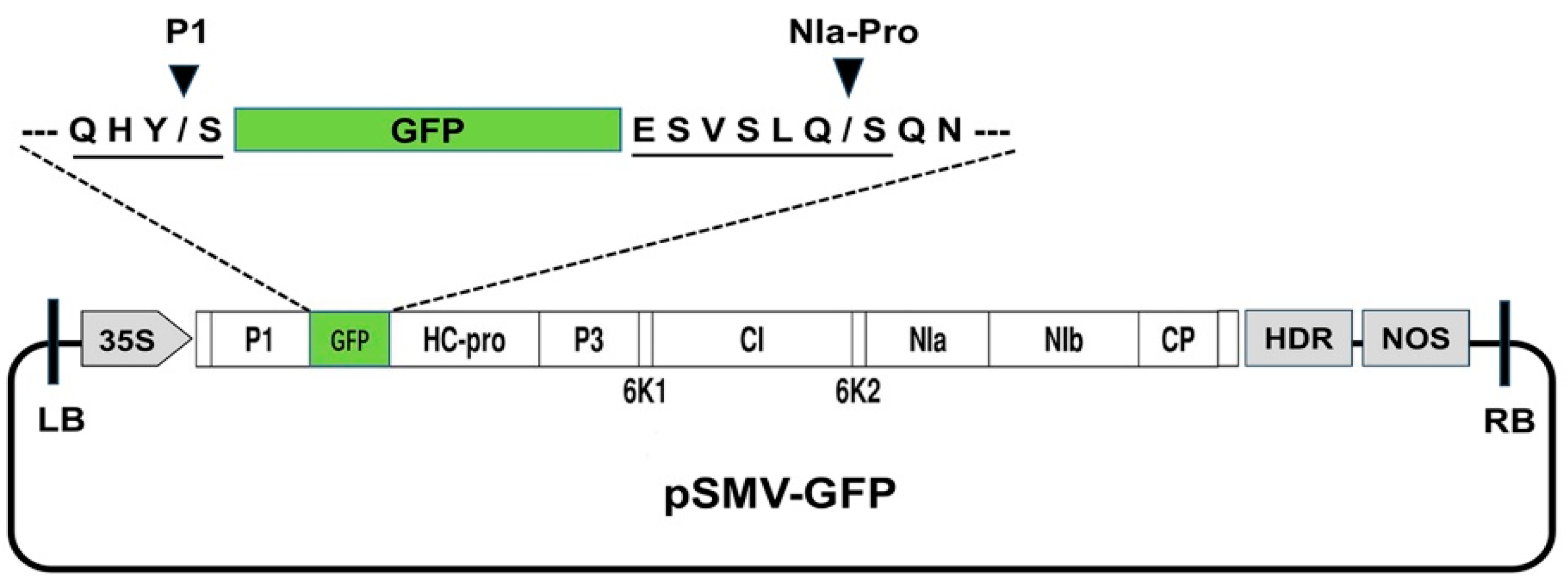
2.2. Construction of the GFP-Tagged SMV Recombinant Virus
2.3. Resistance Assessment Based on Fluorescent Area
2.4. Genome-Wide Association Study
2.5. Linkage Disequilibrium (LD) Decay Distance Analysis
2.6. Candidate Gene Analysis
2.7. Double Antibody Sandwich ELISA (DAS-ELISA)
2.8. qRT-PCR
3. Results
3.1. Construction of the GFP-Tagged SMV Recombinant Virus
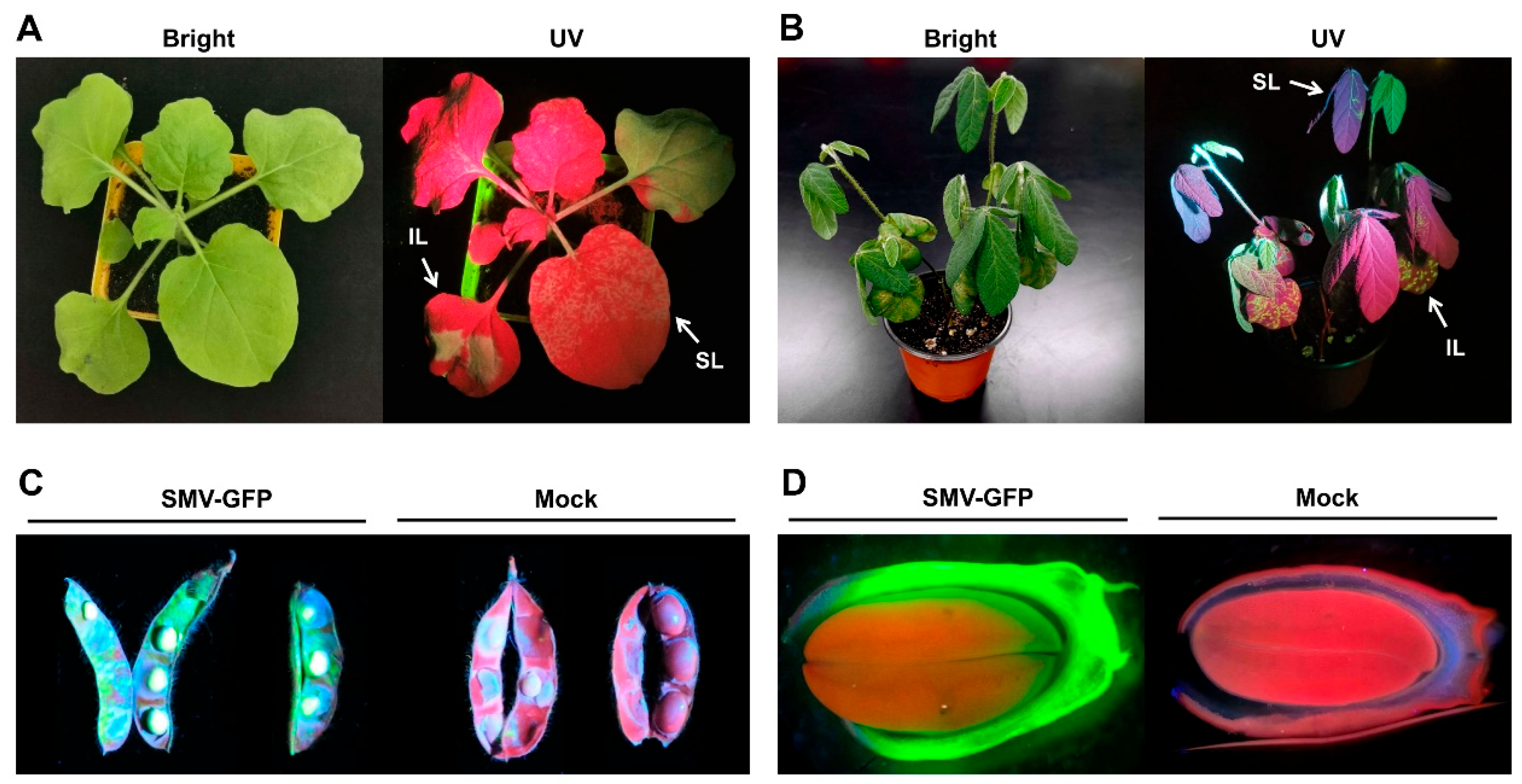
3.2. Infectivity of SMV-GFP on Soybean
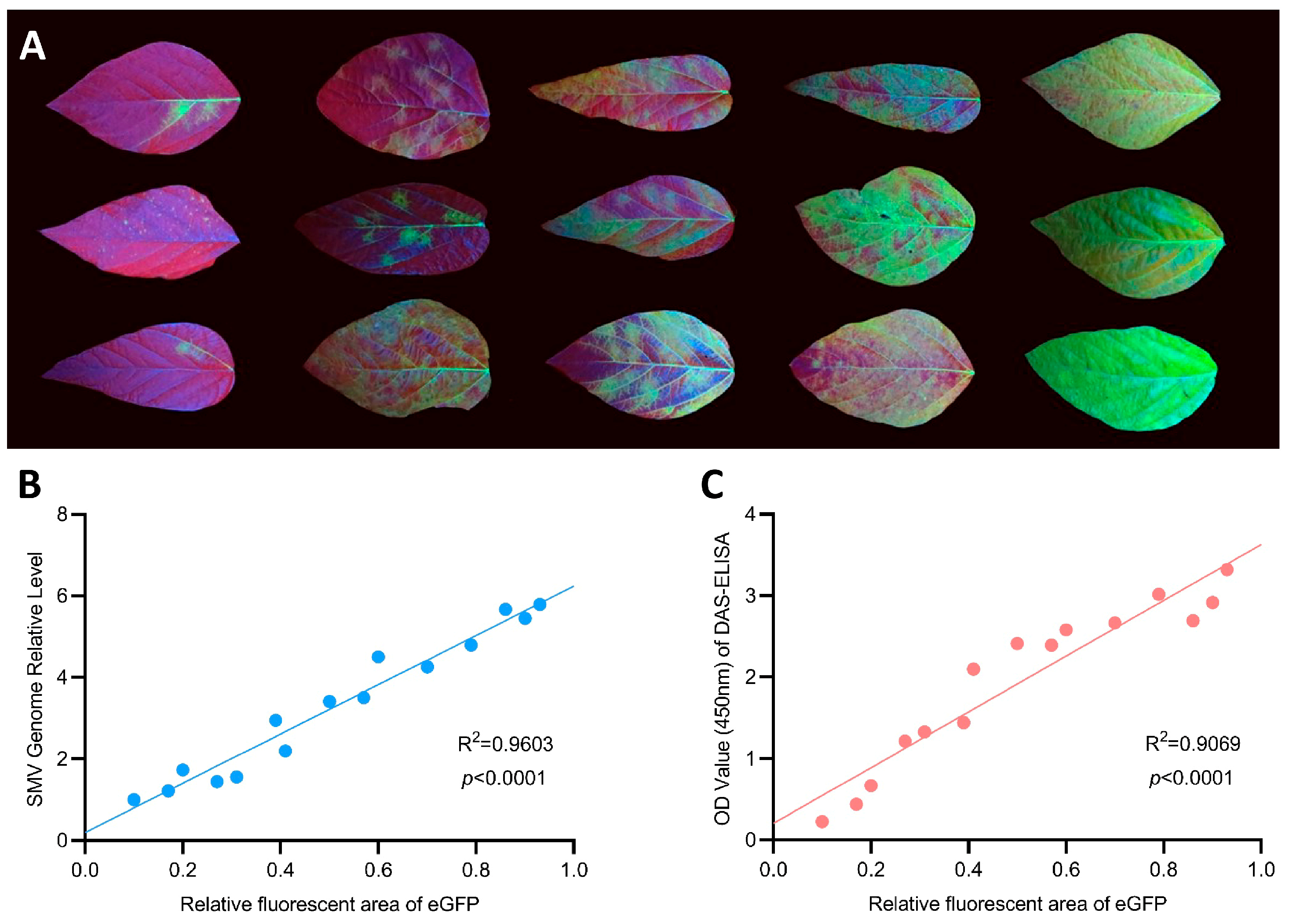
3.3. SMV-GFP Can Be Quantified by Green Fluorescence
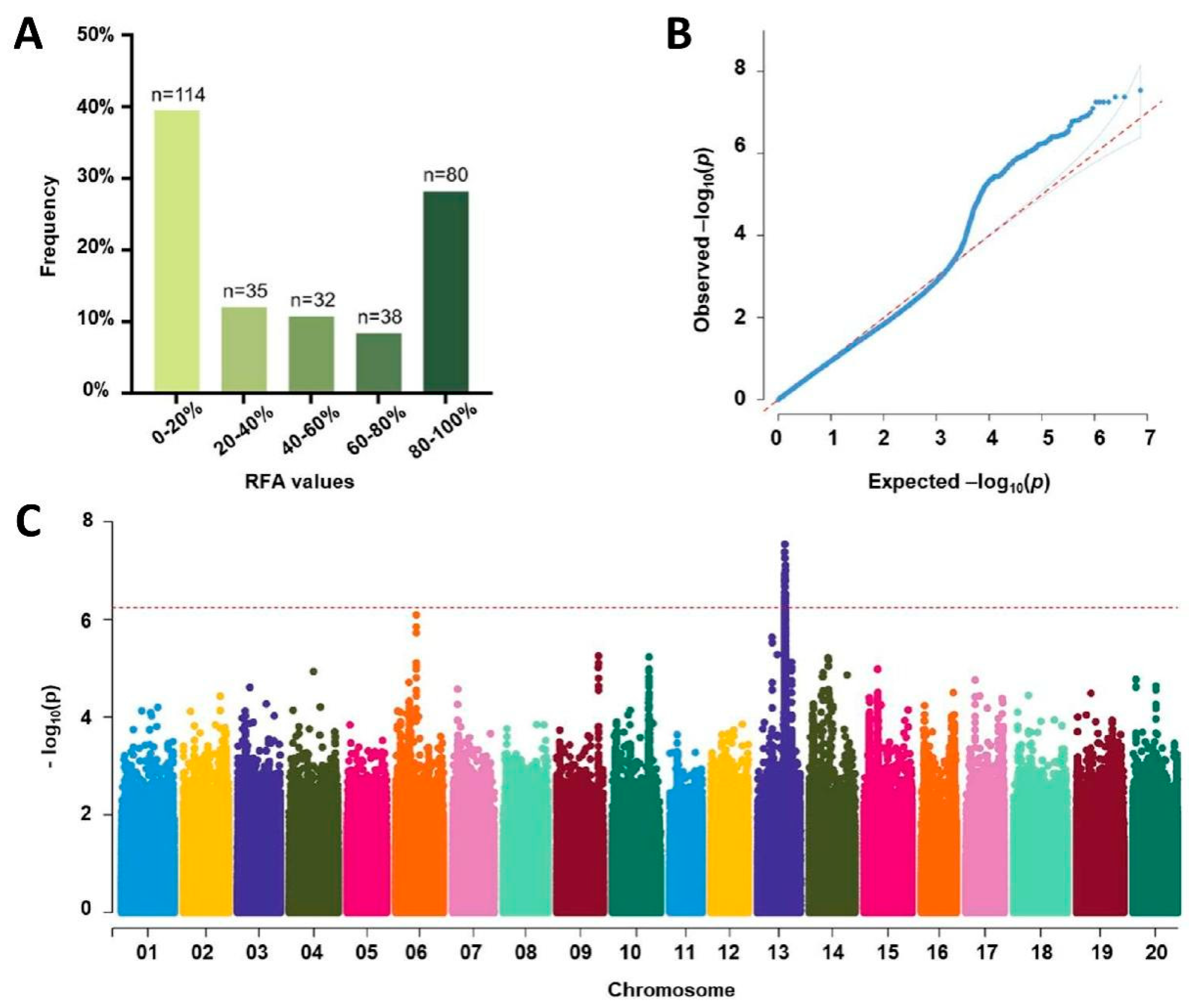
3.4. Phenotypic Analysis of RFA in Soybean Germplasms from Northeast China
3.5. GWAS for SMV Resistance
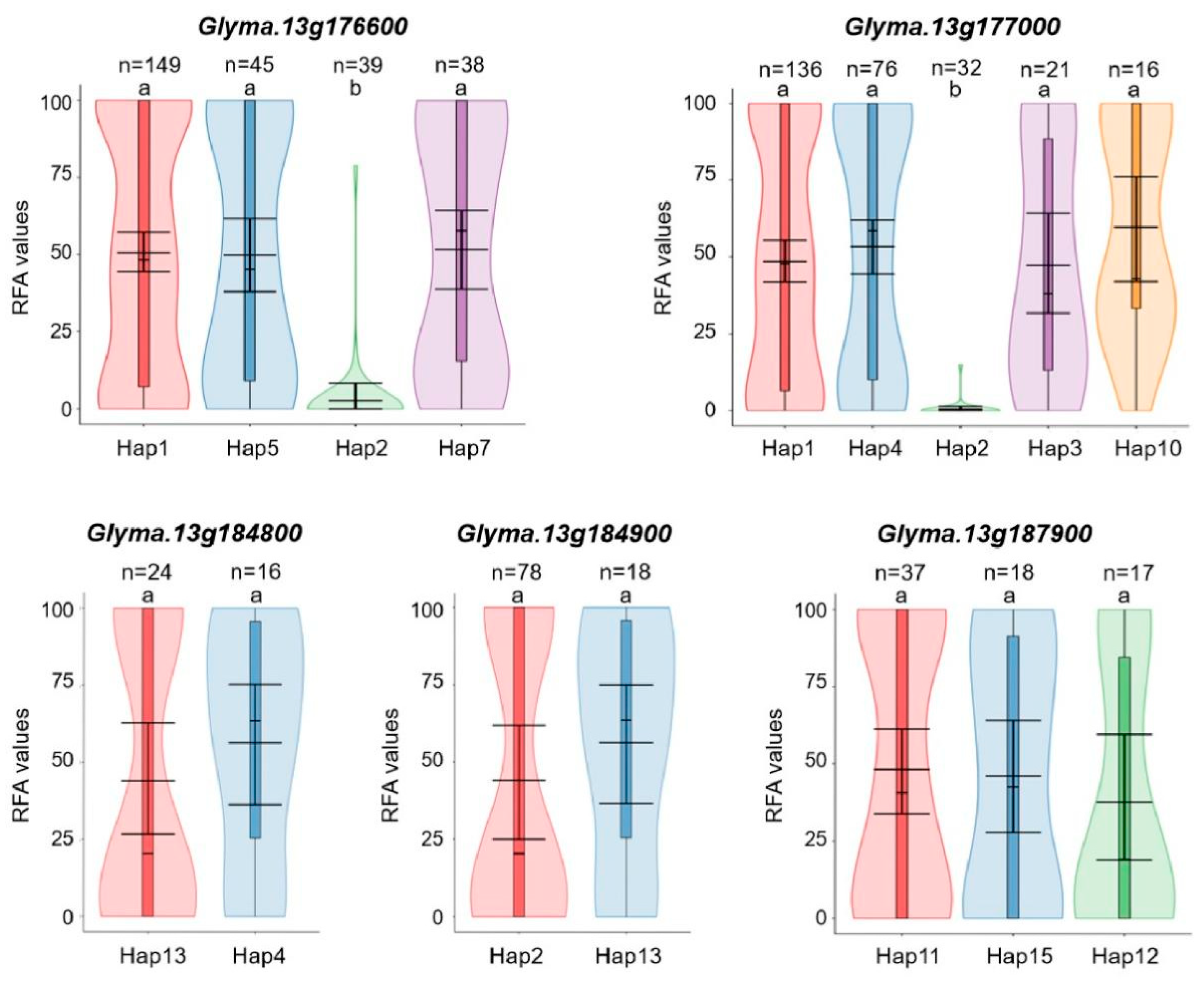
3.6. Candidate Gene Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coradi, P.C.; Müller, A.; Souza, G.A.C.; Steinhaus, J.I.; Wagner, R. Quality of soybean cultivars in the drying and storage processes in real scale and experimental. J. Food Process. Eng. 2020, 43, e13418. [Google Scholar] [CrossRef]
- Widyasari, K.; Alazem, M.; Kim, K. Soybean resistance to Soybean mosaic virus. Plants 2020, 9, 219. [Google Scholar] [CrossRef]
- Hill, J.H.; Whitham, S.A. Control of virus disease in soybean. Adv. Virus Res. 2014, 90, 355–390. [Google Scholar]
- Ross, J.P. Effect of aphid-transmitted Soybean mosaic virus on yields of closely related resistant and susceptible soybean lines. Crop Sci. 1997, 17, 869–872. [Google Scholar] [CrossRef]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Saghai Maroof, M.A. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2018, 19, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Rui, R.; Liu, S.; Karthikeyan, A.; Wang, T.; Niu, H.; Yin, J.; Yang, Y.; Wang, L.; Yang, Q.; Zhi, H.; et al. Fine-mapping and identification of a novel locus Rsc15 underlying soybean resistance to Soybean mosaic virus. Theor. Appl. Genet. 2017, 130, 2395–2410. [Google Scholar] [CrossRef]
- Koning, G.; TeKrony, D.M.; Ghabrial, S.A. Soybean seedcoat mottling: Association with Soybean mosaic virus and Phomopsis spp. Seed Infection. Plant Dis. 2003, 87, 413–417. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Senda, M. Patterning of virus-infected Glycine max seedcoat is associated with suppression of endogenous silencing of chalcone synthase genes. Plant Cell 2004, 16, 807–818. [Google Scholar] [CrossRef]
- Li, K.; Yang, Q.H.; Zhi, H.J.; Gai, J.Y. Identification and distribution of Soybean mosaic virus strains in Southern China. Plant Dis. 2010, 94, 351–357. [Google Scholar] [CrossRef]
- Gao, L.; Wu, Y.; An, J.; Huang, W.; Liu, X.; Xue, Y.; Luan, X.; Lin, F.; Sun, L. Pathogenicity and genome-wide sequence analysis reveals relationships between Soybean mosaic virus strains. Arch. Virol. 2022, 167, 517–529. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Jiang, H.; Ren, R.; Li, C.; Zhi, H.; Chen, S.; Gai, J. Inheritance, fine-mapping, and candidate gene analyses of resistance to Soybean mosaic virus strain SC5 in soybean. Mol. Genet. Genom. 2017, 292, 811–822. [Google Scholar] [CrossRef]
- Hayes, A.J.; Ma, G.R.; Buss, G.R.; Maroof, M.A.S. Molecular marker mapping of RSV4, a gene conferring resistance to all known strains of Soybean mosaic virus. Crop Sci. 2000, 40, 1434–1437. [Google Scholar] [CrossRef]
