Abstract
Disease caused by Plasmodiophora brassicae severely disrupts cruciferous crops by altering root physiology and rhizosphere ecology. While pathogen-induced shifts in rhizosphere microbiomes are documented, the mechanisms linking root exudate reprogramming to microbial community remodeling remain poorly understood. Here, we integrated untargeted metabolomics and 16S rRNA sequencing to investigate how root exudates reshape the rhizosphere microbiome of tumorous stem mustard (Brassica juncea var. tumida) through P. brassicae infection. Metabolomic profiling identified 1718 root exudate metabolites, with flavones (e.g., apigenin 7-O-β-D-rutinoside, VIP > 1.5) and phenolic derivatives (e.g., gastrodin) being selectively enriched in infected plants. P. brassicae infection significantly increased rhizobacterial richness (ACE index, p < 0.05) and restructured the community composition, marked by enrichment of Paenibacillus (LDA score > 3.0). Procrustes analysis revealed tight coupling between microbial community shifts and metabolic reprogramming (M2 = 0.446, p = 0.005), while Spearman correlations implicated pathogen-induced metabolites like geniposidic acid in recruiting beneficial Paenibacillus. Our results reveal that plant hosts dynamically secrete defense-related root metabolites to remodel the rhizosphere microbiome in response to P. brassicae infection. This dual-omics approach elucidates a chemical dialogue mediating plant–microbe–pathogen interactions, offering novel insights for engineering disease-suppressive microbiomes through root exudate manipulation.
1. Introduction
China is the world’s largest producer of Brassicaceae crops, with major cultivated varieties. As a core component of the Chinese diet, Brassicaceae vegetables account for over 30% of total vegetable intake, with per capita annual consumption exceeding 60 kg. China’s Brassicaceae crop exports primarily consist of fresh products and processed goods, with key export commodities, including fresh Chinese cabbage and dehydrated cabbage/cauliflower [1].
However, the sustainable production of Brassicaceae crops faces significant threats from clubroot disease. Clubroot, a devastating disease affecting Brassicaceae crops globally, is caused by Plasmodiophora brassicae Woronin, an obligate biotrophic soil-borne protist. This pathogen infects all 330 genera and 3700 species within Brassicaceae, with notable agricultural hosts, including Brassica napus (oilseed rape), B. rapa subsp. pekinensis (Chinese cabbage), B. rapa subsp. chinensis (bok choy), B. oleracea var. acephala (kale), Raphanus sativus (radish), B. oleracea var. botrytis (cauliflower), and B. juncea var. tumida (tumorous stem mustard) [2].
The pathogen severely disrupts vascular tissue organization and physiological functions, leading to impaired water and nutrient translocation. Consequently, infected plants manifest systemic pathological responses characterized by vascular wilting, growth stunting, and premature senescence [3]. These cumulative physiological disruptions ultimately result in significant yield reductions, causing substantial economic losses in agricultural production.
P. brassicae infection specifically activates abnormal division of cambium cells in the roots and hypocotyls of cruciferous hosts, leading to the formation of gall structures. These hypertrophied galls not only establish a nutrient reservoir for the pathogen [4] but also remodel the vascular system through phloem cell hyperproliferation [5], creating transport channels conducive to carbohydrate acquisition by the pathogen. P. brassicae suppresses host photosynthetic gene expression [6] and starch metabolism pathways [7], forcing continuous sucrose export from leaves to galls. Despite lacking invertase genes to hydrolyze sucrose [8], the pathogen directly assimilates monosaccharides via trehalose metabolism remodeling [9] and sugar transporter systems [5]. The excessive accumulation of sugars (e.g., trehalose, fructose) in galls elevates sugar concentrations in root exudates [4]. Additionally, infection induces the synthesis of phenolic acids and flavonoids as defense compounds [10], which are excreted into the rhizosphere, significantly enriching phenolic content in exudates. Gall formation also triggers epidermal rupture and cellular content leakage, further altering the root exudate composition.
Root exudates comprise diverse compounds, including primary metabolites (e.g., sugars, organic acids, and amino acids) and unique secondary metabolites. Their composition varies with plant species, genotype, developmental stage, root traits, nutrient availability, and environmental conditions [11]. These exudates serve not only as nutritional substrates for rhizosphere microbes but also as a “chemical language” mediating plant–microbe co-evolution [12]. By modulating exudate profiles, plants selectively recruit or inhibit specific microbial taxa, reshaping microbial community structure.
Following P. brassicae infection, host rhizosphere microbial communities undergo significant shifts [13,14]. Microbial diversity correlates strongly with disease severity, and community structure exhibits high sensitivity to pathogen invasion [10,15]. Pathogen–host interactions drive context-dependent microbial changes influenced by plant resistance, pathogenicity, soil properties, and fertilization practices [16,17]. Notably, higher microbial diversity exacerbates clubroot development in susceptible cultivars but not in resistant varieties [18], highlighting the profound impact of P. brassicae on rhizosphere microbiota.
While current research has predominantly characterized microbial community responses to P. brassicae infection [19,20], critical knowledge gaps persist regarding the systematic manipulation of root exudates during pathogen–rhizosphere interactions. We hypothesize that P. brassicae infection induces metabolic reprogramming of root exudates in tumorous stem mustard, and the pathogen-induced changes in root exudates selectively recruit beneficial microbes, thereby remodeling rhizosphere microbiome structure and forming a disease-suppressive microbial community. To address these questions, we employed a dual-omics approach integrating 16S rRNA sequencing and untargeted metabolomics to characterize infection-induced metabolic shifts in tumorous stem mustard root exudates to decipher correlated microbiome restructuring patterns. This work provides novel strategies for breeding resistant cultivars and engineering rhizosphere microbiomes, advancing sustainable disease management in Brassicaceae crops.
