Improving Drought Tolerance in Mungbean (Vigna radiata L. Wilczek): Morpho-Physiological, Biochemical and Molecular Perspectives
Abstract
1. Introduction
2. Morpho-Physiological Trait Variations for Improving Drought Tolerance
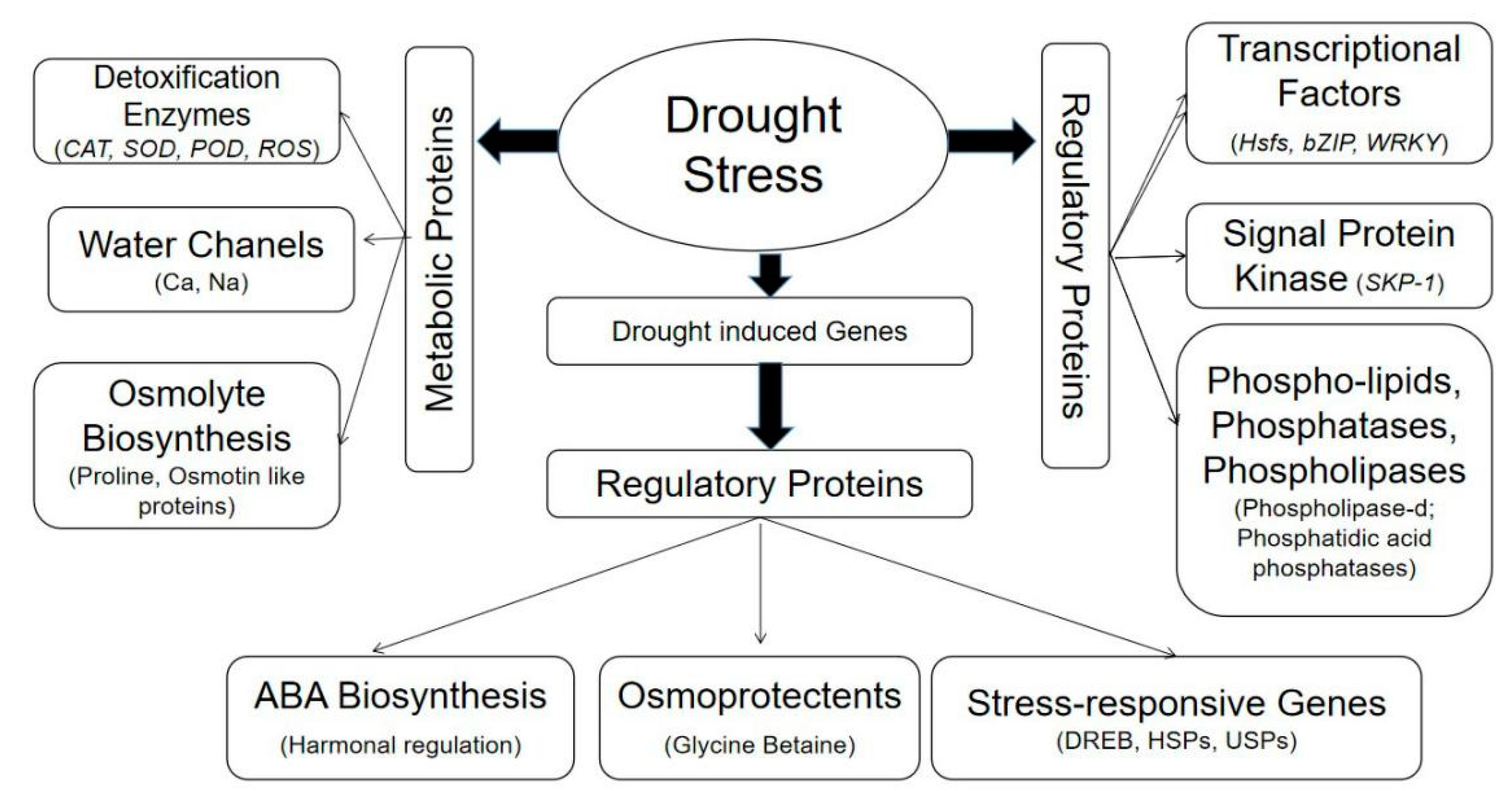
3. Biochemical Traits Modulating Drought Tolerance
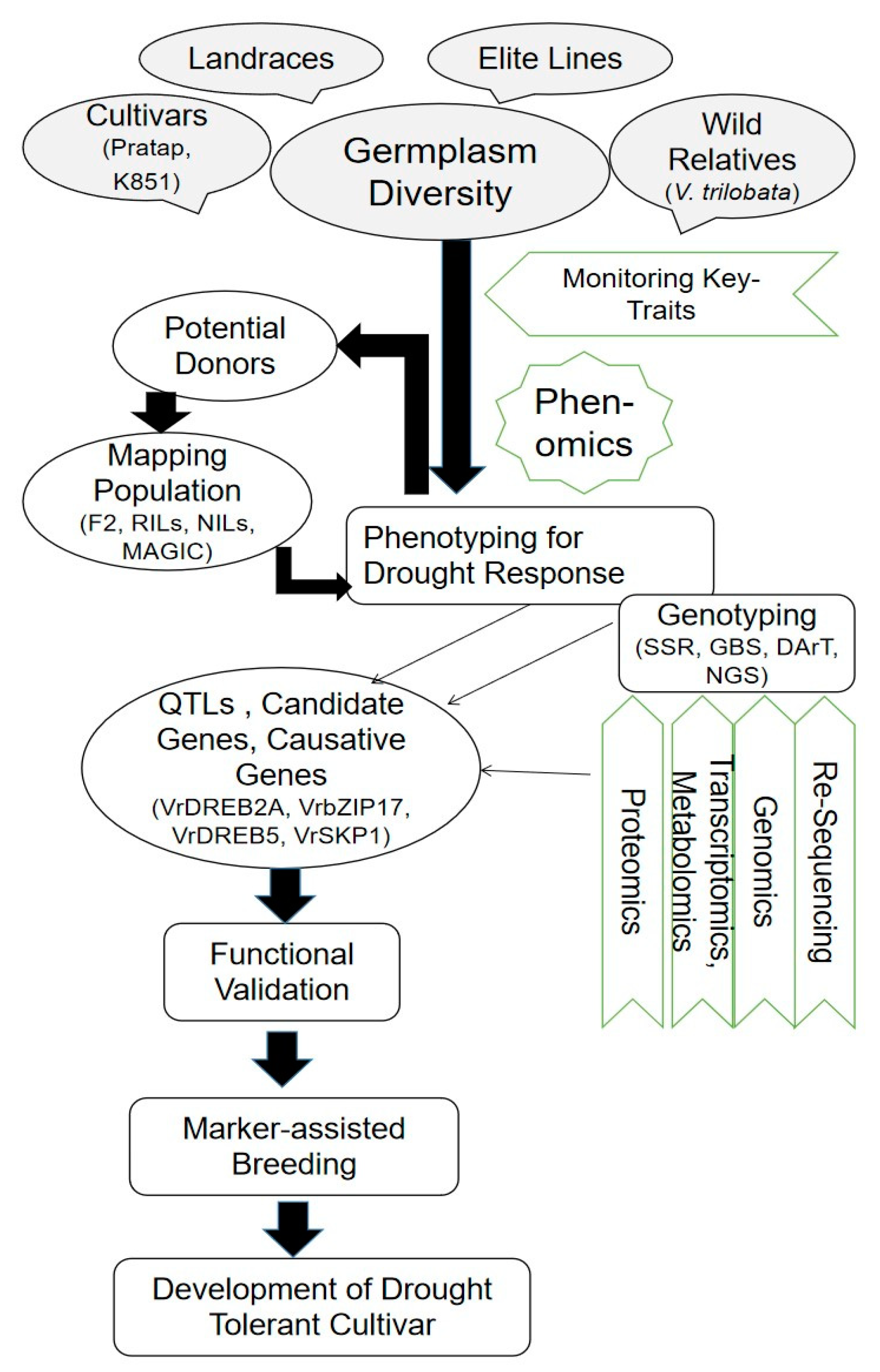
4. Multi-Omics Approaches to Understanding Drought Tolerance
4.1. Genomics Approaches
4.2. Exploring Gene Families and Transcriptional Factors as Drought-Responsive Markers
4.3. The Role of Long Non-Coding RNA (lncRNAs) and Micro-RNA (miRNAs) in Drought Stress
5. Conclusions and Future Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nair, R.M.; Pandey, A.K.; War, A.R.; Hanumantharao, B.; Shwe, T.; Alam, A.; Pratap, A.; Malik, S.R.; Karimi, R.; Mbeyagala, E.K.; et al. Biotic and abiotic constraints in mungbean production—Progress in genetic improvement. Front. Plant Sci. 2019, 10, 1340. [Google Scholar] [CrossRef]
- Pratap, A.; Douglas, C.; Prajapati, U.; Kumari, G.; War, A.R.; Tomar, R.; Pandey, A.K.; Dubey, S. Breeding progress and future challenges: Biotic stresses. In The Mungbean Genome, Compendium of Plant Genomes; Springer: Cham, Switzerland, 2020; pp. 55–80. [Google Scholar]
- Singh, D.P.; Singh, B.B.; Pratap, A. Genetic improvement of mungbean and urdbean and their role in enhancing pulse production in India. Indian J. Genet. Plant Breed. 2016, 76, 550–567. [Google Scholar] [CrossRef]
- Nair, R.M.; Schafleitner, R.; Kenyon, L.; Srinivasan, R.; Easdown, W.; Ebert, A.W.; Hanson, P. Genetic improvement of mungbean. SABRAO J. Breed. Genet. 2012, 44, 177–190. [Google Scholar]
- Allito, B.B.; Nana, E.-M.; Alemneh, A.A. Rhizobia strain and legume genome interaction effects on nitrogen fixation and yield of grain legume: A review. Mol. Soil Biol. 2015, 6, 1–6. [Google Scholar] [CrossRef]
- Lindström, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Favero, V.O.; Carvalho, R.H.; Motta, V.M.; Leite, A.B.C.; Coelho, M.R.R.; Xavier, G.R.; Rumjanek, N.G.; Urquiaga, S. Bradyrhizobium as the only rhizobial inhabitant of mung bean (Vigna radiata) nodules in tropical soils: A strategy based on microbiome for improving biological nitrogen fixation using bio-products. Front. Plant Sci. 2021, 11, 2186. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, E.; Yuan, C.; Zhang, L.; Yousef, A.E. Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl. Environ. Microbiol. 2012, 78, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.M.; Thavarajah, P.; Giri, R.R.; Ledesma, D.; Yang, R.-Y.; Hanson, P.; Easdown, W.; Hughes, J.D. Mineral and phenolic concentrations of mungbean [Vigna radiata (L.) R. Wilczek var. radiata] grown in semi-arid tropical India. J. Food Compos. Anal. 2015, 39, 23–32. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef]
- Arnoldi, A.; Zanoni, C.; Lammi, C.; Boschin, G. The role of grain legumes in the prevention of hypercholesterolemia and hypertension. CRC Crit. Rev. Plant Sci. 2015, 34, 144–168. [Google Scholar] [CrossRef]
- Ebert, A.W. Potential of underutilized traditional vegetables and legume crops to contribute to food and nutritional security, income and more sustainable production systems. Sustainability 2014, 6, 319–335. [Google Scholar] [CrossRef]
- Boelt, B.; Julier, B.; Karagić, Đ.; Hampton, J. Legume seed production meeting market requirements and economic impacts. CRC Crit. Rev. Plant Sci. 2015, 34, 412–427. [Google Scholar] [CrossRef]
- Basu, P.S.; Pratap, A.; Gupta, S.; Sharma, K.; Tomar, R.; Singh, N.P. Physiological traits for shortening crop duration and improving productivity of greengram (Vigna radiata L. Wilczek) under high temperature. Front. Plant Sci. 2019, 10, 1508. [Google Scholar] [CrossRef]
- Nair, R.; Schreinemachers, P. Global status and economic importance of mungbean. In The Mungbean Genome, Compendium of Plant Genomes; Springer: Cham, Switzerland, 2020; pp. 1–8. [Google Scholar]
