Transcriptome Analysis Reveals the Vital Role of ABA Plays in Drought Tolerance of the ABA-Insensitive Alfalfa (Medicago sativa L.)
Abstract
1. Introduction
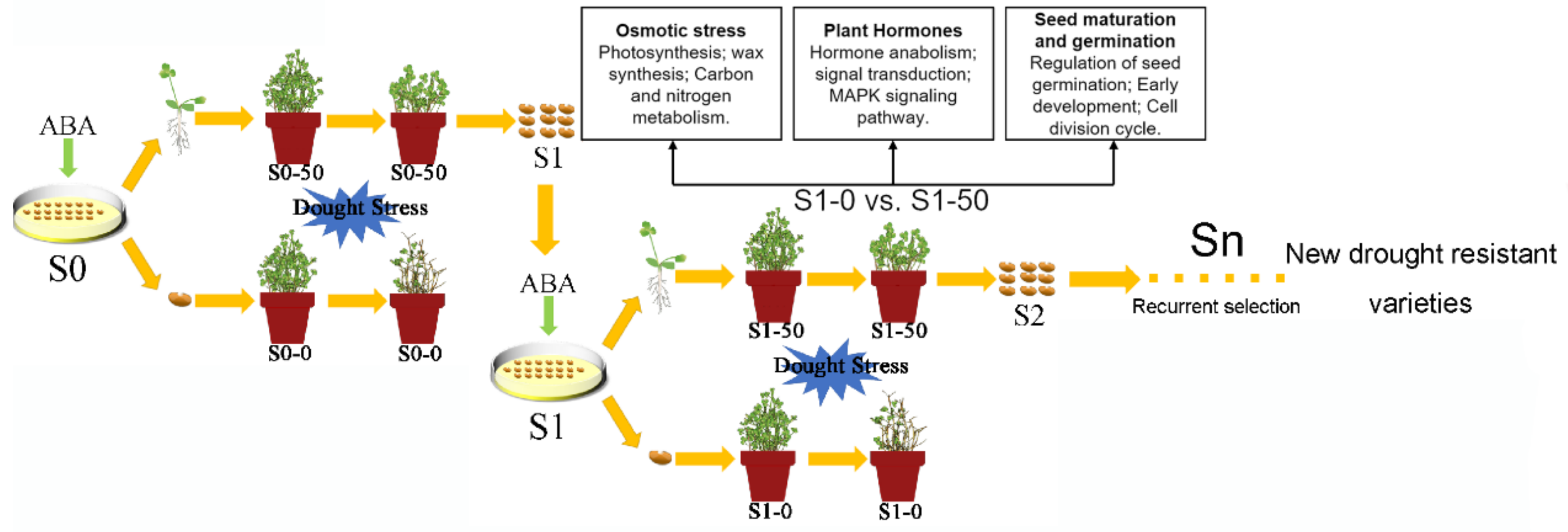
2. Materials and Methods
2.1. Seeds Sterilization and Germination
2.2. Isolated Seeds of ABA Insensitive (S1-50) and Sensitive (S1-0)
2.3. Determination of Water Loss Rate in Alfalfa Leaves
2.4. Drought Tolerance Test
2.5. RNA Isolation
2.6. Library Preparation and Sequencing
2.7. Reference-Based Mapping and Differential Gene Expression Analysis and Annotation
2.8. Quantitative Real-Time PCR
2.9. Statistical Analysis
3. Results
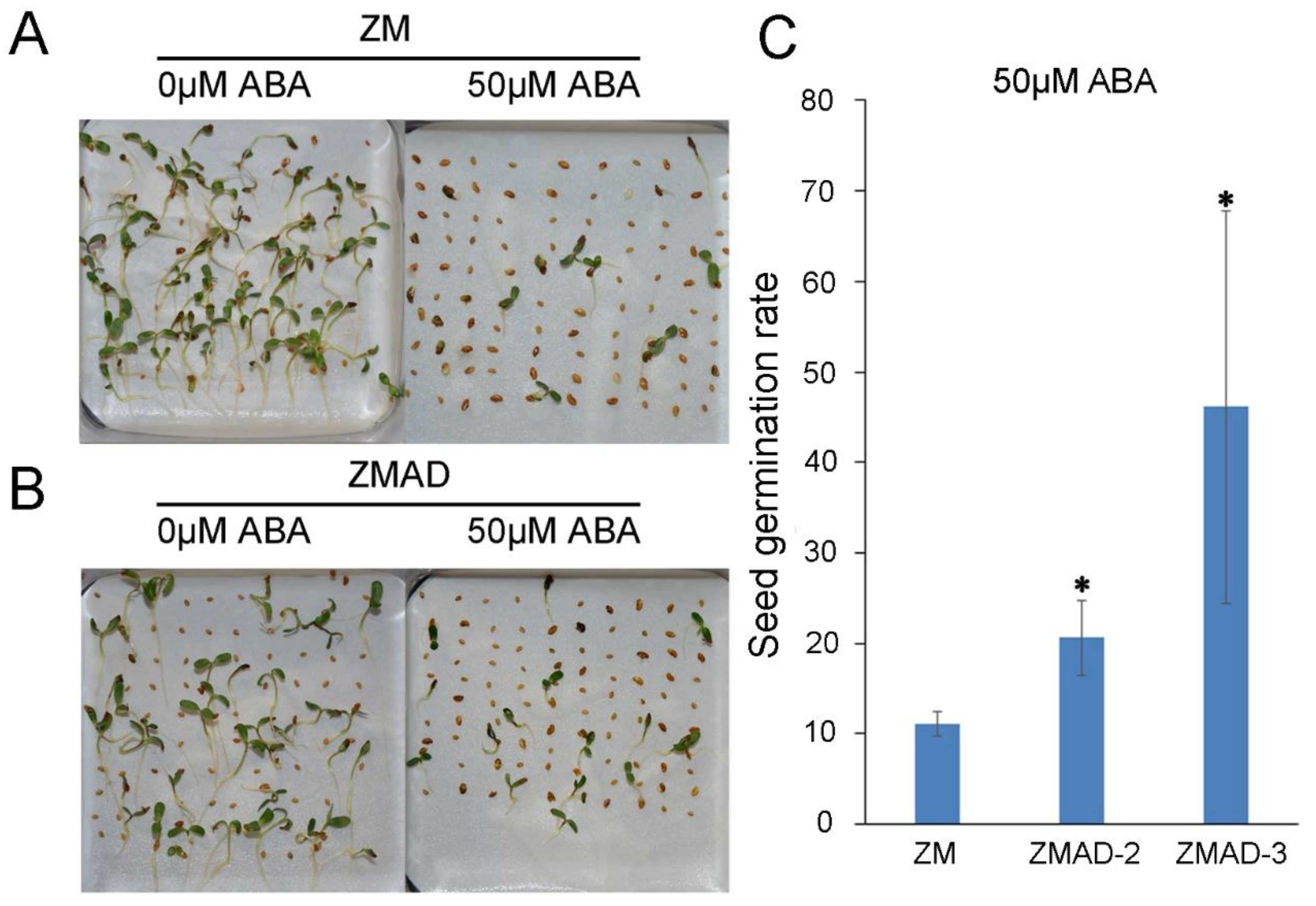
3.1. Seeds Germination of ZM and ZMAD under 0 μM or 50 μM ABA Treatment
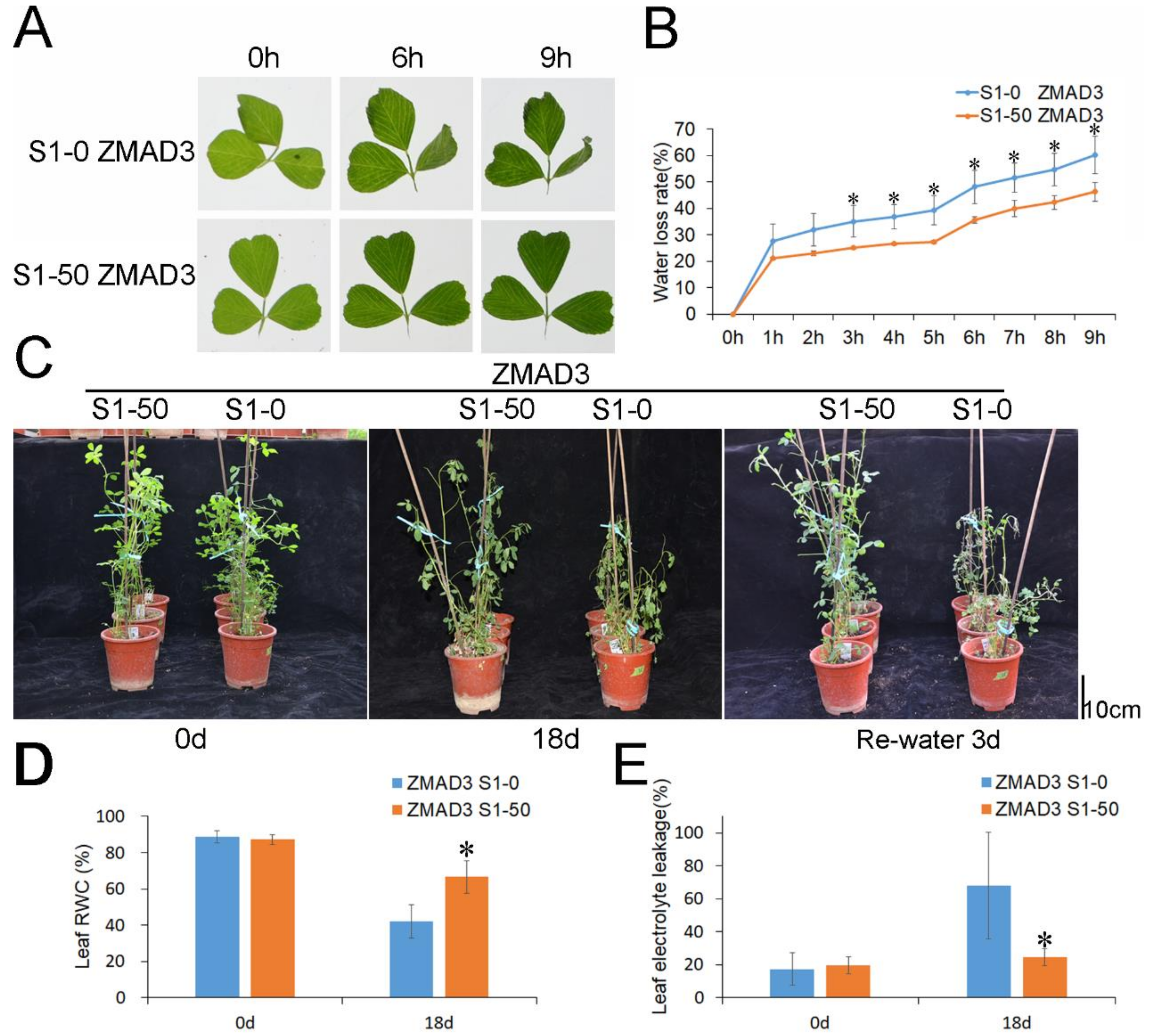
3.2. The S1-50 Alfalfa Plants Showed Better Drought Tolerance Than S1-0 Plants
3.3. Data Quality and Summary of Reads
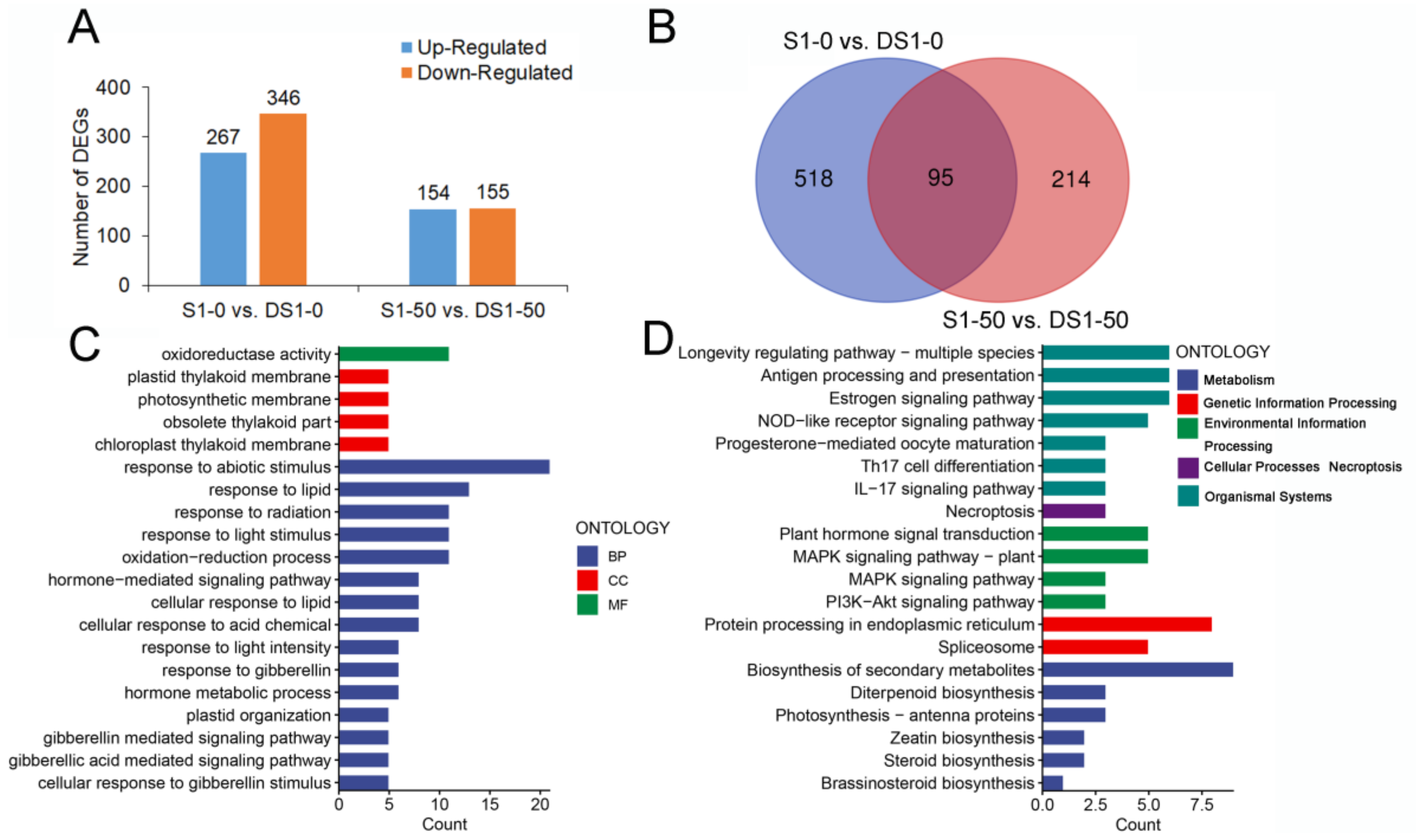
3.4. Deferentially Expressed Genes between S1-0 and S1-50 Plants under Drought Stress Treatment
3.5. DEGs between S1-0 and S1-50 before and after Drought Stress Treatment
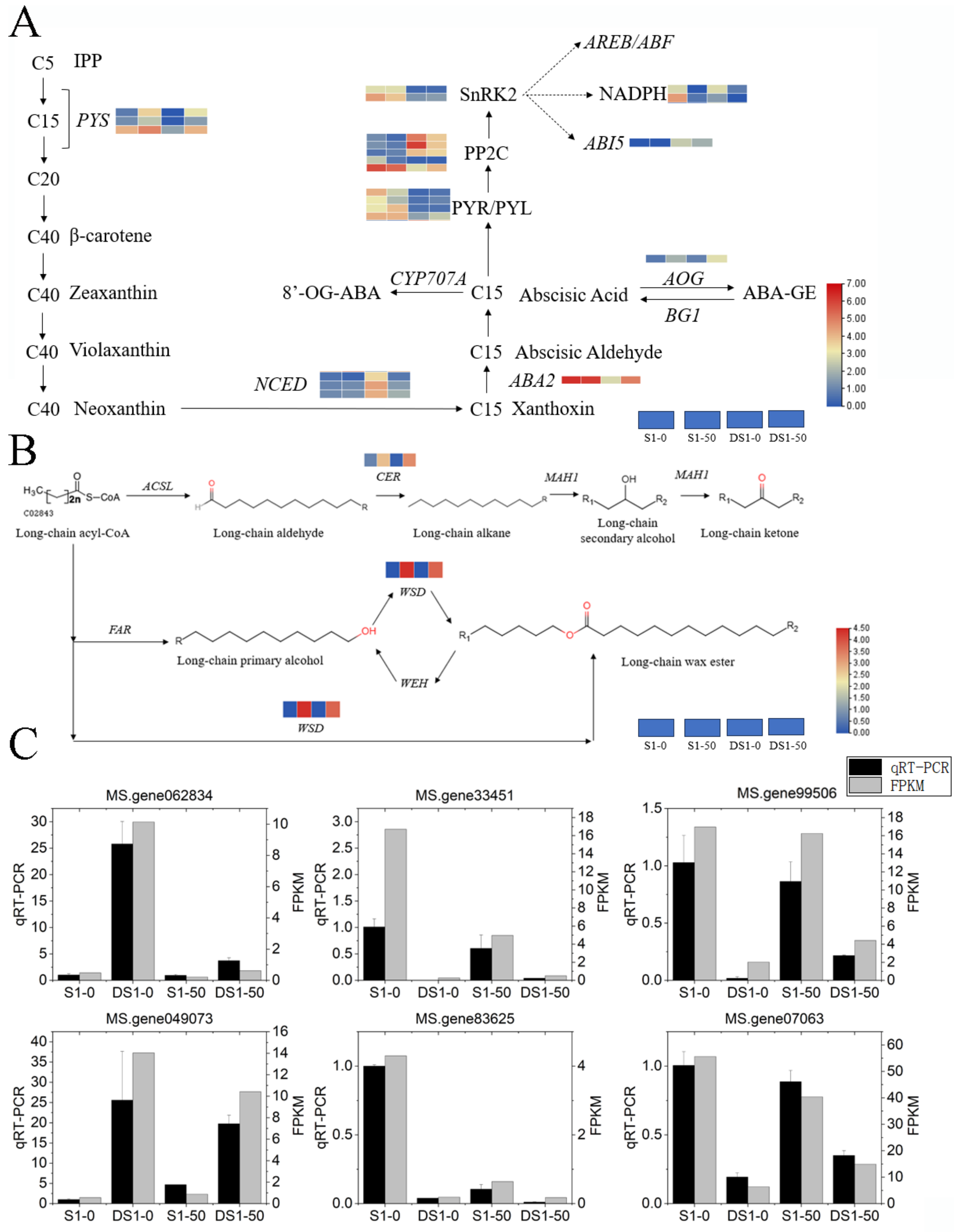
3.6. Integrative Analysis of Drought Tolerance Enhancement through the ABA and Waxes Biosynthesis Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Biazzi, E.; Nazzicari, N.; Pecetti, L.; Brummer, E.C.; Palmonari, A.; Tava, A.; Annicchiarico, P. Genome-Wide Association Mapping and Genomic Selection for Alfalfa (Medicago sativa) Forage Quality Traits. PLoS ONE 2017, 12, e0169234. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of Extreme Weather Disasters on Global Crop Production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.D.; Hannoufa, A.; Acharya, S. Molecular Improvement of Alfalfa for Enhanced Productivity and Adaptability in a Changing Environment. Plant Cell Environ. 2018, 41, 1955–1971. [Google Scholar] [CrossRef]