2. Materials and Methods
2.1. Collection of P. brassicae Resting Spores in Tumorous Stem Mustard
P. brassicae was isolated from symptomatic tumorous stem mustard roots collected from a clubroot disease nursery at Yangtze Normal University. Infected roots exhibiting characteristic gall formations were processed using the following protocol [21]. Roots were surface-sterilized in 70% ethanol for 1 min, followed by three rinses in sterile distilled water to eliminate epiphytic contaminants. Sterilized roots were homogenized in sterile distilled water at a 1:5 (w/v) ratio using a low-speed tissue homogenizer to release pathogen propagules. The homogenate was sequentially filtered through a 100 μm nylon mesh to remove plant debris, yielding a spore-enriched filtrate.
Resting spores were concentrated by mixing the filtrate 1:1 with 60% (w/v) sucrose solution followed by centrifugation (3000× g, 20 min, 4 °C). The pellet containing pure resting spores was re-suspended in sterile water and subjected to three additional wash–centrifugation cycles (3000× g, 10 min) to remove residual sucrose. The purity of resting spores was verified by microscopic examination with ≥95% spore content observed in the final suspension. The final spore suspension was quantified using a hemocytometer, adjusted to a defined concentration, and stored at 4 °C for subsequent inoculation experiments.
2.2. Treatment of Tumorous Stem Mustard by P. brassicae
The tumorous stem mustard cultivar “Yong’an Xiaoye” was used in this study. Following surface sterilization (70% ethanol, 3 min) and three sterile water rinses, seeds were sown in corn field soil (no history of tumorous stem mustard or P. brassicae), with ten seedlings per pot. Experimental treatments commenced at the cotyledon stage were as follows: (1) disease treatment (P): inoculation with a P. brassicae resting spore suspension with concentration of 1.0 × 108 spores/mL); (2) blank control (Ck): application of sterile water [22]. The experiment, conducted in a greenhouse (25 ± 2 °C, 70% RH, 16/8 h light/dark), included three replicates per treatment. Five weeks after inoculation, seedlings were gently uprooted, ensuring whole roots. Root gall formation was recorded for disease incidence and index calculation. Rhizosphere soil adhering to roots was collected for microbial DNA extraction. Roots were then washed thoroughly to prepare for root exudate collection.
2.3. Root Exudate Collection
Root exudates were collected according to the method of de Vries et al. [23]. Roots were meticulously washed to remove soil with minimal damage, and dead roots were removed using sterilized steel tweezers. For exudate collection, ten biological replicates (individual seedling roots) were immersed in 100 mL sterile water contained in 250 mL conical flasks. Incubation proceeded for 12 h at 25 ± 0.5 °C under ambient light conditions in an incubator shaker. The resulting exudate solutions were then filtered through 0.22 μm Millipore membranes. Filtrates were flash-frozen in liquid nitrogen and stored at −80 °C until metabolomic analysis.
2.4. Metabolomic Analysis of Root Exudates
Root exudates underwent untargeted Liquid Chromatography–Mass Spectrometry (LC–MS) metabolomics (Majorbio Bio Pharm Technology Co., Ltd., Shanghai, China). After thawing on ice and brief vortex homogenization (10 s), a 10 mL sample portion was flash-frozen and vacuum-lyophilized. The resulting powder was extracted with 300 μL of 70% methanol spiked with internal standards, vortexed for 3 min, and centrifuged (12,000 rpm, 4 °C, 10 min); supernatants were then vialed for LC-MS. Pooled quality control (QC) samples, analyzed every 10 injections, monitored system performance. Metabolites were identified against the Metware database (Metware Biotechnology, Wuhan, China) and quantified using multiple reaction monitoring (MRM) mode on a triple quadrupole instrument. Multivariate analysis included principal component analysis (PCA) (Bray–Curtis, vegan in R v4.3.2) and partial least squares discriminant analysis (PLS-DA) (ropls in R v4.3.2, permutation-tested with 200 repeats [24]. Differential metabolites were screened based on PLS-DA VIP scores (≥1) and significant fold changes (≥2 or ≤0.5), with results visualized in volcano plots using ggpubr package of R v4.3.2 for pairwise treatment comparisons [25].
2.5. DNA Extraction, Illumina Sequencing, and Analysis
Genomic DNA was isolated from 0.5 g soil aliquots using the FastDNA™ SPIN Kit (MP Biomedicals, Santa Ana, CA, USA). Post-extraction assessments were as follows: the concentration was measured by a NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA) and quality checked via 1% agarose gel electrophoresis. The V3–V4 hypervariable region was amplified using primer pair 338F and 806R [26]. Equimolar purified amplicons were pooled for Illumina MiSeq PE300 sequencing (Majorbio, Shanghai, China). Bioinformatics processing included (1) raw data QC/adapter trimming (fastp v0.19.6) [27]; (2) read merging (FLASH v1.2.11) [28]; (3) chimera filtering (USEARCH v11.0.667) [29]; (4) demultiplexing (QIIME2 v2021.4) [30]; (5) OTU clustering (97% identity, UPARSE) [31]; and (6) taxonomic assignment (RDP Classifier v2.13, SILVA 138 database) [32].
Alpha-diversity indices, including the Abundance-Based Coverage Estimator (ACE), observed species (Sobs), Shannon diversity index, and Simpson dominance index, were calculated to evaluate microbial community richness and evenness. The taxonomic composition was analyzed at the class and genus levels, with results visualized through a circular phylogenetic tree diagram and community structure pie charts to delineate evolutionary relationships and relative abundance distributions.
For β-diversity analysis, non-metric multidimensional scaling (NMDS) analysis based on the Bray–Curtis distance was performed and visualized using the R package vegan (v2.6-4) to identify key variations in rhizosphere bacterial community structure [33]. Significant differences in the bacterial composition between treatment groups were statistically evaluated using permutational multivariate analysis of variance (PERMANOVA; Adonis) and analysis of similarities (ANOSIM) with 999 permutations. Intergroup differential taxa were identified through the following complementary approaches: the Wilcoxon rank-sum test, non-parametric pairwise comparisons between two groups to detect taxa with significant abundance differences (adjusted p < 0.05), and linear discriminant analysis effect size (LEfSe), a multi-class linear discriminant analysis (LDA) method [34] to pinpoint genera with both statistical significance (LDA score > 2.0) and biological relevance across three treatment groups.