- Bangar, P.; Chaudhury, A.; Tiwari, B.; Kumar, S.; Kumari, R.; Bhat, K.V. Morphophysiological and biochemical response of mungbean [Vigna radiata (L.) Wilczek] varieties at different developmental stages under drought stress. Turk. J. Biol. 2019, 43, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Baroowa, B.; Gogoi, N. Effect of induced drought on different growth and biochemical attributes of black gram (Vigna mungo L.) and green gram (Vigna radiata L.). J. Environ. Res. Dev. 2012, 6, 584–593. [Google Scholar]
- Baroowa, B.; Gogoi, N. Biochemical changes in two Vigna spp. during drought and subsequent recovery. Indian J. Plant Physiol. 2013, 18, 319–325. [Google Scholar] [CrossRef]
- Baroowa, B.; Gogoi, N.; Farooq, M. Changes in physiological, biochemical and antioxidant enzyme activities of green gram (Vigna radiata L.) genotypes under drought. Acta Physiol. Plant. 2016, 38, 219. [Google Scholar] [CrossRef]
- Maheswari, M.; Tekula, V.L.; Yellisetty, V.; Sarkar, B.; Yadav, S.K.; Singh, J.; Babu, G.S.; Kumar, A.; Amirineni, S.; Narayana, J.; et al. Functional mechanisms of drought tolerance in maize through phenotyping and genotyping under well watered and water stressed conditions. Eur. J. Agron. 2016, 79, 43–57. [Google Scholar] [CrossRef]
- Hussain, M.; Malik, M.A.; Farooq, M.; Ashraf, M.Y.; Cheema, M.A. Improving drought tolerance by exogenous application of glycinebetaine and salicylic acid in sunflower. J. Agron. Crop Sci. 2008, 194, 193–199. [Google Scholar] [CrossRef]
- Taiz, L.; Zeiger, E. Plant Physiology, 5th ed.; Sinauer Associates: Sunderland, MA, USA, 2010; p. 464. [Google Scholar]
- Zhou, R.; Kong, L.; Wu, Z.; Rosenqvist, E.; Wang, Y.; Zhao, L.; Zhao, T.; Ottosen, C. Physiological response of tomatoes at drought, heat and their combination followed by recovery. Physiol. Plant. 2019, 165, 144–154. [Google Scholar] [CrossRef]
- Zare, M.; Dehghani, B.; Alizadeh, O.; Azarpanah, A. The evaluation of various agronomic traits of mungbean (Vigna radiata L.) genotypes under drought stress and non-stress conditions. Int. J. Farming Allied Sci. 2013, 2, 764–770. [Google Scholar]
- Ranawake, A.L.; Dahanayaka, N.; Amarasingha, U.G.S.; Rodrigo, W.; Rodrigo, U.T.D. Effect of water stress on growth and yield of mung bean (Vigna radiata L.). Trop. Agric. Res. Ext. 2011, 14, 76–79. [Google Scholar] [CrossRef]
- Singh, C.M.; Mishra, S.B.; Pandey, A.; Arya, M. Morphological characterization and discriminant function analysis in mungbean [Vigna radiata (L.) Wilczek] germplasm. Electron. J. Plant Breed. 2014, 5, 87–96. [Google Scholar]
- Mehandi, S.; Singh, I.P.; Bohra, A.; Singh, C.M. Multivariate analysis in green gram [Vigna radiata (L.) Wilczek]. Legum. Res. An Int. J. 2015, 38, 758–762. [Google Scholar] [CrossRef]
- Singh, C.M.; Mishra, S.B.; Pandey, A.; Kumari, K. Branching pattern and harvest index as important selection criteria for improvement of mungbean [Vigna radiata (L.) Wilczek]. Madras Agric. J. 2015, 102, 1–5. [Google Scholar]
- Singh, C.M.; Singh, A.K.; Mishra, S.B.; Pandey, A. Generation mean analysis to estimate the genetic parameters for yield improvement and inheritance of seed colour and lusture in mungbean [Vigna radiata (L.) Wilczek]. Legum. Res. Int. J. 2016, 39, 494–501. [Google Scholar]
- Khan, M.B.; Hussain, M.; Raza, A.; Farooq, S.; Jabran, K. Seed priming with CaCl2 and ridge planting for improved drought resistance in maize. Turk. J. Agric. For. 2015, 39, 193–203. [Google Scholar] [CrossRef]
- Da Silva, E.C.; Nogueira, R.; da Silva, M.A.; de Albuquerque, M.B. Drought stress and plant nutrition. Plant Stress 2011, 5, 32–41. [Google Scholar]
- Bista, D.R.; Heckathorn, S.A.; Jayawardena, D.M.; Mishra, S.; Boldt, J.K. Effects of drought on nutrient uptake and the levels of nutrient-uptake proteins in roots of drought-sensitive and-tolerant grasses. Plants 2018, 7, 28. [Google Scholar] [CrossRef]
- Levitt, J. Chilling, Freezing, and High Temperature Stresses. In Responses of Plants to Environmental Stress; Academic Press: New York, NY, USA, 1980; Volume 2, pp. 23–445. [Google Scholar]
