Abstract
The phytohormone jasmonic acid (JA), a cyclopentane fatty acid, mediates plant responses to abiotic stresses. Abiotic stresses rapidly and dynamically affect JA metabolism and JA responses by upregulating the expression of genes involved in JA biosynthesis and signaling, indicating that JA has a crucial role in plant abiotic stress responses. The crucial role of JA has been demonstrated in many previous studies showing that JA response regulates various plant defense systems, such as removal of reactive oxygen species and accumulation of osmoprotectants. Furthermore, increasing evidence shows that plant tolerance to abiotic stresses is linked to the JA response, suggesting that abiotic stress tolerance can be improved by modulating JA responses. In this review, we briefly describe the JA biosynthetic and signaling pathways and summarize recent studies showing an essential role of JA in plant responses and tolerance to a variety of abiotic stresses, such as drought, cold, salt, and heavy metal stress. Additionally, we discuss JA crosstalk with another key stress hormone, abscisic acid, in plant abiotic stress responses.
1. Introduction
Abiotic stresses, including drought, cold, salt, and heavy metal stress, negatively affect plant growth and productivity by disrupting a variety of biological systems such as reactive oxygen species (ROS) homeostasis, the electron transport system, electrolyte maintenance, organelle integrity, and membrane lipid composition [1,2]. Furthermore, global climate change may increase the severity and frequency of abiotic stresses, threatening food production around the world. For example, it is expected that more than 50% of the world’s arable land will be exposed to drought stress by 2050 [3,4], indicating that development of crops with enhanced abiotic stress tolerance is critical for future sustainable crop production.
Plants have evolved various defenses against abiotic stresses, and the plant defenses include accumulation of osmoprotectants such as proline and trehalose, and activation of ROS scavenging systems [5,6]. Especially, abiotic stresses strongly promote the production and accumulation of ROS, and the increased ROS induces oxidative damages of cellular proteins and lipids, leading to dysfunction of proteins and loss of membrane integrity [1,7]. The activation of plant defense systems is tightly linked to transcription of stress-responsive genes, and cellular ROS activates transcription of stress-responsive antioxidant genes such as SUPEROXIDE DISMUTASE (SOD), CATALASE (CAT) and ASCORBATE PEROXIDASE (APX) through the MITOGEN-ACTIVATED PROTEIN KINASEs (MAPKs) signaling pathway [7,8,9]. The phytohormone jasmonic acid (JA) mediates plant responses to abiotic stresses by regulating transcription of stress-responsive genes including antioxidant genes under stress conditions [10,11,12,13,14]. The essential role of JA was supported by studies showing that the expression of JA biosynthetic and signaling genes is rapidly and dynamically regulated by abiotic stresses, and that overexpression of JA-responsive NAC, MYB, and WRKY transcription factors affects tolerance to abiotic stresses [15,16,17,18]. Therefore, JA-related pathways provide many potential targets for modulating abiotic stress tolerance.
Phytohormones interact with each other to modulate plant physiology and development. The isoprenoid phytohormone abscisic acid (ABA) mediates the plant response to abiotic stresses, and increasing evidence suggests that JA interacts with ABA to modulate the abiotic stress response and tolerance. For example, ABA biosynthetic genes and cellular ABA levels increased in response to JA, and conversely, expression of JA biosynthetic genes and cellular JA levels increased in response to ABA [19,20]. Moreover, the JA-signaling mutant jar1-1 showed a reduced response to ABA [21]. Moreover, the expression of many stress-responsive transcription factors is regulated both by JA and ABA [15,16,17,18]. Although the molecular and genetic mechanisms underlying the crosstalk between JA and ABA remain largely unrevealed, these findings partially support that JA interacts with ABA to modulate the plant response and tolerance to abiotic stresses.
Here, we briefly review the JA biosynthesis and signaling pathways, and discuss the role of JA in the plant response and tolerance to abiotic stresses, focusing on drought, cold, salt, and heavy metal stress. Furthermore, we discuss JA crosstalk with another key stress hormone, ABA, in plant abiotic stress responses.
2. Jasmonic Acid
2.1. JA Biosynthesis
JA is an essential phytohormone involved in the plant response to abiotic and biotic stresses, and JA and its derivatives are collectively referred to as jasmonates. JA was initially isolated as a methyl ester form of JA in Jasminum grandiflorum [22]. JA is classified as a cyclopentane fatty acid. JA is synthesized from α-linolenic acid, a major fatty acid of membranes in plant cells [23,24]. Briefly, JA biosynthesis is catalyzed by a variety of enzymes, such as lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), and oxophytodienoic acid reductase (OPR), which mediate the octadecanoid pathway (Figure 1) [25,26,27,28].
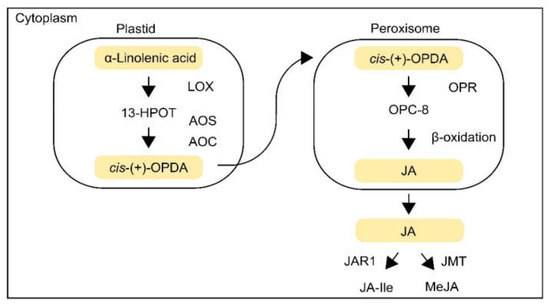
Figure 1.
JA biosynthesis. JA is biosynthesized from α-linolenic acid via the octadecanoid pathway. A variety of enzymes in plastids (lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC)), enzymes in in peroxisomes (OPDA reductase (OPR)), and in the cytoplasm (jasmonate-amido synthetase (JAR1) and jasmonate methyl transferase (JMT)) mediate JA biosynthesis in plants.
In plastids, α-linolenic acid is oxygenated to 13S-hydroperoxyoctadecatrienoic acid (13-HPOT) by the activity of LOX, and 13-HPOT is then converted to cis-(+)-12-oxophytodienoic acid (cis-(+)-OPDA) by AOS and AOC [26,28]. The OPDA is imported into peroxisomes. In peroxisomes, the OPDA is reduced by OPR, and is then shortened to form (+)-7-iso-JA in three rounds of β-oxidation [29]. The (+)-7-iso-JA is then transported to the cytoplasm and further metabolized into methyl jasmonate (MeJA) or JA-isoleucine (JA-Ile) conjugate by jasmonate methyl transferase (JMT) and jasmonate-amido synthetase (JAR1) [30,31]. Details of the JA biosynthetic pathway have been well reviewed in many previous studies [32,33,34,35]. Plants exposed to abiotic stresses increase cellular JA levels by activating the expression of JA biosynthetic genes, suggesting that JA is an essential hormone that mediates plant responses to abiotic stresses [36,37].
2.2. JA Signaling Pathway
Studies showing that exogenous JA treatment rapidly activates the expression of genes involved in plant defense suggested the existence of a JA-specific signaling pathway (Figure 2) [38,39,40]. Indeed, JA regulates the plant response and tolerance to abiotic stresses through the JA-specific signaling pathway, which is composed of the CORONATINE INSENSITIVE 1 (COI1) receptor, JASMONATE ZIM-DOMAIN PROTEIN (JAZ) repressors, and the JA-responsive transcription factor MYC2 [35,41]. JA-Ile interacts with the JA receptor COI1, which is an F-box protein and functions in E3-ubiquitin ligase-mediated proteolysis of JAZs. MYC2 is a key transcription factor in the JA signaling pathway. In 1996, jasmonate insensitive 1 (jin1) mutants, in which JA response is suppressed, were identified in Arabidopsis [42], and a study by Lorenzo et al. (2004) revealed that JIN1 encodes a MYC2 transcription factor [43]. MYC2 promotes the expression of JA-responsive genes by binding to the G-box (5′-CACGTG-3′) in their promoter regions, indicating that activation of MYC2 is essential for the expression of JA responsive genes and induction of the JA response in plants [44]. The COI1–JA-Ile interaction induces ubiquitin-mediated proteolysis of JAZs, which is followed by the release of the MYC2 transcription factor from the JAZ–MYC2 complex, and transcription of JA-responsive genes. The essential roles of MYC2 in the JA signaling pathway have been well demonstrated by knock-out and overexpression in transgenic plants, and a variety of MYC2 orthologs have been identified in crops [45,46,47,48,49]. MYC2 differentially regulates JA-dependent genes [50].