- Luo, D.; Wu, Y.; Liu, J.; Zhou, Q.; Liu, W.; Wang, Y.; Yang, Q.; Wang, Z.; Liu, Z. Comparative Transcriptomic and Physiological Analyses of Medicago sativa L. Indicates That Multiple Regulatory Networks Are Activated during Continuous ABA Treatment. Int. J. Mol. Sci. 2018, 20, 47. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.W. Genetic Engineering to Improve Plant Performance under Drought: Physiological Evaluation of Achievements, Limitations, and Possibilities. J. Exp. Bot. 2013, 64, 83–108. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar Patel, M.; Kumar, N.; Bajpai, A.B.; Siddique, K.H.M. Metabolomics and Molecular Approaches Reveal Drought Stress Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 9108. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, I.M.Y.; Yuan, S.; Yoshihara, S.; Suzaki, T.; Ezura, H.; Miura, K. Stimulation of Tomato Drought Tolerance by PHYTOCHROME A and B1B2 Mutations. Int. J. Mol. Sci. 2023, 24, 1560. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-Cis-Epoxycarotenoid Dioxygenase Gene, Regulates Salt and Water Stress Tolerance and Leaf Senescence in Rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.; Okamoto, M.; Mega, R.; Kanno, Y.; Ohnishi, T.; Seo, M.; Todoroki, Y. Abscinazole-E3M, a Practical Inhibitor of Abscisic Acid 8′-Hydroxylase for Improving Drought Tolerance. Sci. Rep. 2016, 6, 37060. [Google Scholar] [CrossRef]
- Han, Y.; Watanabe, S.; Shimada, H.; Sakamoto, A. Dynamics of the Leaf Endoplasmic Reticulum Modulate β-Glucosidase-Mediated Stress-Activated ABA Production from Its Glucosyl Ester. J. Exp. Bot. 2020, 71, 2058–2071. [Google Scholar] [CrossRef]