- Singh, P.; Mishra, A.K.; Singh, C.M. Genome-wide identification and characterization of Lectin receptor-like kinase (LecRLK) genes in mungbean (Vigna radiata L. Wilczek). J. Appl. Genet. 2021, 62, 223–234. [Google Scholar] [CrossRef]
- Saima, S.; Li, G.; Wu, G. Effects of drought stress on hybrids of Vigna radiata at germination stage. Acta Biol. Hung. 2018, 69, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Islam, M.S.; Sarker, B.C.; Alam, A.; Akhter, M.; Alam, M.J.; Saneoka, H.; Erman, M.; Sabagh, A.E.L. Assessment of water stress tolerance in mungbean induced by polyethyline glycol. bioRxiv 2019, 872663. [Google Scholar] [CrossRef]
- Imtiaz, A.A.; Shahriar, S.A.; Baque, M.A.; Eaty, M.N.K.; Falguni, M.R. Screening of mungbean genotypes under polyethylene glycol (PEG) induced drought stress condition. Annu. Res. Rev. Biol. 2020, 35, 1–12. [Google Scholar] [CrossRef]
- Yadav, S.K.; Lakshmi, N.J.; Singh, V.; Patil, A.; Tiwari, Y.K.; Nagendram, E.; Sathish, P.; Vanaja, M.; Maheswari, M.; Venkateswarlu, B. In vitro screening of Vigna mungo genotypes for PEG induced moisture deficit stress. Indian J. Plant Physiol. 2013, 18, 55–60. [Google Scholar] [CrossRef]
- Ali, Q.; Javed, M.T.; Noman, A.; Haider, M.Z.; Waseem, M.; Iqbal, N.; Waseem, M.; Shah, M.S.; Shahzad, F.; Perveen, R. Assessment of drought tolerance in mung bean cultivars/lines as depicted by the activities of germination enzymes, seedling’s antioxidative potential and nutrient acquisition. Arch. Agron. Soil Sci. 2018, 64, 84–102. [Google Scholar] [CrossRef]
- Carvalho, M.; Matos, M.; Castro, I.; Monteiro, E.; Rosa, E.; Lino-Neto, T.; Carnide, V. Screening of worldwide cowpea collection to drought tolerant at a germination stage. Sci. Hortic. 2019, 247, 107–115. [Google Scholar] [CrossRef]
- Kumar, R.R.; Karajol, K.; Naik, G.R. Effect of polyethylene glycol induced water stress on physiological and biochemical responses in pigeonpea (Cajanus cajan L. Millsp.). Recent Res. Sci. Technol. 2011, 3, 148–152. [Google Scholar]
- Rajamanickam, V.; Ravichandran, L.; Parasuraman, B. Assessment of soybean genotypes for PEG induced drought tolerance at germination and seedling level. Madras Agric. J. 2018, 105, 1–6. [Google Scholar]
- Khazayi, H.; Kafi, M.; Masumi, A. Physiological effects of stress induced by polyethylene glycol on germination of chickpea genotypes. J. Agron. Res. Iran 2008, 2, 453. [Google Scholar]
- Dutta, P.; Bera, A.K. Screening of mungbean genotypes for drought tolerance. Legum. Res. Int. J. 2008, 31, 145–148. [Google Scholar]
- Bibi, A.; Sadaqat, H.A.; Akram, H.M.; Mohammed, M.I. Physiological markers for screening sorghum (Sorghum bicolor) germplasm under water stress condition. Int. J. Agric. Biol. 2010, 12, 451–455. [Google Scholar]
- Ali, M.A.; Abbas, A.; Awan, S.I.; Jabran, K.; Gardezi, S.D.A. Correlated response of various morpho-physiological characters with grain yield in sorghum landraces at different growth phases. J. Anim. Plant Sci. 2011, 21, 671–679. [Google Scholar]
- Rajendran, R.A.; Muthiah, A.R.; Manickam, A.; Shanmugasundaram, P.; Joel, A.J. Indices of drought tolerance in sorghum (Sorghum bicolor L. Moench) genotypes at early stages of plant growth. Res. J. Agric. Biol. Sci. 2011, 7, 42–46. [Google Scholar]
- Aslam, M.; Maqbool, M.A.; Zaman, Q.U.; Latif, M.Z.; Ahmad, R.M. Responses of mungbean genotypes to drought stress at early growth stages. Int. J. Basic Appl. Sci. 2013, 13, 22–27. [Google Scholar]
- Jincya, M.; Prasad, V.; Jeyakumara, P.; Senthila, A.; Manivannanb, N. Evaluation of green gram genotypes for drought tolerance by PEG (polyethylene glycol) induced drought stress at seedling stage. Legum. Res. Int. J. 2021, 44, 684–691. [Google Scholar] [CrossRef]
- Biradar, K.S.; Salimath, P.M.; Ravikumar, R.L. Genetic studies in greengram and association analysis. Karnataka J. Agric. Sci. 2010, 20, 843–844. [Google Scholar]
- Moradi, A.; Ahmadi, A.; Hossain Zadeh, A. The effects of different timings and severity of drought stress on gas exchange parameters of mungbean. Desert 2008, 13, 59–66. [Google Scholar]