2.6. Associations Between Root Exudates and Rhizosphere Bacterial Communities
To resolve relationships between the bacterial community and exudates, we employed two complementary integration approaches: (1) Procrustes analysis aligned the PCA ordinations of differentially abundant bacterial genera and metabolites, and (2) two-way orthogonal partial least squares (O2PLS) modeled covariance patterns between the microbial and metabolomic datasets using multivariate regression. Subsequently, Spearman’s correlation analysis (corrplot package of R v4.3.2) revealed significant pairwise genus-metabolite interactions (|ρ| > 0.4, p < 0.05) [35].
3. Results
3.1. Effects of the P. brassicae Infection on Root Exudate Composition
A total of 1718 metabolites were identified, encompassing amino acids, organic acids, fatty acids, nucleotides, lipids, and other compound classes, with amino acids and carboxylic acids emerging as the predominant categories (Figure S1a). At the metabolic pathway level, amino acid metabolism, the biosynthesis of secondary metabolites, and Xenobiotics biodegradation and metabolism represented the most prominent metabolic pathways (Figure S1b).
PLS-DA analysis revealed distinct clustering patterns between the two experimental groups, with Ck samples occupying the left quadrant and P forming a separate cluster in the right region of the score plot (Figure 1a). VIP score ranking identified apigenin 7-O-β-D-rutinoside and gastrodin as the most discriminatory metabolites enriched in P (VIP > 1.5), whereas Cinncassiol D3, salbutamol, 4-Androstenediol, and Amabiline showed significant accumulation in Ck (VIP > 1.5) (Figure 1b). To delineate the differentially accumulated metabolites, a supervised volcano plot analysis was conducted by integrating rigorous thresholds of fold change (FC ≥ 1.5 or ≤0.67) and VIP. Comparative analysis identified 400 metabolites significantly up-regulated in Ck (vs. 88 in the P group, p < 0.05). Intriguingly, gibberellin A34-catabolite, 20-trihydroxy-leukotriene-B4, and 1,2,3,4-tetrahydronaphthalene exhibited pronounced enrichment in Ck. Significantly, when applying more stringent criteria (p < 0.01, VIP > 2.0), apigenin 7-O-β-D-rutinoside and geniposidic acid were exclusively over-accumulated in the P group (Figure 1c).
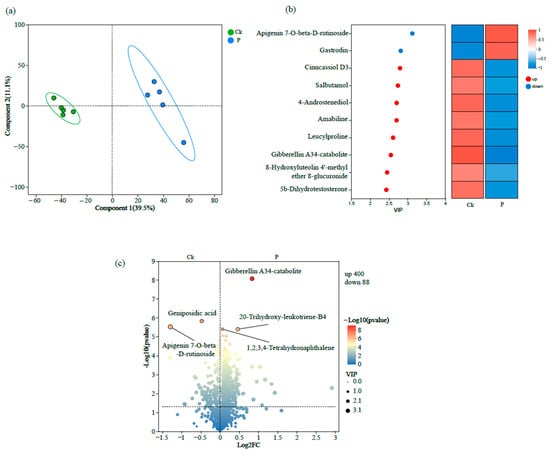
Figure 1.
Multi-dimensional metabolomic analysis of root exudates of tumorous stem mustard either infected with P. brassicae (P) or not infected with P. brassicae (Ck), as represented by (a) partial least squares discriminant analysis (PLS-DA); (b) variable importance in projection scores; and (c) volcano plot of differential metabolites.
3.2. Effects of the P. brassicae Infection on Rhizosphere Bacterial Communities
Alpha-diversity analysis revealed significant differences in community richness between groups, with the ACE index and Sobs values of Ck being substantially lower than those of the P group (p < 0.05) (Figure 2a,b). However, no significant differences were detected in Shannon and Simpson diversity indices between the two groups (Figure 2c,d). Taxonomic profiling at the class level demonstrated that Alphaproteobacteria (24.6% in Ck vs. 26.1% in P) and Gammaproteobacteria (16.7% vs. 16.1%) constituted the predominant bacterial classes, followed by Actinobacteria (10.8% vs. 8.5%) and Bacilli (6.1% vs. 8.0%) (Figure 3a,b). At the genus level, Sphingomonas (7.15% vs. 6.26%) and Bacillus (3.47% vs. 4.07%) emerged as the dominant taxa, with secondary contributions from the Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium (ANPR) complex and Arthrobacter (Figure 3c,d).
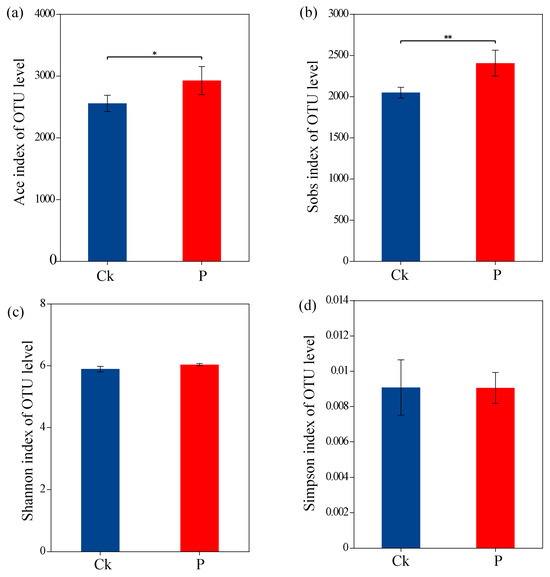
Figure 2.
Alpha-diversity indices of bacterial communities, as determined by (a) Abundance-Based Coverage Estimator (ACE). (b) Number of observed species (Sobs). (c) Shannon index. (d) Simpson index. Asterisks denote statistically significant differences: * p < 0.05; ** p < 0.01.