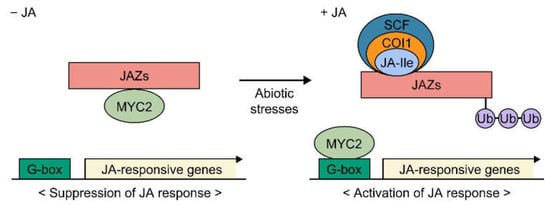
Figure 2.
JA signaling pathway. A schematic of the JA signaling pathway. In the absence of JA, MYC2 activity, which is responsible for transcription of JA-responsive genes, is repressed through direct interaction with JAZs. JA biosynthesis and accumulation induced by abiotic stresses provoke ubiquitin-mediated proteolysis of the JAZs, leading to activation of MYC2 and transcription of JA-responsive genes. SCF, Skp1/Cullin/F-box COI1 complex; Ub, Ubiquitin.
3. JA and Abiotic Stresses
Abiotic stresses induce a variety of cellular disorders such as ROS accumulation, electrolyte leakage, disruption of osmotic homeostasis, and chlorophyll degradation. JA response is closely linked to plant response and tolerance to abiotic stresses, and increasing evidence indicates that JA is deeply involved in the regulation of plant defense systems such as ROS removal and osmoprotectants production. In this review, we discussed the role of JA in abiotic stress response, and the potential of JA to improve abiotic stress tolerance, focusing on drought, cold, salt and heavy metal stress (Figure 3).
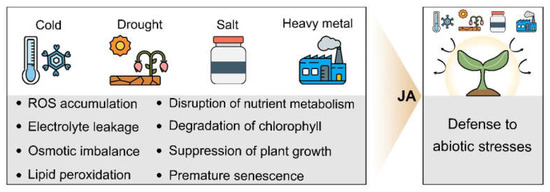
Figure 3.
Abiotic stresses and JA. A schematic of the abiotic stress-induced disorders in plants, and the involvement of JA in plant tolerance to the abiotic stresses such as cold, drought, salt and heavy metal stress.
3.1. JA and Cold Stress
Cold stress is a low-temperature stress including chilling (0–15 °C) and freezing (<0 °C) stress. When Arabidopsis was exposed to cold stress for 12 h, cellular JA levels increased by approximately 4-fold [51]. The expression of genes responsible for JA biosynthesis was upregulated in the cold stress-treated plants, suggesting that cold stress promotes JA production by activating the expression of JA biosynthetic genes. The relationship between cold stress and JA levels was also shown in other crops. Cold stress in rice activates the expression of JA biosynthetic genes such as OsLOX2, OsAOC, OsAOS1, OsAOS2, OsOPR1, and OsOPR7, and consequently, cellular JA levels increased [36]. Artemisia annua also accumulated higher JA levels in response to cold stress, and as expected, the expression of JA biosynthetic genes was activated by cold stress [52]. These results indicated that JA is involved in the plant response and tolerance to cold stress, and the result is that exogenous JA treatment enhanced plant tolerance to cold stress supported this. For example, JA treatment improved cold stress tolerance in orange (Citrus sinensis) [53]. JA-treated oranges displayed higher expression levels of antioxidant genes and lower levels of H2O2 compared with untreated control plants [53]. The effect of JA on cold stress tolerance was also reported in banana (Musa acuminata); JA-treated banana fruit exhibited an enhanced tolerance to cold stress compared with untreated fruit, and the JA treatment increased the expression of genes involved in cold-stress tolerance [54].
The involvement of JA in the plant response and tolerance to cold stress was further demonstrated by studies involving C-repeat-binding factors (CBFs). CBFs encode APETALA2/ETHYLENE-RESPONSIVE FACTOR (AP2/ERF)-type transcription factors, which induce the expression of cold-responsive genes [55,56,57]. Knock-out mutants that lack activity of CBF1 showed a suppressed tolerance to cold stress, whereas CBF1-overexpressing transgenic plants exhibited enhanced tolerance [58,59,60,61]. CBF1 expression is regulated by JA as well as cold stress. Increasing evidence indicates that the JA-dependent regulation of CBF expression is mediated by the physical interaction between JAZs and INDUCER OF CBF EXPRESSION (ICE) proteins. ICEs are basic helix-loop-helix (bHLH) transcription factors and promote the expression of CBFs through direct interaction with CBF promoters [62,63]. Furthermore, JAZs directly interact with ICEs and suppress their transcriptional activity [37]. This finding explains how JA promotes the expression of CBFs in response to cold stress; JA biosynthesis induced by cold stress activates ICEs by degrading JAZs, leading to mass transcription of CBFs.
The role of JA in the plant response and tolerance to cold stress was further supported by studies using knock-out mutants or transgenic plants with modulated JA biosynthesis and response. Knock-out mutants of AOS and LOX2 with defects in JA biosynthesis were hypersensitive to cold stress compared with wild-type plants [37]. The role of JA in cold stress tolerance was also supported by a study using glutamate-like receptors (GLRs) that regulate Ca2+ influx and signaling [64,65,66]. Cold stress increases intracellular Ca2+ levels by promoting Ca2+ influx [67]. Increased Ca2+ induces mass transcription of stress-responsive genes including JA biosynthetic and signaling genes by activating Ca2+-dependent protein kinases (CPKs/CDPKs), which are required for phosphorylation of stress-responsive transcription factors [68,69,70]. In Arabidopsis, overexpression of GLR1.2 and GLR1.3 promoted JA accumulation by activating the expression of JA biosynthetic genes. In addition, GLR1.2- and GLR1.3-overexpressing plants exhibited an increased expression of CBFs and displayed improved tolerance to cold stress. In contrast, glr1.2 and glr1.3 mutants with reduced expression of CBFs were hypersensitive to cold stress [65].
Similar to Arabidopsis, exogenous JA improved cold tolerance in apple by increasing the expression of cold-responsive genes including MdKIN1, MdRD29A, and MdCOR47. A novel B-box (BBX) transcription factor, MdBBX37, that binds MdJAZs and MdICE1 regulates the JA-dependent cold response in apple. The JA-responsive MdBBX37 positively regulates the expression of MdCBFs (which induce the expression of cold-responsive genes) by modulating the interaction between MdJAZs and MdICE1. Additionally, MdBBX37 directly regulates the expression of MdCBF1 and MdCBF4 by binding to their promoters. Consequently, the overexpression of MdBBX37 promoted the expression of cold-responsive genes, leading to improved cold tolerance. In contrast, antisense inhibition of MdBBX37 suppressed the expression of cold-responsive MdCBFs, leading to reduced cold tolerance. MdMYC2 interacts with MdJAZs to induce the expression of JA-responsive genes in apple [71]. Overexpression of MdMYC2 provoked the JA response and activated the expression of MdCBFs, leading to enhanced tolerance to cold stress [72]. However, the effect of MdMYC2 overexpression was nullified by overexpression of MdJAZ1 or MdJAZ4, which interact with MdMYC2 [58]. In tomato, SlF3HL is a cold-responsive gene that encodes 2-oxoglutarate Fe(II)-dependent oxygenase, which is involved in JA biosynthesis [71]. Overexpression of SlF3HL promoted JA biosynthesis and activated the JA response, whereas knock-down of SlF3HL reduced JA accumulation and the JA response. Analysis of electrolyte leakage and ROS accumulation in these plants showed that SlF3HL positively regulates cold tolerance in tomato.