- Sadeghipour, O. The influence of water stress on biomass and harvest index in three mung bean (Vigna radiata (L.) R. Wilczek) cultivars. Asian J. Plant Sci. 2009, 8, 245–249. [Google Scholar] [CrossRef]
- Allahmoradi, P.; Ghobadi, M.; Taherabadi, S.; Taherabadi, S. Physiological aspects of mungbean (Vigna radiata L.) in response to drought stress. In Proceedings of the International Conference on Food Engineering and Biotechnology (IPCBEE) 9, Bangkok, Thailand, 7–9 May 2011; IACSIT Press: Singapore, 2011; pp. 272–275. [Google Scholar]
- Swathi, L. Genetic diversity analysis using morpho-physiological and molecular markers for breeding yield and drought tolerance in mungbean (Vigna radiata (L.) Wilczek). M.Sc. (Ag.) Thesis, Acharya N.G. Ranga Agricultural University, Hyderabad, India, 2013. [Google Scholar]
- Paramesh, M.; Reddy, D.M.; Priya, M.S.; Sumathi, P.; Sudhakar, P.; Reddy, K.H.P. GT biplot analysis for yield and drought related traits in mung bean (Vigna radiata L. Wilczek). Electron. J. Plant Breed. 2016, 7, 538–543. [Google Scholar] [CrossRef]
- Prakash, M.; Sunilkumar, B.; Sathiyanarayanan, G.; Gokulakrishnan, J. Screening for drought tolerance in mungbean. Legum. Res. Int. J. 2017, 40, 423–428. [Google Scholar] [CrossRef]
- Kumar, B.S.; Sathiyanarayanan, G.; Prakash, M. Influence of drought stress on root distribution in mungbean (Vigna radiata (L.) Wilczek). Int. J. Trop. Agric. 2015, 33, 837–843. [Google Scholar]
- Wang, L.F.; Jing, W.U.; Jing, R.L.; Cheng, X.Z.; Wang, S.M. Drought resistance identification of mungbean germplasm resources at seedlings stage. Acta Agron Sin 2015, 41, 145–153. [Google Scholar] [CrossRef]
- Dutta, P.; Bandopadhyay, P.; Bera, A.K. Identification of leaf based physiological markers for drought susceptibility during early seedling development of mungbean. Am. J. Plant Sci. 2016, 7, 1921–1936. [Google Scholar] [CrossRef]
- Kumari, S.; Vaishnav, A.; Jain, S.; Varma, A.; Choudhary, D.K. Induced drought tolerance through wild and mutant bacterial strain Pseudomonas simiae in mung bean (Vigna radiata L.). World J. Microbiol. Biotechnol. 2016, 32, 4. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Kaur, J.; Bains, T.S. Screening of mungbean genotypes for drought tolerance using different water potential levels. J. Adv. Agric. Technol. 2017, 4, 159–164. [Google Scholar] [CrossRef]
- Swathi, L.; Reddy, D.M.; Sudhakar, P.; Vineela, V. Screening of mungbean (Vigna radiata L. Wilczek) genotypes against water stress mediated through polyethylene glycol. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2524–2531. [Google Scholar] [CrossRef]
- Sadeghipour, O. Polyamines protect mung bean [Vigna radiata (L.) Wilczek] plants against drought stress. Biol. Futur. 2019, 70, 71–78. [Google Scholar] [CrossRef]
- Monclus, R.; Dreyer, E.; Villar, M.; Delmotte, F.M.; Delay, D.; Petit, J.-M.; Barbaroux, C.; Le Thiec, D.; Bréchet, C.; Brignolas, F. Impact of drought on productivity and water use efficiency in 29 genotypes of Populus deltoides x Populus nigra. New Phytol. 2006, 169, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Abbate, P.E.; Dardanelli, J.L.; Cantarero, M.G.; Maturano, M.; Melchiori, R.J.M.; Suero, E.E. Climatic and water availability effects on water-use efficiency in wheat. Crop Sci. 2004, 44, 474–483. [Google Scholar] [CrossRef]
- Bowne, J.B.; Erwin, T.A.; Juttner, J.; Schnurbusch, T.; Langridge, P.; Bacic, A.; Roessner, U. Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol. Plant 2012, 5, 418–429. [Google Scholar] [CrossRef]
- Muir, C.D.; Thomas-Huebner, M. Constraint around Quarter-Power Allometric Scaling in Wild Tomatoes (Solanum sect. Lycopersicon; Solanaceae). Am. Nat. 2015, 186, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Drira, M.; Hanin, M.; Masmoudi, K.; Brini, F. Comparison of full-length and conserved segments of wheat dehydrin DHN-5 overexpressed in Arabidopsis thaliana showed different responses to abiotic and biotic stress. Funct. Plant Biol. 2016, 43, 1048–1060. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Jimenez-Berni, J.A.; Bovill, W.D.; Deery, D.M.; James, R.A. High-throughput phenotyping technologies allow accurate selection of stay-green. J. Exp. Bot. 2016, 67, 4919–4924. [Google Scholar] [CrossRef]