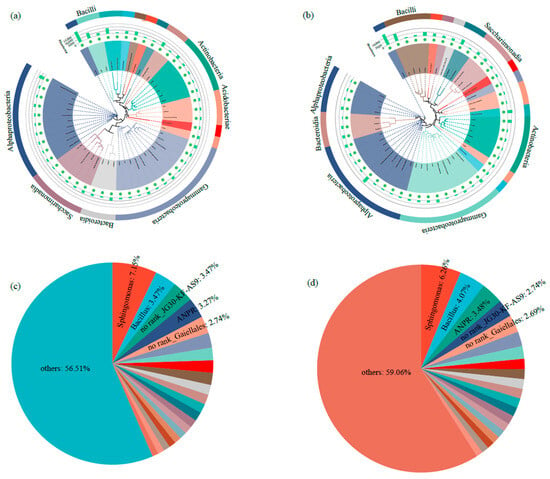
Figure 3.
Composition of rhizosphere bacterial communities in tumorous stem mustard. (a) Composition of rhizosphere bacterial communities at class level in the group without P. brassicae infection (Ck). (b) Composition of rhizosphere bacterial communities at class level with P. brassicae infection. (c) Composition of rhizosphere bacterial communities at genus level for Ck. (d) Composition of rhizosphere bacterial communities at genus level with P. brassicae infection. ANPR represents Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium.
Beta-diversity analysis based on NMDS demonstrated pronounced dissimilarities between groups Ck and P, indicating significant structural divergence in their microbial communities (Figure 4a). Furthermore, inter-group distance box plots (calculated using OTU-level Bray–Curtis dissimilarity) confirmed statistically distinct microbial community structures (p < 0.05, Kruskal–Wallis test) (Figure 4b). Inter-group differential significance testing revealed that the relative abundances of specific bacterial genera, such as Acidibacter, Dyella, Bacillus, Pedobacter, Conexibacter, Azotobacter, Burkholderia-Caballeronia-Paraburkholderia, Gemmatimonas, Sinomonas, and Mesorhizobium, exhibited significant differential abundance between the Ck and P groups (Figure 4c). LEfSe analysis (LDA score threshold > 3.0) identified distinct biomarker taxa: Sinomonas and Acidipila were enriched in Ck, whereas Paenibacillus served as key discriminators for the P group (Figure 4d).
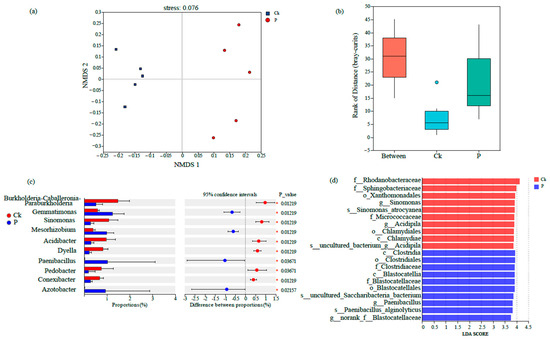
Figure 4.
Beta-diversity of rhizosphere bacterial communities in tumorous stem mustard as determined by (a) non-metric multidimensional scaling (NMDS) analysis, (b) rank distribution of Bray–Curtis dissimilarity distances for plants either infected (P) or not infected (Ck) with P. brassicae, (c) proportional differences of bacterial taxa with statistical significance, (d) linear discriminant analysis (LDA) scores of differential taxa.
3.3. Congruence Between Rhizosphere Bacterial Communities and Root Exudates
Multivariate integration analyses demonstrated significant congruence between rhizosphere microbial communities and root exudate metabolic signatures. Procrustes superimposition revealed robust co-variation patterns across the three experimental groups (M2 = 0.446, Monte Carlo permutation p = 0.005) (Figure S2a). This association was further validated by O2PLS modeling, with the predictive component explaining 63.9% of microbiome variance (R2X = 0.639) and 75.2% of metabolome variance (R2Y = 0.752), indicating tight system-level integration between these ecological compartments (Figure S2b).
In the Ck group, Simkaniaceae showed strong positive correlations with 2,6-Dimethyl-7-octene-1,6-diol 8-O-glucoside, cibaric acid, and geniposidic acid (Figure 5a). In the P group, Paenibacillus was associated with two metabolites, namely, 4-imidazolone-5-propionic acid and geniposidic acid (Figure 5b). While these associations suggest potential metabolic preferences, causal relationships require validation through gnotobiotic assays.
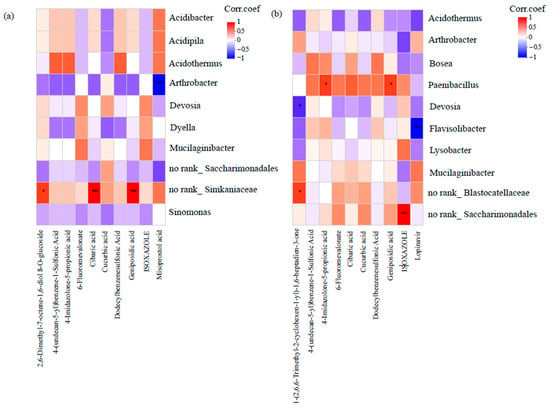
Figure 5.
Heatmap-based analysis of correlation networks between rhizosphere bacterial genera and specific chemicals of root exudates for plants either (a) not infected (Ck) with P. brassicae or (b) infected with P. brassicae (P). Asterisks denote statistically significant differences: * p < 0.05; *** p < 0.001.
4. Discussion
4.1. P. brassicae Infection Alters the Root Exudate Profiles
In this study, P. brassicae infection induced significant alterations in the root exudate profiles of tumorous stem mustard revealed by PLS-DA analysis. These metabolic shifts likely originated from pathogen-mediated disruption of core physiological functions, including down-regulated nutrient partitioning and compromised antioxidant response pathways [36]. The pronounced enrichment of the flavone and flavonol biosynthesis pathway in the P group corroborates the hypothesis that the host plant activates defensive secondary metabolite biosynthesis as a counterstrategy against P. brassicae colonization, indicating targeted mobilization of flavonoid-based defense mechanisms during pathogenesis.