3.2. JA and Drought Stress
Drought stress is a major abiotic stress affecting plant growth and productivity. Drought stress can alter membrane integrity and reduce turgor pressure, and can also reduce gas exchange by inducing the closure of stomata, leading to reduced photosynthetic efficiency [73,74,75]. Studies have shown that JA biosynthesis is promoted in response to drought stress, suggesting that JA is involved in the drought stress response. For example, JA levels in tomato (Solanum lycopersicum) roots exposed to drought for 12 h were approximately 10-fold higher than those in untreated roots, and an increase in cellular JA levels was also observed in tomato leaves [76]. Drought stress-induced JA accumulation was also reported in wheat (Triticum aestivum); cellular JA levels increased by approximately 5-fold in response to drought stress for 24 h [77]. Drought stress induced the expression of JA biosynthesis genes, such as LOX1, AOS1, AOC1, and OPR3, in wheat, indicating that drought stress promotes JA production by activating the expression of JA biosynthetic genes. Recent studies by Kim et al. (2017) and Ogawa et al. (2021) showed that drought stress triggers dynamic metabolic changes that produce acetate to activate JA biosynthesis and response, and exogenous acetate treatment improves drought tolerance [78,79]. These findings indicate a mechanistic connection between drought and JA response, supporting that JA is deeply involved in plant response and tolerance to drought stress.
The involvement of JA in the plant drought response was also shown by analyzing the effect of exogenous JA on drought tolerance. JA-treated soybean (Glycine max) exhibited improved tolerance to drought stress compared with the untreated control plants [80]. Drought induces cellular dehydration, osmotic imbalance, and ROS accumulation. Accumulation of osmoprotectants and removals of ROS are deeply involved in plant defense system to drought stress, and the results that an osmoprotectant proline regulates ROS homeostasis under abiotic stress conditions partially supported this [1,7,81,82]. JA increased accumulation of proline and reduced ROS levels in plants [5,83]. The effect of JA on proline and ROS levels was supported by the results showing that JA regulates transcriptional expression of genes involved in proline metabolism and ROS scavenging system [84,85,86,87]. The effect of exogenous JA on drought tolerance was also shown in other crops such as maize (Zea mays) and pearl millet (Pennisetum glaucum) [86,87]. JA-treated maize and pearl millet showed enhanced tolerance to drought stress by accumulating osmoprotectants such as proline, and by promoting expression of genes that mediate the removal of ROS [86,87].
JAZs are key repressors of the JA signaling pathway, and a study using knock-out or overexpression of JAZs further revealed the crucial role of JA in the plant response and tolerance to drought stress. Fu et al. (2017) showed that OsJAZ1 knock-out rice (Oryza sativa) mutants exhibited improved tolerance to drought stress, whereas OsJAZ1-overexpressing transgenic rice plants displayed reduced tolerance [88]. Knock-out of OsJAZ1 increased the plant survival rate by 8-fold, while overexpression of OsJAZ1 decreased the survival rate by approximately 4-fold compared with wild-type plants grown in drought stress conditions. Transcriptome analysis showed that overexpression of OsJAZ1 suppressed the expression of OsbHLH148 (encoding a key JA-responsive transcription factor) and drought-responsive genes such as OsDREB1A, OsDREB1B, SNAC1, and OsCCD1 [88,89,90,91]. OsbHLH148 is a key transcription factor responsible for the expression of JA-responsive genes in rice and directly interacts with OsJAZs, including OsJAZ1 [92]. Similar to the OsJAZ1 rice mutant, OsbHLH148-overexpressing transgenic plants had upregulated expression of OsDREB1A, B, C, E, and G, which are involved in drought stress tolerance, and as expected, drought stress tolerance was improved in the OsbHLH148-overexpressing transgenic plants. These findings suggested that drought tolerance can be improved by controlling the expression of JA-responsive genes, and several studies have reported success with this approach. Activation of the plant defense system in response to drought stress depends on transcription of stress-responsive genes, and JA-responsive transcription factors are deeply involved in the process [88,89,90,91]. Expression of Grapevine (Vitis amurensis) VaNAC17 is induced by JA and drought. Heterologous overexpression of the VaNAC17 in Arabidopsis increased the expression of genes involved in JA biosynthesis and response and ROS removal under drought conditions. Consequently, cellular JA levels in the VaNAC17-overexpressing transgenic plants increased under drought conditions, compared with wild-type plants while cellular ROS levels were reduced. When drought tolerance of the VaNAC17-overexpressing transgenic plants was tested by measuring the survival rate in drought conditions, the transgenic plants displayed approximately 8-fold higher survival rates than wild-type plants. This finding indicated that VaNAC17 improves drought tolerance by modulating the JA response. In addition, the result that expression of VaNAC17 is also regulated by another key stress hormone ABA suggested a potential interaction between JA and ABA in VaNAC17-mediated drought response [93]. The essential role of JA-responsive transcription factors in drought response and tolerance was also observed in IbMYB116 and GmTGA15. JA activates expression of sweet potato (Ipomoea batatas) IbMYB116 [94]. Heterologous overexpression of IbMYB116 in Arabidopsis upregulated the expression of JA biosynthetic genes, such as LOX, AOS, AOC, and OPR, and promoted JA accumulation and the JA response. The IbMYB116-overexpressing plants exhibited enhanced tolerance to drought stress compared with wild-type plants. In addition, the IbMYB116-expressing plants showed higher expression levels of genes encoding ROS scavenging enzymes and a lower level of hydrogen peroxide (H2O2) than wild-type plants. These results indicated that JA-responsive IbMYB116 promotes plant tolerance to drought stress by modulating the JA response. Another JA-responsive transcription factor, GmTGA15, showed a similar effect on drought tolerance in soybean [95]. Overexpression of GmTGA15 promoted the accumulation of proline and improved tolerance to drought stress in soybean.
3.3. JA and Salt Stress
Salt stress is caused by high concentrations of salts in the soil or water supply, which restricts water uptake and disturbs the osmotic balance, leading to a variety of physiological and developmental disorders, such as ROS accumulation, photosynthetic inhibition, membrane leakage, and growth suppression [96,97,98,99,100]. In Arabidopsis, the cellular JA level increased by 2.8-fold in response to a 24 h treatment with 100 mM NaCl, and the increased JA level activated the JA response. The increase in the JA level by salt stress was also shown in other plants [101]. Cellular JA levels in Medicago truncatula increased by 2.5-fold in response to a 1 h treatment of 200 mM NaCl [102]. Salt-treated M. truncatula displayed higher expression levels of the JA biosynthetic genes MtAOS1 and MtAOS2 than salt-untreated control plants. Similarly, salt stress induced the expression of JA biosynthetic genes and promoted JA accumulation in maize and sweet potato [103,104]. These results showed that JA is deeply involved in the plant response and tolerance to salt stress, and studies on the effect of exogenous JA on salt tolerance supported this. Salt-reduced plant growth and chlorophyll levels in soybean, almond (Prunus dulcis), and pepper (Capsicum annuum) [105,106,107], whereas exogenous JA treatment reduced the negative effect of the salt stress on plant growth and chlorophyll levels, indicating that JA promotes plant tolerance to salt stress.
The role of JA in the salt response and tolerance was also tested by genetic approaches using JA biosynthesis and signaling mutants. In Arabidopsis, LOX3 regulates the production of JA. In normal growth conditions, the germination rate was similar between the wild type and the LOX3 knock-out mutant (lox3). However, in salt stress conditions, the germination rate in the lox3 mutant was 2.5-fold lower than that in wild-type plants [108]. In addition, the lox3 mutant showed approximately 3-fold lower JA levels than wild-type plants under the salt stress conditions, indicating that JA regulates plant tolerance to salt stress. Similarly, a tomato JA-deficient mutant, def-1, was less tolerant to salt stress [109]. Nitrogen accumulation in the def-1 mutant was significantly lower than that in wild-type plants in salt stress conditions.