- Gurumurthy, S.; Sarkar, B.; Vanaja, M.; Lakshmi, J.; Yadav, S.K.; Maheswari, M. Morpho-physiological and biochemical changes in black gram (Vigna mungo L. Hepper) genotypes under drought stress at flowering stage. Acta Physiol. Plant. 2019, 41, 42. [Google Scholar] [CrossRef]
- Anjum, S.A.; Wang, L.C.; Farooq, M.; Hussain, M.; Xue, L.L.; Zou, C.M. Brassinolide application improves the drought tolerance in maize through modulation of enzymatic antioxidants and leaf gas exchange. J. Agron. Crop Sci. 2011, 197, 177–185. [Google Scholar] [CrossRef]
- Din, J.; Khan, S.U.; Ali, I.; Gurmani, A.R. Physiological and agronomic response of canola varieties to drought stress. J. Anim. Plant Sci. 2011, 21, 78–82. [Google Scholar]
- Valliyodan, B.; Ye, H.; Song, L.; Murphy, M.; Shannon, J.G.; Nguyen, H.T. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 2016, 68, 1835–1849. [Google Scholar] [CrossRef]
- Tawfik, K.M. Effect of water stress in addition to potassiomag application on mungbean. Aust. J. Basic Appl. Sci. 2008, 2, 42–52. [Google Scholar]
- Hossain, M.B.; Rahman, M.W.; Rahman, M.N.; Anwar, A.; Hossen, A. Effects of water stress on yield attributes and yield of different mungbean genotypes. Int. J. Sustain. Crop Prod. 2010, 5, 19–24. [Google Scholar]
- Baroowa, B.; Gogoi, N. Morpho-physiological and Yield responses of Black gram (Vigna mungo L.) and Green gram (Vigna radiata L.) genotypes under Drought at different Growth stages. Res. J. Recent Sci. 2016, 5, 43–50. [Google Scholar]
- Ghanbari, M.; Javan, S.M. Study the response of mung bean genotypes to drought stress by multivariate analysis. Int. J. Agric. Innov. Res. 2015, 3, 1298–1302. [Google Scholar]
- Fernandez, G.C.J. Effective selection criteria for assessing plant stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and other Food Crops in Temperature and Water Stress, Shanhua, Taiwan, 13–16 August 1992; pp. 257–270. [Google Scholar]
- Paramesh, M.D.M.; Reddy, M.; SHANTHI, P.; Sudhakar, P.; Narasimhulu, R. Analysis of genetic parameters for morphophysiological traits in mungbean (Vigna radiata L. Wilczek). Indian J. Plant Sci. 2014, 3, 1–6. [Google Scholar]
- Tripathy, S.K.; Mohanty, P.; Jena, M.; Dash, S.; Lenka, D.; Mishra, D.; Nayak, P.K.; Swain, D.; Ranjan, R.; Pradhan, K. Identification of seed storage protein markers for drought tolerance in mungbean. Res. Biotechnol. 2016, 7, 1–7. [Google Scholar] [CrossRef]
- Mandi, S.; Pal, A.K.; Nath, R.; Hembram, S. ROS scavenging and nitrate reductase enzyme activity in mungbean (Vigna radiata L. Wilczek) under drought stress. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1031–1039. [Google Scholar] [CrossRef]
- Fischer, R.A.; Maurer, R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978, 29, 897–912. [Google Scholar] [CrossRef]
- Rosielle, A.A.; Hamblin, J. Theoretical aspects of selection for yield in stress and non-stress environment 1. Crop Sci. 1981, 21, 943–946. [Google Scholar] [CrossRef]
- Fernandez, G.C.J.; Shanmugasundaram, S. The AVRDC mungbean improvement program: The past, present and future. In Mungbean, Proceedings of the Second International Symposium, Bangkok, Thailand, 16–20 November 1987; AVRDC-World Vegetable Center: Tainan City, Taiwan, 1988; pp. 58–70. [Google Scholar]
- Gavuzzi, P.; Rizza, F.; Palumbo, M.; Campanile, R.G.; Ricciardi, G.L.; Borghi, B. Evaluation of field and laboratory predictors of drought and heat tolerance in winter cereals. Can. J. Plant Sci. 1997, 77, 523–531. [Google Scholar] [CrossRef]
- Hashemzehi, M.; Moradgholi, A.; Ghasemi, A. Evaluation of responses of mung bean (Vigna radiata) genotypes to drought stress using different stress tolerance indices. J. Crop Breed. 2013, 5, 112–122. [Google Scholar]