Apigenin 7-O-beta-D-rutinoside, a flavonoid compound, exhibited the highest VIP value, underscoring its pivotal role in the restructured exudate composition. This metabolite, previously associated with antimicrobial activity [37], demonstrated significant enrichment in the P group exudates. Such pathogen-responsive biosynthetic induction aligns with emerging evidence positioning secondary metabolite secretion as a critical determinant of host–pathogen interaction dynamics in clubroot disease resistance [38], thereby reinforcing the functional significance of flavonoid up-regulation in plant defense strategies.
Gastrodin exhibited the second highest VIP score and showed significant enrichment in the P group, indicating its potential involvement in modulating host responses to P. brassicae infection. As extensively documented, gastrodin possesses pleiotropic therapeutic properties, including neuroprotective, anti-inflammatory, antioxidant, anti-apoptotic, and anti-tumor activities [39]. In this study, the up-regulation of gastrodin in P groups may alleviate P. brassicae-induced physiological stress through its established antioxidant properties, potentially disrupting pathogen virulence or enhancing plant defense mechanisms.
Cinncassiol D3, a diterpenoid compound, demonstrates diverse biological activities, including antibacterial, anti-inflammatory, antitumor, and immunomodulatory effects [40]. In this study, it exhibited the highest VIP value within the root exudates of control tumorous stem mustard, suggesting a potential regulatory role in the plant’s developmental processes or pathogen defense mechanisms. Concurrently, salbutamol exhibited the second-highest VIP value in the root exudates, suggesting its significant biological functions within the rhizosphere system of tumorous stem mustard. Salbutamol is a β2-adrenergic receptor agonist used for the aid of bronchospasm in conditions such as chronic obstructive pulmonary and asthma disease [41]. Building on evidence that cabbage roots accumulate salbutamol and transport it symplastically [42], suggesting a potential role in root development or rhizosphere ecology, our study hypothesizes that salbutamol mediates analogous functions in tumorous stem mustard.
Supervised multivariate analysis of volcano plot data revealed distinct metabolic profiles between the Ck and P groups. Notably, geniposidic acid and apigenin 7-O-β-D-rutinoside exhibited significant accumulation in the P group, contributing substantially to the divergence in the root exudate composition. These findings position both metabolites as key drivers of rhizosphere chemical remodeling during P. brassicae infection, potentially serving as biomarkers for host–pathogen interaction dynamics. Geniposidic acid, a monoterpenoid compound with diverse biological activities, has been demonstrated to alleviate oxidative stress in H2O2-treated HaCaT keratinocytes via activation of the Akt/Nrf2/OGG1 signaling pathway [43]. Notably, the observed enrichment of geniposidic acid in P groups suggests its potential role in mitigating physiological stress induced by P. brassicae infection, possibly through analogous antioxidant mechanisms.
Our metabolic profiling did not detect significant elevations in organic acids and amino acids within root exudates of the P group, potentially associated with tumor-like hyperplasia and localized epidermal rupture in infected roots. This temporal discrepancy likely arises from the abbreviated experimental timeline (5 weeks post-inoculation), which may preclude completion of pathogen-induced root lesion developmental processes. In contrast, the Ck group maintained characteristic enrichment in core metabolic pathways—including carbohydrate metabolism, amino acid metabolism, and lipid metabolism—indicating baseline metabolic homeostasis in non-infected plants.
4.2. Rhizosphere Microbial Community Restructuring Under P. brassicae Infection Conditions
Our results demonstrated that P. brassicae infection induced significant reconfiguration of the rhizosphere microbial community structure. At the alpha-diversity level, the P group exhibited markedly higher microbial community richness compared to the control, as evidenced by both the ACE index and observed species number (p < 0.05). The observed enrichment of rhizobacterial diversity in diseased plant rhizospheres aligns with recent findings [44], who reported analogous microbial community shifts in tomato plants infected by root-knot nematodes (Meloidogyne spp.). We hypothesize that this phenomenon may be attributed to nutrient leakage caused by cellular content release from clubroot galls [4]. Specifically, the leakage of sugar and amino acid rich substances may create a nutrient-enriched microenvironment, which could promote bacterial proliferation.
The beta diversity of the rhizosphere bacterial community in the P group differed significantly from that of the control group, demonstrating that pathogen invasion altered the composition of rhizosphere bacterial communities in tumorous stem mustard. Comparative studies of oilseed rape cultivars have revealed distinct rhizosphere bacterial community profiles in response to P. brassicae infection [45], corroborating these findings. The physiological integrity of plant roots has been shown to be disrupted by P. brassicae infection, resulting in modified endophytic bacterial and fungal community structures [19,20]. Such physiological changes induced by pathogen infection may fundamentally alter root exudate profiles, thereby driving the reorganization of rhizosphere bacterial communities. This phenomenon mirrors observations in other soil-borne disease systems, including root-knot nematode (Meloidogyne spp.) infections where similar microbiome perturbations have been documented [44]. Specifically, during root-knot nematode parasitism, activation of the plant’s immune system leads to the biosynthesis and secretion of defense compounds such as tomatine and flavonoids. These metabolites not only serve direct antagonistic functions against pathogens but also systemically modulate host-associated microbiomes [46,47]. However, the molecular mechanisms underlying metabolite-mediated microbiome engineering remain incompletely resolved, necessitating experimental validation.
LEfSe analysis identified distinct microbial biomarkers between groups, with Paenibacillus showing significant enrichment in infected samples. This contrasts with previous reports of Pseudomonas dominance in clubroot-infected oilseed rape rhizospheres [45], suggesting host-specific recruitment of biocontrol agents. The observed divergence in keystone taxa likely reflects (i) phylogenetic differences between tumorous stem mustard and oilseed rape and (ii) geographic variation in soil microbiomes. As a well-characterized plant growth-promoting rhizobacterium, Paenibacillus demonstrates broad-spectrum antagonistic activity against soil-borne pathogens [48], suggesting its potential role in directly suppressing P. brassicae proliferation within the rhizosphere of tumorous stem mustard. Conversely, the Ck group exhibited enrichment of Sinomonas and Acidipila, highlighting their ecological significance in the rhizosphere microenvironment. Sinomonas, commonly inhabiting soil–plant interfaces, has been reported to enhance plant growth through nutrient mobilization and phytohormone modulation [49], a mechanism likely conserved in tumorous stem mustard. Notably, Acidipila, previously identified as a beneficial taxon recruited by tomato with distinct fruit-color phenotypes [44], was similarly enriched here, indicating its potential to enhance plant biomass accumulation even in cruciferous hosts.