Several studies using overexpression systems also supported the role of JA in the plant salt response and tolerance. Heterologous overexpression of peanut (Arachis hypogaea) AhAOC, which is responsible for JA biosynthesis, increased cellular JA levels by 1.5-fold in rice [110]. The transgenic rice exhibited increased expression levels of stress-responsive genes such as OsLEA3, OsMYB2, and OsONAC045, and formed longer roots than wild-type plants under salt stress conditions [110]. This finding was consistent with the result that transgenic Arabidopsis heterologously overexpressing the maize JA biosynthetic gene ZmOPR1 showed improved salt tolerance compared with wild-type plants [111]. Similar to the overexpression of AhAOC and ZmOPR1, overexpression of Amur grape (Vitis amurensis) VaNAC26 improved salt stress tolerance in Arabidopsis [112]. The transgenic Arabidopsis plants heterologously overexpressing salt stress-responsive VaNAC26 showed an increased expression of genes responsible for JA biosynthesis and accumulated approximately 3-fold higher levels of JA than wild-type plants. These transgenic plants displayed around 4-fold higher survival rates than wild-type plants when grown under salt stress conditions.
3.4. JA and Heavy Metal Stress
Heavy metals include metals and metalloids that have been associated with potential toxicity or ecotoxicity, and heavy metal stress is caused by excessive accumulation and absorption of heavy metals. Increasing evidence suggests that JA is involved in the plant response to heavy metal stress. In Arabidopsis, cellular JA levels rapidly increased in response to treatment with copper (Cu) or cadmium (Cd) [113,114]. For example, JA levels increased by 4-fold and 6-fold in response to a 7 h treatment with 100 μM Cu or 100 μM Cd, respectively [115]. Heavy metal-induced JA production was also reported in crops such as pepper and rice. When pepper was exposed to 50 mg L−1 Cd for 48 h, cellular JA levels increased by approximately 2-fold [116]. Similarly, a 6 h treatment of 100 mM Cu increased JA levels by 9-fold in rice [117]. These results indicated that heavy metal stress promotes the accumulation of JA, and the finding that the expression of JA-biosynthetic or JA-responsive genes is activated by heavy metal stress supported this. The involvement of JA in heavy metal stress was also demonstrated in several studies analyzing the effect of exogenous JA on plants exposed to heavy metals. Cd reduced root growth and chlorophyll b levels in pepper, and exogenous JA treatment reduced these negative effects [116]. Lead (Pb) reduced the levels of chlorophyll a and carotenoids in Wolffia arrhiza, and an exogenous JA treatment recovered the Pb-induced reduction of chlorophyll a and carotenoids by reducing Pb absorption and accumulation [118,119].
Genetic approaches using JA biosynthetic and signaling mutants further demonstrated the role of JA in heavy metal stress. In Arabidopsis, the JA biosynthetic mutant aos was hypersensitive to Cd [120]. Cellular accumulation of Cd and the Cd-induced albino phenotype were more severe in the aos mutant than in wild-type plants. In another study, the Arabidopsis lox1-1 mutant, which lacks the activity of LOX1, was less tolerant to Cd than the wild type. In addition, Cd-induced accumulation of H2O2 was accelerated in the lox1-1 mutant [121]. Similar to the JA biosynthetic mutants, the JA signaling mutant jar1-1, in which the JA response is severely compromised, was also hypersensitive to heavy metal stress. Selenium (Se) reduces root growth in Arabidopsis, and the inhibitory effect of Se on root growth was more dramatic in the jar1-1 mutant than in the wild type [122]. The role of JA in heavy metal tolerance was also shown in crops such as tomato. The tomato spr2 mutant is a JA-deficient mutant [123]. When wild-type and spr2 plants were exposed to the same concentration of Cd, the spr2 mutant exhibited more severe phenotypes in Cd-induced growth inhibition and water loss than in wild-type control plants [124]. Unlike JA biosynthetic or signaling mutants, transgenic plants overexpressing JA biosynthetic genes with increased JA levels and response exhibited improved tolerance to heavy metal stress. For example, overexpression of cotton (Gossypium hirsutum) GhAOS activated JA biosynthesis and improved tolerance to heavy metal stress [125]. The survival rate of GhAOS-overexpressing transgenic plants under Cu-treated conditions was approximately 2-fold higher than that of wild-type plants. Similarly, overexpression of wheat TaAOS also improved plant tolerance to heavy metal stress caused by zinc (Zn) [126]. In Zn-treated conditions, the chlorophyll level in tobacco (Nicotiana tabacum) heterologously overexpressing TaAOS was around 3-fold higher than that in wild-type control plants. These findings showed that JA plays an essential role in plant defense to heavy metal stress, and suggested that heavy metal tolerance can be improved by modulating the JA response.
4. Crosstalk between JA and ABA
ABA is another key regulator determining the plant response and tolerance to abiotic stresses, and numerous studies reporting that ABA biosynthesis is activated by abiotic stresses, and plant tolerance to abiotic stresses is tightly linked to the ABA response support the essential role of ABA in stress tolerance [127,128,129]. Briefly, the biosynthesis of ABA includes the production of the ABA precursor xanthoxin in plastids and conversion of xanthoxin to ABA in the cytoplasm [129,130]. Production of xanthoxin is mediated by the enzymatic activity of 9-cis-epoxycarotenoid dioxygenase (NCED). NCED expression and NCED activity are tightly linked to ABA biosynthesis and the ABA response, indicating that NCED-mediated xanthoxin synthesis is a crucial step in ABA biosynthesis [128,131,132,133,134,135,136]. Cellular ABA activates the expression of ABA-responsive genes and provokes the ABA response through ABA-specific signaling pathway composed of ABA receptors, phosphatases, kinases, and transcription factors [137,138]. ABA signaling is initiated by the interaction between ABA and the ABA receptor PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL). The interaction activates the kinase activity of SnRK2, which is responsible for phosphorylation and activation of the ABA-responsive transcription factors (ABFs).
Increasing evidence suggests that JA interacts with ABA to modulate the plant response and tolerance to abiotic stresses. In soybean and tomato, ABA levels increased by approximately 8-fold and 2.3-fold in response to a 24 hr treatment with JA, respectively, and ABA accumulation was promoted in citrus fruits in response to JA treatment [77,139]. These results indicated that JA promotes ABA biosynthesis, and the finding that expression of ABA biosynthetic genes is activated by JA supported this. Similarly, ABA affects JA biosynthesis and accumulation. In Arabidopsis, a 3 h treatment of ABA activated the expression of JA biosynthetic genes and increased JA levels by approximately 2-fold [131]. The promotion of JA accumulation by ABA treatment was also shown in rice [140]. These findings indicated that ABA promotes JA biosynthesis, and a recent study showing that ABA activated the biosynthesis of JA via a SAPK10–bZIP72–AOC pathway supported this [140]. Collectively, these studies suggest that JA and ABA synergistically interact to modulate the plant response and tolerance to abiotic stresses.