- Hayes, A.J.; Maroof, M.A.S. Targeted resistance gene mapping in soybean using modified AFLPs. Theor. Appl. Genet. 2000, 100, 1279–1283. [Google Scholar] [CrossRef]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef]
- Suh, S.J.; Bowman, B.C.; Jeong, N.; Yang, K.; Kastl, C.; Tolin, S.A.; Maroof, M.A.S.; Jeong, S.C. The Rsv3 locus conferring resistance to Soybean mosaic virus is associated with a cluster of coiled-coil nucleotide-binding leucine-rich repeat genes. Plant Genome. 2011, 4, 55–64. [Google Scholar] [CrossRef]
- Klepadlo, M.; Chen, P.Y.; Shi, A.N.; Mason, R.E.; Korth, K.L.; Srivastava, V.; Wu, C.J. Two tightly linked genes for Soybean mosaic virus resistance in soybean. Crop Sci. 2017, 57, 1844–1853. [Google Scholar] [CrossRef]
- Fu, S.; Zhan, Y.; Zhi, H.; Gai, J.; Yu, D. Mapping of SMV resistance gene Rsc-7 by SSR markers in soybean. Genetica 2006, 128, 63–69. [Google Scholar] [CrossRef]
- Li, C.; Adhimoolam, K.; Yuan, Y.; Yin, J.L.; Ren, R.; Yang, Y.Q.; Zhi, H.J. Identification of candidate genes for resistance to Soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci. 2017, 68, 156–166. [Google Scholar] [CrossRef]
- Li, H.C.; Zhi, H.J.; Gai, J.Y.; Guo, D.Q.; Wang, Y.W.; Li, K.; Bai, L.; Yang, H. Inheritance and gene mapping of resistance to Soybean mosaic virus strain SC14 in soybean. J. Integr. Plant Biol. 2006, 48, 1466–1472. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Liu, N.; Yang, Z.; Zheng, G.; Zhi, H. Fine mapping and identification of the soybean RSC4 resistance candidate gene to Soybean mosaic virus. Plant Breed. 2011, 130, 653–659. [Google Scholar] [CrossRef]