- Ali, A.; Kim, J.K.; Jan, M.; Khan, H.A.; Khan, I.U.; Shen, M.; Park, J.; Lim, C.J.; Hussain, S.; Baek, D.; et al. Rheostatic Control of ABA Signaling through HOS15-Mediated OST1 Degradation. Mol. Plant 2019, 12, 1447–1462. [Google Scholar] [CrossRef]
- Pizzio, G.A.; Mayordomo, C.; Lozano-Juste, J.; Garcia-Carpintero, V.; Vazquez-Vilar, M.; Nebauer, S.G.; Kaminski, K.P.; Ivanov, N.V.; Estevez, J.C.; Rivera-Moreno, M.; et al. PYL1- and PYL8-like ABA Receptors of Nicotiana Benthamiana Play a Key Role in ABA Response in Seed and Vegetative Tissue. Cells 2022, 11, 795. [Google Scholar] [CrossRef]
- Li, Y.; Ding, Y.; Qu, L.; Li, X.; Lai, Q.; Zhao, P.; Gao, Y.; Xiang, C.; Cang, C.; Liu, X.; et al. Structure of the Arabidopsis Guard Cell Anion Channel SLAC1 Suggests Activation Mechanism by Phosphorylation. Nat. Commun. 2022, 13, 2511. [Google Scholar] [CrossRef] [PubMed]
- Mimata, Y.; Munemasa, S.; Nakamura, T.; Nakamura, Y.; Murata, Y. Extracellular Malate Induces Stomatal Closure via Direct Activation of Guard-Cell Anion Channel SLAC1 and Stimulation of Ca2+ Signalling. New Phytol. 2022, 236, 852–863. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, B.; Gao, S.; Xu, K. Grafting Improves Tomato Drought Tolerance through Enhancing Photosynthetic Capacity and Reducing ROS Accumulation. Protoplasma 2019, 256, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Ye, W.; Matsushima, D.; Rhaman, M.S.; Munemasa, S.; Okuma, E.; Nakamura, Y.; Biswas, M.S.; Mano, J.; Murata, Y. Reactive Carbonyl Species Function as Signal Mediators Downstream of H2O2 Production and Regulate [Ca2+]Cyt Elevation in ABA Signal Pathway in Arabidopsis Guard Cells. Plant Cell Physiol. 2019, 60, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Postiglione, A.E.; Muday, G.K. The Role of ROS Homeostasis in ABA-Induced Guard Cell Signaling. Front. Plant Sci. 2020, 11, 968. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato. Int. J. Mol. Sci. 2020, 21, 159. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Prasad, S.S.; Mitra, J.; Siddiqui, N.; Sahoo, L.; Kobayashi, Y.; Koyama, H. How Do Plants Remember Drought? Planta 2022, 256, 7. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.-L.; Yue, X.-F.; Min, Z.; Wang, X.-H.; Fang, Y.-L.; Zhang, J.-X. VvNAC17, a Novel Stress-Responsive Grapevine (Vitis vinifera L.) NAC Transcription Factor, Increases Sensitivity to Abscisic Acid and Enhances Salinity, Freezing, and Drought Tolerance in Transgenic Arabidopsis. Plant Physiol. Biochem. PPB 2020, 146, 98–111. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, D.; Yan, J.; Wang, K.; Lin, S.; Zhang, W. ABA-Insensitivity of Alfalfa (Medicago sativa L.) during Seed Germination Associated with Plant Drought Tolerance. Environ. Exp. Bot. 2022, 203, 105069. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Yan, J.; Wang, K.; Luo, H.; Zhang, W. MiR319 Mediated Salt Tolerance by Ethylene. Plant Biotechnol. J. 2019, 17, 2370–2383. [Google Scholar] [CrossRef]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-Aware Chromosome-Level Genome Assembly and Efficient Transgene-Free Genome Editing for the Autotetraploid Cultivated Alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef]
- Fang, L.; Liu, T.; Li, M.; Dong, X.; Han, Y.; Xu, C.; Li, S.; Zhang, J.; He, X.; Zhou, Q.; et al. MODMS: A Multi-Omics Database for Facilitating Biological Studies on Alfalfa (Medicago sativa L.). Hortic. Res. 2024, 11, uhad245. [Google Scholar] [CrossRef]
- Yang, C.; Wang, H.; Ouyang, Q.; Chen, G.; Fu, X.; Hou, D.; Xu, H. Deficiency of Auxin Efflux Carrier OsPIN1b Impairs Chilling and Drought Tolerance in Rice. Plants 2023, 12, 4058. [Google Scholar] [CrossRef] [PubMed]