The synergistic interaction between JA and ABA has also been investigated by genetic approaches. ABA levels in JA biosynthetic mutant aos were lower than those in wild-type plants under drought conditions, and JA levels in ABA biosynthetic mutant aba2 were lower than those in wild-type plants [141]. JA-responsive MYC2 is a key regulator of the JA signaling pathway. Knock-out Arabidopsis mutants that lack the activity of MYC2 (atmyc2) showed suppressed expression of JA-responsive genes and reduced tolerance to abiotic stresses [50]. The expression of ABA-responsive genes DEHYDRATION 22 (RD22) and ALCOHOL DEHYDROGENASE 1 (ADH1) was reduced in atmyc2 mutants. Together with the result that the expression of JA-responsive MYC2 is regulated by ABA [142], these findings suggested the interaction between JA and ABA in the abiotic stress response and tolerance. As described in the section of JA and drought stress, OsbHLH148, is a key transcription factor controlling JA and drought response in rice. Similar to the expression of AtMYC2, the expression of OsbHLH148 was upregulated by JA and ABA, and the upregulation was further increased by co-treatment of JA and ABA. Moreover, OsbHLH148 affects expression of ABA-responsive genes such as OsDREBs as well as JA-responsive genes such as OsJAZs [92]. Crosstalk of JA and ABA was also shown in stomatal closure. JA induces stomatal closure in wild-type plants. However, the effect of JA was not observed in ABA biosynthetic mutant aba2 [143], and a study using jin1/atmyc2 and ost1 mutant plants supported this [144,145]. The jin1 mutant was hyposensitive to ABA, and the effect of exogenous ABA on stomatal closure was reduced in the jin1 mutant compared with wild-type plants. In addition, exogenous ABA maintained the water content in wild-type plants under abiotic stress conditions, but the effect of ABA was reduced or lacking in the jin1 mutant [145]. OPEN STOMATA 1 (OST1) encoding a serine–threonine protein kinase is a key regulator of ABA-mediated stomatal closure [146]. Unlike wild-type plants, JA-induced stomatal closure was abolished or significantly impaired in two alleles of OST1 knock-out mutants (ost1-2 and srk2e) [144]. The interaction between JA and ABA in abiotic stress response was also observed in a study using NCED-overexpressing transgenic plants [147]. NCED encodes a 9-cis-epoxycarotenoid dioxygenase, a key ABA biosynthetic enzyme. Overexpression of the VaNCED1, which was isolated from a drought-tolerant cultivar of Vitis amurensis improved drought tolerance in a drought-sensitive cultivar of Vitis vinifera. The overexpression of VaNCED1 activated the expression of JA biosynthetic genes such as AOC and OPR3, and increased the accumulation of JA as well as ABA levels. Recent studies reported that JA-signaling proteins directly interact with ABA signaling proteins. For example, a study by Aleman et al. (2016) showed the interaction between MYC2 and ABA receptor PYL6, and a study by Sun et al. (2020) showed the interaction between JAZ1 and PYL4 [148,149]. ABIs function as key transcription factors in ABA signaling, and it was also shown that JAZs interact with ABIs such as ABI3 and ABI5 [131,150,151]. Although the molecular mechanisms underlying the crosstalk between JA and ABA remain largely unknown, these findings support the crosstalk between JA and ABA, and suggest that direct interaction between JA and ABA signaling components is deeply involved in the JA-ABA crosstalk.
5. Future Perspectives
Extensive studies have identified and characterized the components mediating phytohormone biosynthetic and signaling pathways, including JA, and have provided important clues to understand and modulate the hormonal responses involved in plant growth and physiology. Here, we described the biosynthetic and signaling pathways of JA, and reviewed studies showing the role of JA in the plant response and tolerance to abiotic stresses. The essential role of JA in the abiotic stress tolerance is partially supported by crosstalk with another key stress hormone, ABA, suggesting that plant tolerance to abiotic stresses can be improved by modulating JA biosynthetic and signaling processes.
Despite the essential role of JA in plant response and tolerance to abiotic stresses, the molecular mechanisms underlying this process remain largely unknown. A variety of cellular signaling pathways such as Ca2+ and ROS signaling are deeply involved in plant stress response. Increasing evidence has shown that JA response is affected by Ca2+ and ROS levels, and conversely, the cellular level of Ca2+ and ROS are also affected by JA response [21,68,69,152,153,154,155]. These findings suggest that JA signaling is connected with other stress signaling networks, proposing that the connection is deeply involved in the JA-mediated plant response and tolerance to abiotic stresses. Further genetic and molecular studies will expand our understanding of how plants respond and adapt to abiotic stress conditions, and how JA signaling is connected in other stress signaling networks in this process.
Author Contributions
Y.Y. and G.J. designed the review; H.K. and S.S. access information; H.K. and S.S. wrote the article with contributions of Y.Y. and G.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was carried out with the support of the BioGreen21 Agri-Tech Innovation Program (Project No. PJ01567301) Rural Development Administration, Republic of Korea, and the National Research Foundation of Korea Grant funded by the Korean Government (MOE) [NRF-2019R1A2C1007103]. This work was also supported by “Regional Innovation Strategy (RIS)” through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (MOE).
Conflicts of Interest
The authors declare no conflict of interest.
References
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [Green Version]
- Bajji, M.; Kinet, J.-M.; Lutts, S. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 2002, 36, 61–70. [Google Scholar] [CrossRef]
- Marthandan, V.; Geetha, R.; Kumutha, K.; Renganathan, V.G.; Karthikeyan, A.; Ramalingam, J. Seed Priming: A Feasible Strategy to Enhance Drought Tolerance in Crop Plants. Int. J. Mol. Sci. 2020, 21, 8258. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Sirhindi, G.; Mir, M.A.; Abd-Allah, E.F.; Ahmad, P.; Gucel, S. Jasmonic acid modulates the physio-biochemical attributes, antioxidant enzyme activity, and gene expression in Glycine max under nickel toxicity. Front. Plant Sci. 2016, 7, 591. [Google Scholar] [CrossRef] [Green Version]
- Kaur, G.; Asthir, B. Proline: A key player in plant abiotic stress tolerance. Biol. Plant. 2015, 59, 609–619. [Google Scholar] [CrossRef]
- Nadarajah, K.K. ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Azarabadi, S.; Abdollahi, H.; Torabi, M.; Salehi, Z.; Nasiri, J. ROS generation, oxidative burst and dynamic expression profiles of ROS-scavenging enzymes of superoxide dismutase (SOD), catalase (CAT) and ascorbate peroxidase (APX) in response to Erwinia amylovora in pear (Pyrus communis L.). Eur. J. Plant Pathol. 2017, 147, 279–294. [Google Scholar] [CrossRef]
- Jiang, Y.; Tang, X.; Zhou, B.; Sun, T.; Chen, H.; Zhao, X.; Wang, Y. The ROS-mediated pathway coupled with the MAPK-p38 signalling pathway and antioxidant system plays roles in the responses of Mytilus edulis haemocytes induced by BDE-47. Aquat. Toxicol. 2017, 187, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef] [Green Version]
- Farhangi-Abriz, S.; Ghassemi-Golezani, K. Jasmonates: Mechanisms and functions in abiotic stress tolerance of plants. Biocatal. Agric. Biotechnol. 2019, 20, 101210. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [Green Version]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef] [Green Version]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The Role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Shen, X.; Guo, X.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.; Li, T. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Kidokoro, S.; Ito, Y.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; Nakashima, K. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genom. 2010, 284, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Ollas, C.; Dodd, I.C. Physiological impacts of ABA–JA interactions under water-limitation. Plant Mol. Biol. 2016, 91, 641–650. [Google Scholar] [CrossRef]