4.3. Root Exudates Regulate Rhizosphere Microbes
Root exudates, rich in carbohydrates, organic acids, and amino acids, serve as key drivers shaping rhizosphere microbiome assembly by providing nutrient niches [50,51]. These exudates not only mediate plant–microbe crosstalk critical for maintaining plant health but also sustain microbial diversity [52]. Procrustes analysis revealed a significant correlation between the metabolome profiles and rhizosphere microbiome (M2 = 0.446, p = 0.005), indicating close interactions between root exudates and microbial communities.
To elucidate the chemotactic interactions between rhizosphere microbiota and root exudate components, correlation heatmap analysis revealed significant positive associations (r > 0.65, p < 0.05) between Paenibacillus abundance and two key metabolites: geniposidic acid and 4-imidazolone-5-propionic acid, suggesting that these compounds may promote or recruit Paenibacillus. This mechanism parallels the well-documented “cry-for-help” strategy employed by plants under pathogen attack, where roots release specific signals into the soil to recruit beneficial microbes [52]. Supporting evidence comes from studies of Eucommia ulmoides, in which Paenibacillus as an endophytic bacterium showed significant positive correlations with geniposidic acid [53], indicating that this metabolite may serve as an attractant or growth promoter for these bacteria. In our study, tumorous stem mustard appears to utilize geniposidic acid as a signaling molecule (functioning as a “chemical language”) to attract Paenibacillus. This mechanism is functionally analogous to the benzoxazinoid-mediated recruitment of Pseudomonas putida by maize for suppression of Fusarium graminearum infections [54].
4-Imidazolone-5-propionic acid has been characterized in previous studies as a pivotal histidine pathway intermediate. Its degradation to L-formiminoglutamic acid in B. subtilis is recognized as essential for bacterial growth [55]. Our findings demonstrate that 4-imidazolone-5-propionic acid serves as a key metabolic mediator in the Brassica–Paenibacillus system through the mechanisms of nitrogen provisioning and acts as a specialized nitrogen source via histidine degradation pathways. Future studies could employ in vitro supplementation experiments to assess the effects of this metabolite on Paenibacillus growth.
The secretion of geniposidic acid and 4-imidazolone-5-propionic acid by tumorous stem mustard exemplifies a sophisticated public goods investment strategy for rhizosphere microbiome engineering. These metabolites function as signaling molecules and nutritional substrates, meeting classic public goods criteria through their diffusible nature and consumable properties [56].
Root exudates play a pivotal role in shaping rhizosphere microbial communities through the targeted recruitment of beneficial microbes, with significant applications in plant disease control and crop productivity enhancement [57]. Similarly, wheat enhances the concentration of organic compounds that mediate root exudation and promote plant growth-promoting rhizobacteria colonization, thereby increasing crop yield [58]. In this study, root exudates from tumorous stem mustard—specifically geniposidic acid and 4-imidazolone-5-propionic acid—demonstrate significant potential for recruiting beneficial microorganisms to combat P. brassicae infection.
5. Conclusions
Our findings reveal that P. brassicae infection triggers significant metabolic reprogramming in tumorous stem mustard root exudates, characterized by the selective enrichment of defense-related compounds, particularly flavones and phenolic derivatives. This pathogen-induced phytochemical remodeling mediates functional restructuring of the rhizosphere microbiome through chemotaxis-driven recruitment of beneficial microorganisms. Particularly noteworthy is the enhanced enrichment of Paenibacillus, which exhibits direct antagonistic activity against P. brassicae. Future studies should prioritize exogenous metabolite trials involving the systematic application of identified differential metabolites to evaluate dose-dependent effects on rhizosphere microbiota diversity and clubroot disease incidence. This multilevel approach will clarify whether root-exudate-mediated microbiome engineering can complement current fumigation-based control strategies against soil-borne pathogens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15081944/s1, Figure S1: The compound and pathway classification of root exudates. (a) Compound classification. (b) Pathway classification; Figure S2: Multi-omics integration analysis of rhizosphere microbiome–metabolome interactions. (a) Procrustes analysis. (b) Two-way orthogonal partial least squares (O2PLS) analysis.
Author Contributions
Writing—original draft, funding acquisition, project administration, writing—review and editing: D.W.; conceptualization, writing—original draft, data curation, investigation: J.L.; methodology, visualization, investigation, formal analysis: Z.C.; software, methodology, investigation and validation: X.Q.; resources, visualization, software, methodology: J.S.; writing—original draft, methodology, investigation and software: X.X.; formal analysis, methodology, investigation and validation: L.L. (Luyun Luo); formal analysis, software, data curation, resources: L.L. (Ling Li); writing—original draft, writing—review and editing, conceptualization, supervision: X.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Chongqing Municipality Natural Science Foundation Innovation and Development Joint Fund (CSTB2022NSCQ-LZX0005) and Chongqing Talent Plan “Lump-sum Funding System” Project (CQYC20220302547).