- Arif, M.; Li, Z.; Luo, Q.; Li, L.; Shen, Y.; Men, S. The BAG2 and BAG6 Genes Are Involved in Multiple Abiotic Stress Tolerances in Arabidopsis Thaliana. Int. J. Mol. Sci. 2021, 22, 5856. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.Z.; Ali, A.; Ali, W.; Aasim, M.; Khan, T.; Khan, Z.; Munir, I. Heterologous Expression of Bacterial Dehydrin Gene in Arabidopsis Thaliana Promotes Abiotic Stress Tolerance. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2023, 29, 1239–1246. [Google Scholar] [CrossRef]
- Jain, N.; Khurana, P.; Khurana, J.P. AtTLP2, a Tubby-like Protein, Plays Intricate Roles in Abiotic Stress Signalling. Plant Cell Rep. 2023, 42, 235–252. [Google Scholar] [CrossRef] [PubMed]
- Adie, B.A.T.; Pérez-Pérez, J.; Pérez-Pérez, M.M.; Godoy, M.; Sánchez-Serrano, J.-J.; Schmelz, E.A.; Solano, R. ABA Is an Essential Signal for Plant Resistance to Pathogens Affecting JA Biosynthesis and the Activation of Defenses in Arabidopsis. Plant Cell 2007, 19, 1665–1681. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple Exposures to Drought “train” Transcriptional Responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.; Bendix, C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; de Oliveira, M.C.N.; Harmon, F.G.; Nepomuceno, A. Diurnal Oscillations of Soybean Circadian Clock and Drought Responsive Genes. PLoS ONE 2014, 9, e86402. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The Physiology of Plant Responses to Drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Dwivedi, A.K.; Singh, V.; Anwar, K.; Pareek, A.; Jain, M. Integrated Transcriptome, Proteome and Metabolome Analyses Revealed Secondary Metabolites and Auxiliary Carbohydrate Metabolism Augmenting Drought Tolerance in Rice. Plant Physiol. Biochem. 2023, 201, 107849. [Google Scholar] [CrossRef] [PubMed]