- Mafakheri, K.H.; Bihamta, M.R.; Abbasi, A.R. Screening for drought stress tolerance in cowpea genotypes (Vigna unguiculata L.). Iran. J. Pulses Res. 2015, 6, 123–138. [Google Scholar]
- Kumar, R.; Singh, C.M.; Arya, M.; Kumar, R.; Mishra, S.B.; Singh, U.K.; Paswan, S. Investigating stress indices to discriminate the physiologically efficient heat tolerant genotypes of mungbean [Vigna radiata (L.) Wilczek]. Legum. Res. Int. J. 2020, 43, 43–49. [Google Scholar]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. In Sustainable Agriculture; Springer: Dordrecht, The Netherlands, 2009; pp. 153–188. [Google Scholar]
- Jaleel, C.A.; Manivannan, P.; Wahid, A.; Farooq, M.; Al-Juburi, H.J.; Somasundaram, R.; Panneerselvam, R. Drought stress in plants: A review on morphological characteristics and pigments composition. Int. J. Agric. Biol. 2009, 11, 100–105. [Google Scholar]
- Ahmad, A.; Selim, M.; Alderfasi, A.; Afzal, M. Effect of drought stress on mungbean (Vigna radiata L.) under arid climatic conditions of Saudi Arabia. In Ecosystems and Sustainable Development; WIT Press: Southampton, UK, 2015; pp. 185–193. [Google Scholar]
- Rafiei, S.M.; Asgharipur, M.R. Yield reaction and morphological characteristics of some mung bean genotypes to drought stress. Int. J. Mod. Agric. 2009, 5, 67–76. [Google Scholar]
- Alderfasi, A.A.; Alzarqaa, A.A.; AL-Yahya, F.A.; Roushdy, S.S.; Dawabah, A.A.; Alhammad, B.A. Effect of combined biotic and abiotic stress on some physiological aspects and antioxidant enzymatic activity in mungbean (Vigna radiate L.). Afr. J. Agric. Res. 2017, 12, 700–705. [Google Scholar]
- Burke, J.J. Evaluation of source leaf responses to water-deficit stresses in cotton using a novel stress bioassay. Plant Physiol. 2007, 143, 108–121. [Google Scholar] [CrossRef]
- Narina, S.S.; Pathak, S.C.; Bhardwaj, H.L. Chlorophyll fluorescence to evaluate pigeonpea breeding lines and mungbean for drought tolerance. J. Agric. Sci. 2014, 6, 238. [Google Scholar] [CrossRef][Green Version]
- Rahbarian, R.; Khavari-Nejad, R.; Ganjeali, A.; Bagheri, A.; Najafi, F. Drought stress effects on photosynthesis, chlorophyll fluorescence and water relations in tolerant and susceptible chickpea (Cicer arietinum L.) genotypes. Acta Biol. Crac. S. Bot. 2011, 53, 47–56. [Google Scholar] [CrossRef]
- Fu, J.; Huang, B. Involvement of antioxidants and lipid peroxidation in the adaptation of two cool-season grasses to localized drought stress. Environ. Exp. Bot. 2001, 45, 105–114. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Ren, J.; Sun, L.N.; Zhang, Q.Y.; Song, X.S. Drought tolerance is correlated with the activity of antioxidant enzymes in Cerasus humilis seedlings. Biomed. Res. Int. 2016, 2016, 9851095. [Google Scholar] [CrossRef]
- Sanghani, J.M.; Mandaviya, M.K.; Sanghani, A.O.; Raval, S.S.; Golakiya, B.A. Analysis of genetic diversity among mung bean (Vigna radiata L.) genotypes through different biochemical markers. Indian J. Agric. Biochem. 2015, 28, 24–28. [Google Scholar]
- Sairam, R.K.; Dharmar, K.; Lekshmy, S.; Chinnusamy, V. Expression of antioxidant defense genes in mung bean (Vigna radiata L.) roots under water-logging is associated with hypoxia tolerance. Acta Physiol. Plant. 2011, 33, 735–744. [Google Scholar] [CrossRef]
- Nahar, K.; Hasanuzzaman, M.; Alam, M.M.; Fujita, M. Glutathione-induced drought stress tolerance in mung bean: Coordinated roles of the antioxidant defence and methylglyoxal detoxification systems. AoB Plants 2015, 7, plv069. [Google Scholar] [CrossRef]
- Sengupta, D.; Kannan, M.; Reddy, A.R. A root proteomics-based insight reveals dynamic regulation of root proteins under progressive drought stress and recovery in Vigna radiata (L.) Wilczek. Planta 2011, 233, 1111–1127. [Google Scholar] [CrossRef] [PubMed]
- Nahar, K.; Hasanuzzaman, M.; Alam, M.M.; Fujita, M. Roles of exogenous glutathione in antioxidant defense system and methylglyoxal detoxification during salt stress in mung bean. Biol. Plant. 2015, 59, 745–756. [Google Scholar] [CrossRef]