- Wang, D.; Ma, Y.; Yang, Y.; Liu, N.; Li, C.; Song, Y.; Zhi, H. Fine mapping and analyses of R (SC8) resistance candidate genes to Soybean mosaic virus in soybean. Theor. Appl. Genet. 2011, 122, 555–565. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, D.G.; Li, H.C.; Zheng, G.J.; Yang, Y.Q.; Li, H.W.; Zhi, H.J. Fine mapping of the R-SC14Q locus for resistance to Soybean mosaic virus in soybean. Euphytica 2011, 181, 127–135. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, D.; Zhang, H.; Shen, Y.; Yang, Y.; Li, K.; Wang, L.; Yang, Y.; Zhi, H.; Morris, B. Fine mapping of the RSC8 locus and expression analysis of candidate SMV resistance genes in soybean. Plant Breed. 2016, 135, 701–706. [Google Scholar] [CrossRef]
- Jiang, H.; Jia, H.; Hao, X.; Li, K.; Gai, J. Mapping locus RSC11K and predicting candidate gene resistant to Soybean mosaic virus SC11 through linkage analysis combined with genome resequencing of the parents in soybean. Genomics 2022, 114, 110387. [Google Scholar] [CrossRef]
- Usovsky, M.; Chen, P.; Li, D.; Wang, A.; Shi, A.; Zheng, C.; Shakiba, E.; Lee, D.; Vieira, C.C.; Lee, Y.C.; et al. Decades of genetic research on Soybean mosaic virus resistance in soybean. Viruses 2022, 14, 1122. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Ren, R.; Wang, T.; Gao, L.; Song, P.; Yang, Y.; Zhi, H.; Li, K. Development of comprehensive serological techniques for sensitive, quantitative and rapid detection of Soybean mosaic virus. Int. J. Mol. Sci. 2022, 23, 9457. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Ding, X.; Li, K.; Liao, W.; Zhong, Y.; Ren, R.; Liu, Z.; Karthikeyan, A.; Zhi, H. Characterization of Soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars. Theor. Appl. Genet. 2015, 128, 1489–1505. [Google Scholar] [CrossRef]