- Suhita, D.; Raghavendra, A.S.; Kwak, J.M.; Vavasseur, A. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate-and abscisic acid-induced stomatal closure. Plant Physiol. 2004, 134, 1536–1545. [Google Scholar] [CrossRef] [Green Version]
- Demole, E.; Lederer, E.; Mercier, D. Isolement et détermination de la structure du jasmonate de méthyle, constituant odorant caractéristique de l’essence de jasmin. Helv. Chim. Acta 1962, 45, 675–685. [Google Scholar] [CrossRef]
- Jang, G.; Yoon, Y.; Choi, Y.D. Crosstalk with jasmonic acid integrates multiple responses in plant development. Int. J. Mol. Sci. 2020, 21, 305. [Google Scholar] [CrossRef] [Green Version]
- Cook, R.; Lupette, J.; Benning, C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 2021, 10, 706. [Google Scholar] [CrossRef]
- Wiszniewski, A.A.; Smith, S.M.; Bussell, J.D. Conservation of two lineages of peroxisomal (Type I) 3-ketoacyl-CoA thiolases in land plants, specialization of the genes in Brassicaceae, and characterization of their expression in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 6093–6103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariutto, M.; Duby, F.; Adam, A.; Bureau, C.; Fauconnier, M.-L.; Ongena, M.; Thonart, P.; Dommes, J. The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 2011, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Schilmiller, A.L.; Koo, A.J.; Howe, G.A. Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol. 2007, 143, 812–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenzel, I.; Hause, B.; Miersch, O.; Kurz, T.; Maucher, H.; Weichert, H.; Ziegler, J.; Feussner, I.; Wasternack, C. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Schilmiller, A.L.; Liu, G.; Lee, G.I.; Jayanty, S.; Sageman, C.; Vrebalov, J.; Giovannoni, J.J.; Yagi, K.; Kobayashi, Y. Role of β-oxidation in jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 2005, 17, 971–986. [Google Scholar] [CrossRef] [Green Version]
- Melotto, M.; Mecey, C.; Niu, Y.; Chung, H.S.; Katsir, L.; Yao, J.; Zeng, W.; Thines, B.; Staswick, P.; Browse, J. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine-and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008, 55, 979–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staswick, P.E.; Tiryaki, I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 2004, 16, 2117–2127. [Google Scholar] [CrossRef] [Green Version]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, P.; Rasool, S.; Gul, A.; Sheikh, S.A.; Akram, N.A.; Ashraf, M.; Kazi, A.; Gucel, S. Jasmonates: Multifunctional roles in stress tolerance. Front. Plant Sci. 2016, 7, 813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Strnad, M. Jasmonates: News on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. Int. J. Mol. Sci. 2018, 19, 2539. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of CBF expression–c-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wager, A. Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Front. Plant Sci. 2012, 3, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, M.; Kisseleva, L.; Sawa, S.; Furukawa, T.; Komatsu, S.; Koshiba, T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 2004, 45, 550–559. [Google Scholar] [CrossRef] [Green Version]
- Reymond, P.; Farmer, E.E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1998, 1, 404–411. [Google Scholar] [CrossRef]
- Ali, M.; Baek, K.-H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.; Bell, E.; Mullet, J.E. Two methyl jasmonate-insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 1996, 111, 525–531. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa, P.; Browse, J. The Arabidopsis JAZ2 promoter contains a G-Box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 2012, 53, 330–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Zhang, T.; Geng, S.; Scott, P.B.; Li, H.; Chen, S. Comparative proteomics and metabolomics of JAZ7-mediated drought tolerance in Arabidopsis. J. Proteom. 2019, 196, 81–91. [Google Scholar] [CrossRef]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef] [Green Version]
- Major, I.T.; Yoshida, Y.; Campos, M.L.; Kapali, G.; Xin, X.F.; Sugimoto, K.; de Oliveira Ferreira, D.; He, S.Y.; Howe, G.A. Regulation of growth–defense balance by the JASMONATE ZIM-DOMAIN (JAZ)-MYC transcriptional module. New Phytol. 2017, 215, 1533–1547. [Google Scholar] [CrossRef] [Green Version]
- Goossens, J.; Swinnen, G.; Vanden Bossche, R.; Pauwels, L.; Goossens, A. Change of a conserved amino acid in the MYC 2 and MYC 3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 2015, 206, 1229–1237. [Google Scholar] [CrossRef]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Guo, Z.; Li, H.; Wang, M.; Onac, E.; Zhou, J.; Xia, X.; Shi, K.; Yu, J.; Zhou, Y. Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol. 2016, 170, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Wang, H.; Chen, Y.; Zhu, S.; Chen, M.; Lan, X.; Chen, G.; Liao, Z. Cold stress improves the production of artemisinin depending on the increase in endogenous jasmonate. Biotechnol. Appl. Biochem. 2017, 64, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Habibi, F.; Ramezanian, A.; Rahemi, M.; Eshghi, S.; Guillén, F.; Serrano, M.; Valero, D. Postharvest treatments with γ-aminobutyric acid, methyl jasmonate, or methyl salicylate enhance chilling tolerance of blood orange fruit at prolonged cold storage. J. Sci. Food Agric. 2019, 99, 6408–6417. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.L.; Wang, J.N.; Shan, W.; Fan, J.G.; Kuang, J.F.; Wu, K.Q.; Li, X.P.; Chen, W.X.; He, F.Y.; Chen, J.Y. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2013, 36, 30–51. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef]
- Wang, P.; Chen, X.; Guo, Y.; Zheng, Y.; Yue, C.; Yang, J.; Ye, N. Identification of CBF transcription factors in tea plants and a survey of potential CBF target genes under low temperature. Int. J. Mol. Sci. 2019, 20, 5137. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.-K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBF s in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [Green Version]
- Fursova, O.V.; Pogorelko, G.V.; Tarasov, V.A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 2009, 429, 98–103. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.-H.; Hong, X.; Agarwal, M.; Zhu, J.-K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef]
- Zheng, Y.; Luo, L.; Wei, J.; Chen, Q.; Yang, Y.; Hu, X.; Kong, X. The glutamate receptors AtGLR1. 2 and AtGLR1. 3 increase cold tolerance by regulating jasmonate signaling in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 506, 895–900. [Google Scholar] [CrossRef]
- Kang, J.; Turano, F.J. The putative glutamate receptor 1.1 (AtGLR1. 1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 6872–6877. [Google Scholar] [CrossRef] [Green Version]
- Carpaneto, A.; Ivashikina, N.; Levchenko, V.; Krol, E.; Jeworutzki, E.; Zhu, J.-K.; Hedrich, R. Cold transiently activates calcium-permeable channels in Arabidopsis mesophyll cells. Plant Physiol. 2007, 143, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisahn, J.; Herde, O.; Willmitzer, L.; Peña-Cortés, H. Analysis of the transient increase in cytosolic Ca2+ during the action potential of higher plants with high temporal resolution: Requirement of Ca2+ transients for induction of jasmonic acid biosynthesis and PINII gene expression. Plant Cell Physiol. 2004, 45, 456–459. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.-P.; Guo, Y.; Sun, Y.; Sun, D.-Y.; Wang, X.-J. Influx of extracellular Ca2+ involved in jasmonic-acid-induced elevation of [Ca2+] cyt and JR1 expression in Arabidopsis thaliana. J. Plant Res. 2006, 119, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Yang, T.; Poovaiah, B. Calcium signaling-mediated plant response to cold stress. Int. J. Mol. Sci. 2018, 19, 3896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, T.; Wang, Y.; Wang, Q.; Dang, N.; Wang, L.; Liu, C.; Zhu, J.; Zhan, X. The tomato 2-oxoglutarate-dependent dioxygenase gene SlF3HL is critical for chilling stress tolerance. Hortic. Res. 2019, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple BT2 protein negatively regulates jasmonic acid-triggered leaf senescence by modulating the stability of MYC2 and JAZ2. Plant Cell Environ. 2021, 44, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, V.; Shahzad, B.; Ramakrishnan, M.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Yadav, P.; Khanna, K. Photosynthetic response of plants under different abiotic stresses: A review. J. Plant Growth Regul. 2020, 39, 509–531. [Google Scholar] [CrossRef]