- Zarbakhsh, S.; Shahsavar, A.R. Exogenous γ-Aminobutyric Acid Improves the Photosynthesis Efficiency, Soluble Sugar Contents, and Mineral Nutrients in Pomegranate Plants Exposed to Drought, Salinity, and Drought-Salinity Stresses. BMC Plant Biol. 2023, 23, 543. [Google Scholar] [CrossRef] [PubMed]
- Gou, C.; Huang, Q.; Rady, M.M.; Wang, L.; Ihtisham, M.; El-Awady, H.H.; Seif, M.; Alazizi, E.M.Y.; Eid, R.S.M.; Yan, K.; et al. Integrative Application of Silicon and/or Proline Improves Sweet Corn (Zea mays L. Saccharata) Production and Antioxidant Defense System under Salt Stress Condition. Sci. Rep. 2023, 13, 18315. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.-S.; Ge, N.; Wang, Q.-Y.; Zhao, L.-T.; Chen, C.; Chen, J.-W. Genome-Wide Identification and Characterization of Members of the LEA Gene Family in Panax Notoginseng and Their Transcriptional Responses to Dehydration of Recalcitrant Seeds. BMC Genom. 2023, 24, 126. [Google Scholar] [CrossRef]
- Lv, A.; Su, L.; Fan, N.; Wen, W.; Wang, Z.; Zhou, P.; An, Y. Chloroplast-Targeted Late Embryogenesis Abundant 1 Increases Alfalfa Tolerance to Drought and Aluminum. Plant Physiol. 2023, 193, 2750–2767. [Google Scholar] [CrossRef]
- Zhu, P.; Li, R.; Fan, W.; Xia, Z.; Li, J.; Wang, C.; Zhao, A. A Mulberry 9-Cis-Epoxycarotenoid Dioxygenase Gene MaNCED1 Is Involved in Plant Growth Regulation and Confers Salt and Drought Tolerance in Transgenic Tobacco. Front. Plant Sci. 2023, 14, 1228902. [Google Scholar] [CrossRef]
- Sharma, S.; Villamor, J.G.; Verslues, P.E. Essential Role of Tissue-Specific Proline Synthesis and Catabolism in Growth and Redox Balance at Low Water Potential1[W][OA]. Plant Physiol. 2011, 157, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A Dual Function of SnRK2 Kinases in the Regulation of SnRK1 and Plant Growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef]