- Jin, S.; Tian, H.; Ti, M.; Song, J.; Hu, Z.; Zhang, Z.; Xin, D.; Chen, Q.; Zhu, R. Genetic analysis of soybean flower size phenotypes based on computer vision and genome-wide association studies. Int. J. Mol. Sci. 2024, 25, 7622. [Google Scholar] [CrossRef]
- Sun, K.; Zhao, D.; Liu, Y.; Huang, C.; Zhang, W.; Li, Z. Rapid construction of complex plant RNA virus infectious cDNA clones for Agroinfection using a yeast-E. coli-Agrobacterium shuttle vector. Viruses 2017, 9, 332. [Google Scholar] [CrossRef]
- Zaulda, F.A.; Yang, S.H.; Han, J.P.; Mlotshwa, S.; Dorrance, A.; Qu, F. A cowpea severe mosaic virus-based vector simplifies virus induced gene silencing and foreign protein expression in soybean. Plant Methods 2022, 18, 116. [Google Scholar] [CrossRef]
- Yin, L.L.; Zhang, H.H.; Tang, Z.S.; Xu, J.Y.; Yin, D.; Zhang, Z.W.; Yuan, X.H.; Zhu, M.J.; Zhao, S.H.; Li, X.Y.; et al. rMVP: A memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.M.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Li, M.X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.T.; Huang, S.Y.; Yang, T.; Tang, Y.P.; Yang, S.B.; Aisimutuola, P.; et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef]
- Qin, J.; Song, Q.; Shi, A.; Li, S.; Zhang, M.; Zhang, B. Genome-wide association mapping of resistance to Phytophthora sojae in a soybean [Glycine max (L.) Merr.] germplasm panel from maturity groups IV and V. PLoS ONE 2017, 12, e0184613. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huang, T.; Ruan, Z.; Tu, R.; He, Y.; Liu, Q.; Cheng, S.; He, G.; Shen, X. A MACPF protein OsCAD1 balances plant growth and immunity through regulating salicylic acid homeostasis in rice. Plant Cell Environ. 2025, 6. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, F.; Huang, X. Characterization of an Arabidopsis thaliana DUF761-contaning protein with a potential role in development and defense responses. Theor. Exp. Plant Phys. 2019, 31, 303–316. [Google Scholar] [CrossRef]
- Seo, J.; Lee, H.; Kim, K. Systemic gene delivery into soybean by simple rub-inoculation with plasmid DNA of a Soybean mosaic virus-based vector. Arch. Virol. 2009, 154, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Baek, E.; Yoon, J.; Palukaitis, P. The use of a Tobacco mosaic virus-based expression vector system in chrysanthemum. Plant Pathol. J. 2017, 33, 429–433. [Google Scholar] [CrossRef]
- Kawakubo, A.; Gallois, J.; Nakahara, K.S. Monitoring systemic infection by cucumber mosaic virus using a small fluorescent protein iLOV in plants. J. Gen. Plant Pathol. 2023, 89, 47–52. [Google Scholar] [CrossRef]
- Kwon, S.; Lee, Y.; Cho, Y.; Byun, H.; Seo, J. Engineering of stable infectious cDNA constructs of a fluorescently tagged tomato chlorosis virus. Virology 2024, 593, 110010. [Google Scholar] [CrossRef]
- El-Gebaly, F.E.; Ckurshumova, W.; Liu, J.; Fefer, M.; Krysmanski, E.C.; Cameron, R.K. Development of a rapid screen to identify formulations that enhance plant resistance to viral infection. Physiol. Mol. Plant Pathol. 2025, 139, 102783. [Google Scholar] [CrossRef]
- Revers, F.; García, J.A. Molecular biology of potyviruses. Adv. Virus Res. 2015, 92, 101–199. [Google Scholar]