Data Availability Statement
All data supporting this study are included in the manuscript and Supplementary Files. Additional inquiries should be directed to the corresponding author(s). The raw 16S rRNA reads have been uploaded to the NCBI Sequence Read Archive (SRA) database with the accession number PRJNA1262643.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- National Development and Reform Commission/Ministry of Agriculture. National vegetable industry development plan. China Veg. 2012, 3, 1–2. [Google Scholar]
- Zamani-Noor, N.; Brand, S.; Söchting, H.P. Effect of pathogen virulence on pathogenicity, host range and reproduction of Plasmodiophora brassicae, the causal agent of clubroot disease. Plant Dis. 2022, 106, 57–64. [Google Scholar] [CrossRef]
- Xu, X.; Wu, C.; Zhang, F.; Yao, J.; Fan, L.; Liu, Z.; Yao, Y. Comprehensive review of Plasmodiophora brassicae: Pathogenesis, pathotype diversity, and integrated control methods. Front. Microbiol. 2025, 16, 1531393. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, R.; Truman, W.; Blicharz, S. Genius architect or clever thief-how Plasmodiophora brassicae reprograms host development to establish a pathogen-oriented physiological sink. Mol. Plant Microbe Interact. 2019, 32, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Olszak, M.; Truman, W.; Stefanowicz, K.; Sliwinska, E.; Ito, M.; Walerowski, P. Transcriptional profiling identifies critical steps of cell cycle reprogramming necessary for Plasmodiophora brassicae-driven gall formation in arabidopsis. Plant J. 2019, 97, 715–729. [Google Scholar] [CrossRef]
- Irani, S.; Trost, B.; Waldner, M.; Nayidu, N.; Tu, J.; Kusalik, A.J.; Todd, C.D.; Wei, Y.; Bonham-Smith, P.C. Transcriptome analysis of response to Plasmodiophora brassicae infection in the arabidopsis shoot and root. BMC Genom. 2018, 19, 23. [Google Scholar] [CrossRef]
- Hwang, S.F.; Strelkov, S.E.; Feng, J.; Gossen, B.D.; Howard, R.J. Plasmodiophora brassicae: A review of an emerging pathogen of the canadian canola (Brassica napus) crop. Mol. Plant Pathol. 2012, 13, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Fogelqvist, J.; Knaust, A. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 2015, 5, 11153. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Strelkov, S.E.; Links, M.G.; Clarke, W.E.; Robinson, S.J.; Djavaheri, M.; Malinowski, R.; Haddadi, P.; Kagale, S.; Parkin, I.A.; et al. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host brassica spp. BMC Genom. 2016, 17, 272. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, J.; Sun, X.; Huang, L.; Sheng, Y.; Zhang, Q. Comparative metagenomic analysis reveals rhizosphere microbiome assembly and functional adaptation changes caused by clubroot disease in chinese cabbage. Microorganisms 2024, 12, 1370. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, S.; Jiang, C.; Wu, C.; Gao, M.; Wang, Q. A review of root exudates and rhizosphere microbiome for crop production. Environ. Sci. Pollut. Res. 2021, 28, 54497–54510. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Chai, A.L.; Lin, Z.L.; Shi, Y.X.; Xie, X.W.; Li, L. Deciphering differences in microbial community diversity between clubroot-diseased and healthy soils. Microorganisms 2024, 12, 251. [Google Scholar] [CrossRef]
- Wu, W.X.; Huang, X.Q.; Zhang, L.; Yang, X.X.; Li, H.Z.; Liu, Y. Crucifer clubroot disease changes the microbial community structure of rhizosphere soil. Acta Ecol. Sin. 2020, 40, 1532–1541. [Google Scholar] [CrossRef]
- Lebreton, L.; Guillerm-Erckelboudt, A.Y.; Gazengel, K.; Linglin, J.; Ourry, M.; Glory, P. Temporal dynamics of bacterial and fungal communities during the infection of brassica rapa roots by the protist Plasmodiophora brassicae. PLoS ONE 2019, 14, e0204195. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Elvia, J.; Galindo-González, L.; Fredua-Agyeman, R.; Hwang, S.F.; Strelkov, S.E. Clubroot-induced changes in the root and rhizosphere microbiome of susceptible and resistant canola. Plants 2024, 13, 1880. [Google Scholar] [CrossRef]
- Gazengel, K.; Aigu, Y.; Lariagon, C.; Humeau, M.; Gravot, A.; Manzanares-Dauleux, M.J. Nitrogen supply and host-plant genotype modulate the transcriptomic profile of Plasmodiophora brassicae. Front. Microbiol. 2021, 12, 701067. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, X.; Sarenqimuge, S.; von Tiedemann, A. The soil bacterial community regulates germination of Plasmodiophora brassicae resting spores rather than root exudates. PloS Pathog. 2023, 19, e1011175. [Google Scholar] [CrossRef]
- Tian, X.; Wang, D.; Mao, Z.; Pan, L.; Liao, J.; Cai, Z. Infection of Plasmodiophora brassicae changes the fungal endophyte community of tumourous stem mustard roots as revealed by high-throughput sequencing and culture-dependent methods. PLoS ONE 2019, 14, e0214975. [Google Scholar] [CrossRef]
- Wang, D.; Sun, T.; Zhao, S.; Pan, L.; Liu, H.; Tian, X. Physiological change alters endophytic bacterial community in clubroot of tumorous stem mustard infected by Plasmodiophora brassicae. BMC Microbiol. 2020, 20, 244. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.; Wang, Y.; Fang, B.; Ge, W.; Wang, X.; Zou, J.; Ji, R. Transcriptome analysis of chinese cabbage infected with Plasmodiophora brassicae in the primary stage. Sci. Rep. 2024, 14, 26180. [Google Scholar] [CrossRef]
- Ji, R.; Zhao, L.; Xing, M.; Shen, X.; Bi, Q.; Peng, S. Infection of Plasmodiophora brassicae in chinese cabbage. Genet. Mol. Res. 2014, 13, 10976–10982. [Google Scholar] [CrossRef]