- Wang, J.; Li, C.; Li, L.; Gao, L.; Hu, G.; Zhang, Y.; Reynolds, M.P.; Zhang, X.; Jia, J.; Mao, X.; et al. DIW1 Encoding a Clade I PP2C Phosphatase Negatively Regulates Drought Tolerance by De-Phosphorylating TaSnRK1.1 in Wheat. J. Integr. Plant Biol. 2023, 65, 1918–1936. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Ahmad, N.; Wang, Y.; Ge, H.; Wang, Y.; Liu, W.; Li, X.; Wang, N.; Wang, F.; et al. CePP2C19 Confers Tolerance to Drought by Regulating the ABA Sensitivity in Cyperus Esculentus. BMC Plant Biol. 2023, 23, 524. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Schorderet, M.; Ryser, U.; Buchala, A.; Kolattukudy, P.; Métraux, J.P.; Nawrath, C. Transgenic Arabidopsis Plants Expressing a Fungal Cutinase Show Alterations in the Structure and Properties of the Cuticle and Postgenital Organ Fusions. Plant Cell 2000, 12, 721–738. [Google Scholar] [CrossRef]
- Rahman, T.; Shao, M.; Pahari, S.; Venglat, P.; Soolanayakanahally, R.; Qiu, X.; Rahman, A.; Tanino, K. Dissecting the Roles of Cuticular Wax in Plant Resistance to Shoot Dehydration and Low-Temperature Stress in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 1554. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, J.; Meng, W.; Zeng, L.; Sun, L. Genome-Wide Analysis of the WSD Family in Sunflower and Functional Identification of HaWSD9 Involvement in Wax Ester Biosynthesis and Osmotic Stress. Front. Plant Sci. 2022, 13, 975853. [Google Scholar] [CrossRef]
- Li, T.; Sun, Y.; Liu, T.; Wu, H.; An, P.; Shui, Z.; Wang, J.; Zhu, Y.; Li, C.; Wang, Y.; et al. TaCER1-1A Is Involved in Cuticular Wax Alkane Biosynthesis in Hexaploid Wheat and Responds to Plant Abiotic Stresses. Plant Cell Environ. 2019, 42, 3077–3091. [Google Scholar] [CrossRef] [PubMed]

| Sample | Treatment | Biological Replicate | Total Reads | Total Mapped | Mapping Rate (%) |