- Sharma, A.; Zheng, B. Melatonin mediated regulation of drought stress: Physiological and molecular aspects. Plants 2019, 8, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Huang, Z. Effects of endogenous abscisic acid, jasmonic acid, polyamines, and polyamine oxidase activity in tomato seedlings under drought stress. Sci. Hortic. 2013, 159, 172–177. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Xie, J.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Abscisic acid and jasmonic acid are involved in drought priming-induced tolerance to drought in wheat. Crop. J. 2021, 9, 120–132. [Google Scholar] [CrossRef]
- Kim, J.-M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 2017, 3, 1–7. [Google Scholar]
- Ogawa, D.; Suzuki, Y.; Yokoo, T.; Katoh, E.; Teruya, M.; Muramatsu, M.; Ma, J.F.; Yoshida, Y.; Isaji, S.; Ogo, Y. Acetic-acid-induced jasmonate signaling in root enhances drought avoidance in rice. Sci. Rep. 2021, 11, 6280. [Google Scholar] [CrossRef]
- Riemann, M.; Dhakarey, R.; Hazman, M.; Miro, B.; Kohli, A.; Nick, P. Exploring Jasmonates in the Hormonal Network of Drought and Salinity Responses. Front. Plant Sci. 2015, 6, 1077. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 2020, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Cruz de Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef] [Green Version]
- Fugate, K.K.; Lafta, A.M.; Eide, J.D.; Li, G.; Lulai, E.C.; Olson, L.L.; Deckard, E.L.; Khan, M.F.; Finger, F.L. Methyl jasmonate alleviates drought stress in young sugar beet (Beta vulgaris L.) plants. J. Agron. Crop. Sci. 2018, 204, 566–576. [Google Scholar] [CrossRef]
- Anjum, S.; Wang, L.; Farooq, M.; Khan, I.; Xue, L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J. Agron. Crop. Sci. 2011, 197, 296–301. [Google Scholar] [CrossRef]
- Cao, S.; Cai, Y.; Yang, Z.; Zheng, Y. MeJA induces chilling tolerance in loquat fruit by regulating proline and γ-aminobutyric acid contents. Food Chem. 2012, 133, 1466–1470. [Google Scholar] [CrossRef]
- Awan, S.A.; Khan, I.; Rizwan, M.; Zhang, X.; Brestic, M.; Khan, A.; El-Sheikh, M.A.; Alyemeni, M.N.; Ali, S.; Huang, L. Exogenous abscisic acid and jasmonic acid restrain polyethylene glycol-induced drought by improving the growth and antioxidative enzyme activities in pearl millet. Physiol. Plant. 2021, 172, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Abdelgawad, Z.; Khalafaallah, A.A.; Abdallah, M. Impact of methyl jasmonate on antioxidant activity and some biochemical aspects of maize plant grown under water stress condition. Agric. Sci. 2014, 5, 1077. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Wu, H.; Ma, S.; Xiang, D.; Liu, R.; Xiong, L. OsJAZ1 attenuates drought resistance by regulating JA and ABA signaling in rice. Front. Plant Sci. 2017, 8, 2108. [Google Scholar] [CrossRef] [Green Version]
- Jing, P.; Zou, J.; Kong, L.; Hu, S.; Wang, B.; Yang, J.; Xie, G. OsCCD1, a novel small calcium-binding protein with one EF-hand motif, positively regulates osmotic and salt tolerance in rice. Plant Sci. 2016, 247, 104–114. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [Green Version]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Fang, L.; Zhu, Z.; Zhang, L.; Sun, X.; Wang, Y.; Wang, Q.; Li, S.; Xin, H. The transcription factor VaNAC17 from grapevine (Vitis amurensis) enhances drought tolerance by modulating jasmonic acid biosynthesis in transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 621–634. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, H.; He, S.; Zhai, H.; Zhao, N.; Xing, S.; Wei, Z.; Liu, Q. A novel sweetpotato transcription factor gene IbMYB116 enhances drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Fang, X.; Yuan, X.; Zhang, Y.; Li, H.; Zhou, Y.; Cui, X. Overexpression of Transcription Factor GmTGA15 Enhances Drought Tolerance in Transgenic Soybean Hairy Roots and Arabidopsis Plants. Agronomy 2021, 11, 170. [Google Scholar] [CrossRef]
- Li, J.; Essemine, J.; Shang, C.; Zhang, H.; Zhu, X.; Yu, J.; Chen, G.; Qu, M.; Sun, D. Combined proteomics and metabolism analysis unravels prominent roles of antioxidant system in the prevention of alfalfa (Medicago sativa L.) against salt stress. Int. J. Mol. Sci. 2020, 21, 909. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Ghoulam, C.; Foursy, A.; Fares, K. Effects of salt stress on growth, inorganic ions and proline accumulation in relation to osmotic adjustment in five sugar beet cultivars. Environ. Exp. Bot. 2002, 47, 39–50. [Google Scholar] [CrossRef]
- Wang, Y.; Nii, N. Changes in chlorophyll, ribulose bisphosphate carboxylase-oxygenase, glycine betaine content, photosynthesis and transpiration in Amaranthus tricolor leaves during salt stress. J. Hortic. Sci. Biotechnol. 2000, 75, 623–627. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [Green Version]
- Prerostova, S.; Dobrev, P.I.; Gaudinova, A.; Hosek, P.; Soudek, P.; Knirsch, V.; Vankova, R. Hormonal dynamics during salt stress responses of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea. Plant Sci. 2017, 264, 188–198. [Google Scholar] [CrossRef]
- De Domenico, S.; Taurino, M.; Gallo, A.; Poltronieri, P.; Pastor, V.; Flors, V.; Santino, A. Oxylipin dynamics in Medicago truncatula in response to salt and wounding stresses. Physiol. Plant. 2019, 165, 198–208. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Zhai, H.; Li, Y.; Wang, X.; Liu, Q.; He, S. Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci. Rep. 2017, 7, 40819. [Google Scholar] [CrossRef] [Green Version]
- Shahzad, A.; Pitann, B.; Ali, H.; Qayyum, M.; Fatima, A.; Bakhat, H. Maize genotypes differing in salt resistance vary in jasmonic acid accumulation during the first phase of salt stress. J. Agron. Crop. Sci. 2015, 201, 443–451. [Google Scholar] [CrossRef]
- Tavallali, V.; Karimi, S. Methyl jasmonate enhances salt tolerance of almond rootstocks by regulating endogenous phytohormones, antioxidant activity and gas-exchange. J. Plant Physiol. 2019, 234, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Rezai, S.; Orojloo, M.; Bidabadi, S.S.; Soleimanzadeh, M. Possible Role of Methyl Jasmonate in Protection to NaCl-Induced Salt Stress in Pepper cv. “Green Hashemi”. Int. J. Agric. Crop. Sci. 2013, 6, 1235. [Google Scholar]
- Yoon, J.Y.; Hamayun, M.; Lee, S.-K.; Lee, I.-J. Methyl jasmonate alleviated salinity stress in soybean. J. Crop. Sci. Biotechnol. 2009, 12, 63–68. [Google Scholar] [CrossRef]
- Ding, H.; Lai, J.; Wu, Q.; Zhang, S.; Chen, L.; Dai, Y.-S.; Wang, C.; Du, J.; Xiao, S.; Yang, C. Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response. Plant Sci. 2016, 244, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Abouelsaad, I.; Renault, S. Enhanced oxidative stress in the jasmonic acid-deficient tomato mutant def-1 exposed to NaCl stress. J. Plant Physiol. 2018, 226, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, Y.; Wang, S.; Li, H. Cloning and characterization of peanut allene oxide cyclase gene involved in salt-stressed responses. Genet. Mol. Res. 2015, 14, 2331–2340. [Google Scholar] [CrossRef]