- de Vries, F.T.; Williams, A.; Stringer, F.; Willcocks, R.; McEwing, R.; Langridge, H.; Straathof, A.L. Changes in root-exudate-induced respiration reveal a novel mechanism through which drought affects ecosystem carbon cycling. New Phytol. 2019, 224, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, J. Untargeted metabolomic analysis of metabolites related to body dysmorphic disorder (bdd). Funct. Integr. Genom. 2023, 23, 70. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. Ggtranscript: An r package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Balzerani, F.; Blasco, T.; Pérez-Burillo, S.; Francino, M.P.; Rufián-Henares, J.Á.; Valcarcel, L.V.; Planes, F.J. Q2-metnet: Qiime2 package to analyse 16s rrna data via high-quality metabolic reconstructions of the human gut microbiota. Bioinformatics 2024, 40, btae455. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cole, J.R. Updated rdp taxonomy and rdp classifier for more accurate taxonomic classification. Microbiol. Resour. Announc. 2024, 13, e0106323. [Google Scholar] [CrossRef]
- Dixon, P. Vegan, a package of r functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Friendly, M. Corrgrams: Exploratory displays for correlation matrices. Am. Stat. 2002, 56, 316–324. [Google Scholar] [CrossRef]
- Malinowski, R.; Smith, J.A.; Fleming, A.J.; Scholes, J.D.; Rolfe, S.A. Gall formation in clubroot-infected arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J. 2012, 71, 226–238. [Google Scholar] [CrossRef]
- Nazaruk, J.; Gudej, J. Flavonoid compounds from the flowers of Cirsium rivulare (jacq.) all. Acta Pol. Pharm. 2003, 60, 87–89. [Google Scholar]
- Yang, K.; Fu, R.; Feng, H.; Jiang, G.; Finkel, O.; Sun, T.; Liu, M.; Huang, B.; Li, S.; Wang, X.; et al. Rin enhances plant disease resistance via root exudate-mediated assembly of disease-suppressive rhizosphere microbiota. Mol. Plant. 2023, 16, 1379–1395. [Google Scholar] [CrossRef]
- El Menyiy, N.; Elouafy, Y.; Moubachir, R.; Abdnim, R.; Benali, T.; Taha, D.; Khalid, A.; Abdalla, A.N.; Hamza, S.M.A.; El-Shazly, M.; et al. Chemistry, biological activities, and pharmacological properties of gastrodin: Mechanism insights. Chem. Biodivers. 2024, 21, e202400402. [Google Scholar] [CrossRef]
- Liu, S.; Yang, L.; Zheng, S.; Hou, A.; Man, W.; Zhang, J.; Wang, S.; Wang, X.; Yu, H.; Jiang, H. A review: The botany, ethnopharmacology, phytochemistry, pharmacology of Cinnamomi cortex. RSC Adv. 2021, 11, 27461–27497. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.; Vale, N. Salbutamol in the management of asthma: A review. Int. J. Mol. Sci. 2022, 23, 14207. [Google Scholar] [CrossRef]
- Herklotz, P.A.; Gurung, P.; Vanden Heuvel, B.; Kinney, C.A. Uptake of human pharmaceuticals by plants grown under hydroponic conditions. Chemosphere 2010, 78, 1416–1421. [Google Scholar] [CrossRef]
- Cheng, S.; Jia, H.; Zhang, Y.; Zhou, J.; Chen, X.; Wu, L.; Wang, J. Geniposidic acid from Eucommia ulmoides oliver staminate flower tea mitigates cellular oxidative stress via activating akt/nrf2 signaling. Molecules 2022, 27, 8568. [Google Scholar] [CrossRef]
- Kudjordjie, E.N.; Santos, S.S.; Topalović, O.; Vestergård, M. Distinct changes in tomato-associated multi-kingdom microbiomes during meloidogyne incognita parasitism. Environ. Microbiome. 2024, 19, 53. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Wu, W.; Huang, X.; Yang, X.; Yu, Y.; Zhang, Z.; Hu, Z.; Zhou, X.; Zhou, K.; Liu, Y.L. Characterization of rhizosphere bacterial communities in oilseed rape cultivars with different susceptibility to Plasmodiophora brassicae infection. Front. Plant Sci. 2025, 15, 1496770. [Google Scholar] [CrossRef]
- Kudjordjie, E.N.; Sapkota, R.; Nicolaisen, M. Arabidopsis assemble distinct root-associated microbiomes through the synthesis of an array of defense metabolites. PLoS ONE 2021, 16, e0259171. [Google Scholar] [CrossRef] [PubMed]
- Sikder, M.M.; Vestergård, M.; Kyndt, T.; Topalović, O.; Kudjordjie, E.N.; Nicolaisen, M. Genetic disruption of arabidopsis secondary metabolite synthesis leads to microbiome-mediated modulation of nematode invasion. ISME J. 2022, 1, 12. [Google Scholar] [CrossRef] [PubMed]
- Dobrzyński, J.; Naziębło, A. Paenibacillus as a biocontrol agent for fungal phytopathogens: Is p. Polymyxa the only one worth attention? Microb. Ecol. 2024, 87, 134. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, J.; Su, Z.; Chen, Q.; Li, J.; Zhao, J.; Xuan, W.; Miao, Y.; Zhang, J.; Zhang, R. Sinomonas gamaensis neau-hv1 remodels the iaa14-arf7/19 interaction to promote plant growth. New Phytol. 2025, 245, 2016–2037. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Afridi, M.S.; Kumar, A.; Javed, M.A.; Dubey, A.; de Medeiros, F.H.V.; Santoyo, G. Harnessing root exudates for plant microbiome engineering and stress resistance in plants. Microbiol. Res. 2024, 279, 127564. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, K.; Li, P.; Wan, D.; Liu, J.; Yi, X.; Peng, Y. Characteristics of endophytic bacteria and active ingredients in the eucommiae cortex from different origins. Front. Microbiol. 2023, 14, 1164674. [Google Scholar] [CrossRef]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef]
- Su, H.; Sheng, X.; Liu, Y. Exploring the substrate specificity and catalytic mechanism of imidazolonepropionase (huti) from Bacillus subtilis. Phys. Chem. Chem. Phys. 2016, 18, 27928–27938. [Google Scholar] [CrossRef]
- Smith, P.; Schuster, M. Public goods and cheating in microbes. Curr Biol. 2019, 29, R442–R447. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Srivastava, A.K.; Rajput, V.D.; Chauhan, P.K.; Bhojiya, A.A.; Jain, D.; Chaubey, G.; Dwivedi, P.; Sharma, B.; Minkina, T. Root exudates: Mechanistic insight of plant growth promoting rhizobacteria for sustainable crop production. Front. Microbiol. 2022, 13, 916488. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).