- Bao, W.; Yan, T.; Deng, X.; Wuriyanghan, H. Synthesis of full-length cDNA infectious clones of Soybean mosaic virus and functional identification of a key amino acid in the silencing suppressor HC-Pro. Viruses 2020, 12, 886. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, Y.-N.; Zhang, C.; Liu, X.-T.; Liu, C.-C.; Guo, R.; Niu, K.-X.; Zhu, A.-Q.; Yang, J.-Y.; Chen, J.-Q.; et al. Molecular mapping of the gene(s) conferring resistance to Soybean mosaic virus and bean common mosaic virus in the soybean cultivar raiden. Theor. Appl. Genet. 2019, 132, 3101–3114. [Google Scholar] [CrossRef]
- Zheng, G.J.; Yang, Y.Q.; Ying, M.A.; Yang, X.F.; Chen, S.Y.; Rui, R.E.; Wang, D.G.; Yang, Z.L.; Zhi, H.J. Fine mapping and candidate gene analysis of resistance gene RSC3Q to Soybean mosaic virus in Qihuang 1. J. Integr. Agric. 2014, 13, 2608–2615. [Google Scholar] [CrossRef]
- Ma, Y.; Li, H.; Wang, D.; Liu, N.; Zhi, H. Molecular mapping and marker assisted selection of Soybean mosaic virus resistance gene RSC12 in soybean. Legume Genomes Genet. 2010, 8, 41–46. [Google Scholar]
- Liu, S.; Cheng, Y.; Ma, Q.; Li, M.; Jiang, Z.; Xia, Q.; Nian, H. Fine mapping and genetic analysis of resistance genes, Rsc18, against Soybean mosaic virus. J. Integr. Agric. 2022, 21, 644–653. [Google Scholar] [CrossRef]
- Karthikeyan, A.; Li, K.; Li, C.; Yin, J.; Li, N.; Yang, Y.; Song, Y.; Ren, R.; Zhi, H.; Gai, J. Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus SC20 in soybean. Theor. Appl. Genet. 2018, 131, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, S.; Sogame, M.; Hata, M.; Singkaravanit-Ogawa, S.; Piślewska-Bednarek, M.; Onozawa-Komori, M.; Nishiuchi, T.; Hiruma, K.; Saitoh, H.; Terauchi, R.; et al. Dysfunction of Arabidopsis MACPF domain protein activates programmed cell death via tryptophan metabolism in MAMP-triggered immunity. Plant J. 2017, 89, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Jian, H.; Wei, F.; Gu, L.; Hu, T.; Lv, X.; Guo, X.; Lu, J.; Ma, L.; Wang, H.; et al. Phylogenetic analysis of the membrane attack complex/perforin domain-containing proteins in Gossypium and the role of GhMACPF26 in cotton under cold stress. Front. Plant Sci. 2021, 12, 68227. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, T.D.; Thamm, A.M.; de Crecy-Lagard, V.; Hanson, A.D. Proteins of unknown biochemical function: A persistent problem and a roadmap to help overcome it. Plant Physiol. 2015, 169, 1436–1442. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Su, H.; Gao, Y.; Tian, H.; Hao, Y.; Hu, Y.; Zhu, M.; Chen, Q.; Xin, D.; Song, S. Genome-Wide Association Study of Soybean Mosaic Virus Resistance with a GFP-Based Rapid Evaluation System. Agronomy 2025, 15, 1960. https://doi.org/10.3390/agronomy15081960
Zhou J, Su H, Gao Y, Tian H, Hao Y, Hu Y, Zhu M, Chen Q, Xin D, Song S. Genome-Wide Association Study of Soybean Mosaic Virus Resistance with a GFP-Based Rapid Evaluation System. Agronomy. 2025; 15(8):1960. https://doi.org/10.3390/agronomy15081960
Chicago/Turabian StyleZhou, Jiaying, Hao Su, Yunlai Gao, Huilin Tian, Yun Hao, Yuxi Hu, Mingze Zhu, Qingshan Chen, Dawei Xin, and Shuang Song. 2025. "Genome-Wide Association Study of Soybean Mosaic Virus Resistance with a GFP-Based Rapid Evaluation System" Agronomy 15, no. 8: 1960. https://doi.org/10.3390/agronomy15081960
APA StyleZhou, J., Su, H., Gao, Y., Tian, H., Hao, Y., Hu, Y., Zhu, M., Chen, Q., Xin, D., & Song, S. (2025). Genome-Wide Association Study of Soybean Mosaic Virus Resistance with a GFP-Based Rapid Evaluation System. Agronomy, 15(8), 1960. https://doi.org/10.3390/agronomy15081960