- Gu, D.; Liu, X.; Wang, M.; Zheng, J.; Hou, W.; Wang, G.; Wang, J. Overexpression of ZmOPR1 in Arabidopsis enhanced the tolerance to osmotic and salt stress during seed germination. Plant Sci. 2008, 174, 124–130. [Google Scholar] [CrossRef]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojuederie, O.B.; Babalola, O.O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, C.; Oliver, D.J. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 1998, 10, 1539–1550. [Google Scholar] [CrossRef] [Green Version]
- Maksymiec, W.; Wianowska, D.; Dawidowicz, A.L.; Radkiewicz, S.; Mardarowicz, M.; Krupa, Z. The level of jasmonic acid in Arabidopsis thaliana and Phaseolus coccineus plants under heavy metal stress. J. Plant Physiol. 2005, 162, 1338–1346. [Google Scholar] [CrossRef]
- Yan, Z.; Chen, J.; Li, X. Methyl jasmonate as modulator of Cd toxicity in Capsicum frutescens var. fasciculatum seedlings. Ecotoxicol. Environ. Saf. 2013, 98, 203–209. [Google Scholar] [CrossRef]
- Rakwal, R.; Tamogami, S.; Kodama, O. Role of jasmonic acid as a signaling molecule in copper chloride-elicited rice phytoalexin production. Biosci. Biotechnol. Biochem. 1996, 60, 1046–1048. [Google Scholar] [CrossRef]
- Bali, S.; Jamwal, V.L.; Kaur, P.; Kohli, S.K.; Ohri, P.; Gandhi, S.G.; Bhardwaj, R.; Al-Huqail, A.A.; Siddiqui, M.H.; Ahmad, P. Role of P-type ATPase metal transporters and plant immunity induced by jasmonic acid against Lead (Pb) toxicity in tomato. Ecotoxicol. Environ. Saf. 2019, 174, 283–294. [Google Scholar] [CrossRef]
- Piotrowska, A.; Bajguz, A.; Godlewska-Żyłkiewicz, B.; Czerpak, R.; Kamińska, M. Jasmonic acid as modulator of lead toxicity in aquatic plant Wolffia arrhiza (Lemnaceae). Environ. Exp. Bot. 2009, 66, 507–513. [Google Scholar] [CrossRef]
- Lei, G.J.; Sun, L.; Sun, Y.; Zhu, X.F.; Li, G.X.; Zheng, S.J. Jasmonic acid alleviates cadmium toxicity in Arabidopsis via suppression of cadmium uptake and translocation. J. Integr. Plant Biol. 2020, 62, 218–227. [Google Scholar] [CrossRef]
- Keunen, E.; Remans, T.; Opdenakker, K.; Jozefczak, M.; Gielen, H.; Guisez, Y.; Vangronsveld, J.; Cuypers, A. A mutant of the Arabidopsis thaliana LIPOXYGENASE1 gene shows altered signalling and oxidative stress related responses after cadmium exposure. Plant Physiol. Biochem. 2013, 63, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Van Hoewyk, D.; Takahashi, H.; Inoue, E.; Hess, A.; Tamaoki, M.; Pilon-Smits, E.A. Transcriptome analyses give insights into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiol. Plant. 2008, 132, 236–253. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Hernández, C.; López, M.G.; Délano-Frier, J.P. Reduced levels of volatile emissions in jasmonate-deficient spr2 tomato mutants favour oviposition by insect herbivores. Plant Cell Environ. 2006, 29, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ma, Q.; Xu, X.; Li, G.; Hao, L. Tomato jasmonic acid-deficient mutant spr2 seedling response to cadmium stress. J. Plant Growth Regul. 2016, 35, 603–610. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, H.; Xin, Q. Improvement of copper tolerance of Arabidopsis by transgenic expression of an allene oxide cyclase gene, GhAOC1, in upland cotton (Gossypium hirsutum L.). Crop. J. 2015, 3, 343–352. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-H.; Wang, Y.-G.; Wang, S.-P.; Li, H.-J.; Xin, Q.-G. Improved zinc tolerance of tobacco by transgenic expression of an allene oxide synthase gene from hexaploid wheat. Acta Physiol. Plant. 2014, 36, 2433–2440. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Rodrigo, M.-J.; Alquezar, B.; Zacarías, L. Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J. Exp. Bot. 2006, 57, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Seiler, C.; Harshavardhan, V.T.; Rajesh, K.; Reddy, P.S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef] [Green Version]
- Burbidge, A.; Grieve, T.M.; Jackson, A.; Thompson, A.; McCarty, D.R.; Taylor, I.B. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 1999, 17, 427–431. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI 5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Priya, R.; Siva, R. Analysis of phylogenetic and functional diverge in plant nine-cis epoxycarotenoid dioxygenase gene family. J. Plant Res. 2015, 128, 519–534. [Google Scholar] [CrossRef]
- Sun, L.; Sun, Y.; Zhang, M.; Wang, L.; Ren, J.; Cui, M.; Wang, Y.; Ji, K.; Li, P.; Li, Q. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 2012, 158, 283–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Leng, P.; Zhang, G.; Li, X. Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 2009, 166, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef]
- Sirko, A.; Wawrzyńska, A.; Brzywczy, J.; Sieńko, M. Control of ABA Signaling and Crosstalk with Other Hormones by the Selective Degradation of Pathway Components. Int. J. Mol. Sci. 2021, 22, 4638. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- De Ollas, C.; Hernando, B.; Arbona, V.; Gómez-Cadenas, A. Jasmonic acid transient accumulation is needed for abscisic acid increase in citrus roots under drought stress conditions. Physiol. Plant. 2013, 147, 296–306. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336. [Google Scholar] [CrossRef]
- Brossa, R.; López-Carbonell, M.; Jubany-Marí, T.; Alegre, L. Interplay between abscisic acid and jasmonic acid and its role in water-oxidative stress in wild-type, ABA-deficient, JA-deficient, and ascorbate-deficient Arabidopsis plants. J. Plant Growth Regul. 2011, 30, 322–333. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of endogenous abscisic acid in methyl jasmonate-induced stomatal closure in Arabidopsis. Plant Physiol. 2011, 156, 430–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Adachi, Y.; Nakamura, Y.; Munemasa, S.; Mori, I.C.; Murata, Y. Involvement of OST1 protein kinase and PYR/PYL/RCAR receptors in methyl jasmonate-induced stomatal closure in Arabidopsis guard cells. Plant Cell Physiol. 2016, 57, 1779–1790. [Google Scholar] [CrossRef]
- Yastreb, T.; Kolupaev, Y.E.; Lugovaya, A.; Dmitriev, A. Formation of adaptive reactions in Arabidopsis thaliana wild-type and mutant jin1 plants under action of abscisic acid and salt stress. Cytol. Genet. 2017, 51, 325–330. [Google Scholar] [CrossRef]
- Acharya, B.R.; Jeon, B.W.; Zhang, W.; Assmann, S.M. Open Stomata 1 (OST 1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 2013, 200, 1049–1063. [Google Scholar] [CrossRef]
- He, R.; Zhuang, Y.; Cai, Y.; Agüero, C.B.; Liu, S.; Wu, J.; Deng, S.; Walker, M.A.; Lu, J.; Zhang, Y. Overexpression of 9-cis-epoxycarotenoid dioxygenase cisgene in grapevine increases drought tolerance and results in pleiotropic effects. Front. Plant Sci. 2018, 9, 970. [Google Scholar] [CrossRef]
- Aleman, F.; Yazaki, J.; Lee, M.; Takahashi, Y.; Kim, A.Y.; Li, Z.; Kinoshita, T.; Ecker, J.R.; Schroeder, J.I. An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: A putative link of ABA and JA signaling. Sci. Rep. 2016, 6, 28941. [Google Scholar] [CrossRef]
- Sun, K.; Xue, X.; Liu, N.; Zhu, Z.; Li, H. A point-to-point protein–protein interaction assay reveals the signaling interplays among plant hormones and environmental cues. Plant Direct 2020, 4, e00228. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665. [Google Scholar] [CrossRef]
- Pan, J.; Hu, Y.; Wang, H.; Guo, Q.; Chen, Y.; Howe, G.A.; Yu, D. Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 2020, 32, 3846–3865. [Google Scholar] [CrossRef]
- Maruta, T.; Inoue, T.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 2011, 180, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Vadassery, J.; Reichelt, M.; Hause, B.; Gershenzon, J.; Boland, W.; Mithöfer, A. CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 2012, 159, 1159–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Iwabuchi, M.; Matsui, M.; Hirochika, H.; Oda, K. Role of the rice transcription factor JAmyb in abiotic stress response. J. Plant Res. 2013, 126, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Fichman, Y.; Devireddy, A.R.; Sengupta, S.; Azad, R.K.; Mittler, R. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 13810–13820. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).