- Sai, C.B.; Chidambaranathan, P. Reproductive stage drought tolerance in blackgram is associated with role of antioxidants on membrane stability. Plant Physiol. Rep. 2019, 24, 399–409. [Google Scholar] [CrossRef]
- Ghallab, K.H.; Ekram, A.M.; Afiah, S.A.; Ahmed, S.M. Characterization of some superior mungbean genotypes on the agronomic and biochemical genetic levels. Egypt. J. Desert Res. 2007, 57, 1–11. [Google Scholar]
- Ramanjulu, S.; Bartels, D. Drought-and desiccation-induced modulation of gene expression in plants. Plant. Cell Environ. 2002, 25, 141–151. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. CRC Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Wohlbach, D.J.; Quirino, B.F.; Sussman, M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 2008, 20, 1101–1117. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Yagoob, H.; Yagoob, M. The effects of water deficit stress on protein yield of mung bean genotypes. PJAS 2014, 2, 30–35. [Google Scholar]
- Kabbadj, A.; Makoudi, B.; Mouradi, M.; Pauly, N.; Frendo, P.; Ghoulam, C. Physiological and biochemical responses involved in water deficit tolerance of nitrogen-fixing Vicia faba. PLoS ONE 2017, 12, e0190284. [Google Scholar] [CrossRef]
- Ocampo, E.; Robles, R. Drought tolerance in mungbean. I. Osmotic adjustment in drought stressed mungbean. Philipp. J. Crop Sci. 2000, 25, 1–5. [Google Scholar]
- Fahramand, M.; Mahmoody, M.; Keykha, A.; Noori, M.; Rigi, K. Influence of abiotic stress on proline, photosynthetic enzymes and growth. Int. Res. J. Appl. Basic Sci. 2014, 8, 257–265. [Google Scholar]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Wehmeyer, N.; Vierling, E. The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 2000, 122, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef]
- Yuliasti, Y.; Reflinur, R. Evaluation of mungbean mutant lines to drought stress and their genetic relationships using SSR markers. Atom Indones. 2015, 41, 161–167. [Google Scholar] [CrossRef]
- Liu, C.; Wu, J.; Wang, L.; Fan, B.; Cao, Z.; Su, Q.; Zhang, Z.; Wang, Y.; Tian, J.; Wang, S. Quantitative trait locus mapping under irrigated and drought treatments based on a novel genetic linkage map in mungbean (Vigna radiata L.). Theor. Appl. Genet. 2017, 130, 2375–2393. [Google Scholar] [CrossRef]
- Sholihin, H.D.M. Molecular mapping of drought resistance in mungbean (Vigna radiata): 1. Linkage map in mungbean using AFLP markers. J. Bioteknol. Pertan. 2002, 7, 17–24. [Google Scholar]
- Miyagi, M.; Humphry, M.; Ma, Z.Y.; Lambrides, C.J.; Bateson, M.; Liu, C.J. Construction of bacterial artificial chromosome libraries and their application in developing PCR-based markers closely linked to a major locus conditioning bruchid resistance in mungbean (Vigna radiata L. Wilczek). Theor. Appl. Genet. 2004, 110, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-M.; Ku, H.-M.; Schafleitner, R.; Bains, T.S.; George Kuo, C.; Liu, C.-A.; Nair, R.M. The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 2013, 192, 205–216. [Google Scholar]
- Chotechung, S.; Somta, P.; Chen, J.; Yimram, T.; Chen, X.; Srinives, P. A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 2016, 129, 1673–1683. [Google Scholar] [CrossRef]
- Schafleitner, R.; Huang, S.; Chu, S.; Yen, J.; Lin, C.; Yan, M.; Krishnan, B.; Liu, M.; Lo, H.; Chen, C.; et al. Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis. BMC Plant Biol. 2016, 16, 159. [Google Scholar]
- Humphry, M.E.; Magner, T.; McIntyre, C.L.; Aitken, E.A.B.; Liu, C.J. Identification of a major locus conferring resistance to powdery mildew (Erysiphe polygoni DC) in mungbean (Vigna radiata L. Wilczek) by QTL analysis. Genome 2003, 46, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Kasettranan, W.; Somta, P.; Srinives, P. Mapping of quantitative trait loci controlling powdery mildew resistance in Mungbean (Vigna radiata (L.) Wilczek). J. Crop Sci. Biotechnol. 2010, 13, 155–161. [Google Scholar] [CrossRef]