|---|---|---|---|---|---|
| S1-0 | Control | 1 | 44,542,818 | 43,091,372 | 96.74 |
| 2 | 44,707,562 | 43,175,678 | 96.57 | ||
| 3 | 110,570,548 | 106,908,754 | 96.69 | ||
| DS1-0 | Drought (18 d) | 1 | 44,868,990 | 43,253,798 | 96.40 |
| 2 | 46,082,556 | 43,690,094 | 94.81 | ||
| 3 | 44,924,794 | 42,946,096 | 95.60 | ||
| S1-50 | Control | 1 | 44,114,614 | 42,721,328 | 96.84 |
| 2 | 45,108,312 | 43,688,472 | 96.85 | ||
| 3 | 46,831,492 | 45,535,672 | 97.23 | ||
| DS1-50 | Drought (18 d) | 1 | 46,928,416 | 45,547,974 | 97.06 |
| 2 | 44,681,506 | 43,377,766 | 97.08 | ||
| 3 | 45,269,416 | 43,987,334 | 97.17 | ||
| Total | 608,631,024 | 587,924,338 | 96.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, M.; Xu, Z.; Liu, Y.; Liu, Y.; Liu, J.; Zhang, W. Transcriptome Analysis Reveals the Vital Role of ABA Plays in Drought Tolerance of the ABA-Insensitive Alfalfa (Medicago sativa L.). Agronomy 2024, 14, 406. https://doi.org/10.3390/agronomy14030406
Xu M, Xu Z, Liu Y, Liu Y, Liu J, Zhang W. Transcriptome Analysis Reveals the Vital Role of ABA Plays in Drought Tolerance of the ABA-Insensitive Alfalfa (Medicago sativa L.). Agronomy. 2024; 14(3):406. https://doi.org/10.3390/agronomy14030406
Chicago/Turabian StyleXu, Mingzhi, Zhenpeng Xu, Yanrong Liu, Yaling Liu, Jinghui Liu, and Wanjun Zhang. 2024. "Transcriptome Analysis Reveals the Vital Role of ABA Plays in Drought Tolerance of the ABA-Insensitive Alfalfa (Medicago sativa L.)" Agronomy 14, no. 3: 406. https://doi.org/10.3390/agronomy14030406
APA StyleXu, M., Xu, Z., Liu, Y., Liu, Y., Liu, J., & Zhang, W. (2024). Transcriptome Analysis Reveals the Vital Role of ABA Plays in Drought Tolerance of the ABA-Insensitive Alfalfa (Medicago sativa L.). Agronomy, 14(3), 406. https://doi.org/10.3390/agronomy14030406