- Sai, C.B.; Nagarajan, P.; Raveendran, M.; Rabindran, R.; Senthil, N. Understanding the inheritance of Mungbean Yellow Mosaic Virus (MYMV) resistance in mungbean (Vigna radiata L. Wilczek). Mol. Breed. 2017, 37, 63. [Google Scholar] [CrossRef]
- Kitsanachandee, R.; Somta, P.; Chatchawankanphanich, O.; Akhtar, K.P.; Shah, T.M.; Nair, R.M.; Bains, T.S.; Sirari, A.; Kaur, L.; Srinives, P. Detection of quantitative trait loci for Mungbean Yellow Mosaic India Virus (MYMIV) resistance in mungbean (Vigna radiata (L.) Wilczek) in India and Pakistan. Breed. Sci. 2013, 63, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.K.M.M.; Somta, P.; Srinives, P.; Mahbubul Alam, A.K.M.; Somta, P.; Srinives, P. Identification and confirmation of quantitative trait loci controlling resistance to mungbean yellow mosaic disease in mungbean [Vigna radiata (L.) Wilczek]. Mol. Breed. 2014, 34, 1497–1506. [Google Scholar] [CrossRef]
- Singh, C.M.; Singh, P.; Pratap, A.; Pandey, R.; Purwar, S.; Douglas, C.A.; Baek, K.-H.; Mishra, A.K. Breeding for enhancing Legumovirus resistance in mungbean: Current understanding and future directions. Agronomy 2019, 9, 622. [Google Scholar] [CrossRef]
- Chankaew, S.; Isemura, T.; Naito, K.; Ogiso-Tanaka, E.; Tomooka, N.; Somta, P.; Kaga, A.; Vaughan, D.A.; Srinives, P. QTL mapping for salt tolerance and domestication-related traits in Vigna marina subsp. oblonga, a halophytic species. Theor. Appl. Genet. 2014, 127, 691–702. [Google Scholar] [CrossRef]
- Varshney, R.K.; Gaur, P.M.; Chamarthi, S.K.; Krishnamurthy, L.; Tripathi, S.; Kashiwagi, J.; Samineni, S.; Singh, V.K.; Thudi, M.; Jaganathan, D. Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome 2013, 6, 1–9. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Nayak, S.N.; Gaur, P.M.; Kashiwagi, J.; Krishnamurthy, L.; Jaganathan, D.; Koppolu, J.; Bohra, A.; Tripathi, S. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 2014, 127, 445–462. [Google Scholar] [CrossRef]
- Abdelsalam, N.R.; Abdel-Megeed, A.; Ali, H.M.; Salem, M.Z.M.; Al-Hayali, M.F.A.; Elshikh, M.S. Genotoxicity effects of silver nanoparticles on wheat (Triticum aestivum L.) root tip cells. Ecotoxicol. Environ. Saf. 2018, 155, 76–85. [Google Scholar] [CrossRef]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Sorrells, M. Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Peterson, G.W. Genetic diversity analysis with 454 pyrosequencing and genomic reduction confirmed the eastern and western division in the cultivated barley gene pool. Plant Genome 2011, 4, 226–237. [Google Scholar] [CrossRef]
- Lu, F.; Lipka, A.E.; Glaubitz, J.; Elshire, R.; Cherney, J.H.; Casler, M.D.; Buckler, E.S.; Costich, D.E. Switchgrass genomic diversity, ploidy, and evolution: Novel insights from a network-based SNP discovery protocol. PLoS Genet 2013, 9, e1003215. [Google Scholar] [CrossRef]
- Fu, Y.-B.; Cheng, B.; Peterson, G.W. Genetic diversity analysis of yellow mustard (Sinapis alba L.) germplasm based on genotyping by sequencing. Genet. Resour. Crop Evol. 2014, 61, 579–594. [Google Scholar] [CrossRef]
- Spindel, J.; Begum, H.; Akdemir, D.; Virk, P.; Collard, B.; Redoña, E.; Atlin, G.; Jannink, J.-L.; McCouch, S.R. Genomic selection and association mapping in rice (Oryza sativa): Effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical Rice breeding Lines. PLoS Genet. 2015, 11, e1004982. [Google Scholar]
- Thudi, M.; Upadhyaya, H.D.; Rathore, A.; Gaur, P.M.; Krishnamurthy, L.; Roorkiwal, M.; Nayak, S.N.; Chaturvedi, S.K.; Basu, P.S.; Gangarao, N.V.P.R.; et al. Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS ONE 2014, 9, e96758. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, L.; Li, J.; Chen, J.; Liu, C. Genome-wide identification, characterization, and expression profiling of the legume BZR transcription factor gene family. Front. Plant Sci. 2018, 9, 1332. [Google Scholar] [CrossRef] [PubMed]
- Narum, S.R.; Buerkle, C.A.; Davey, J.W.; Miller, M.R.; Hohenlohe, P.A. Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 2013, 22, 2841. [Google Scholar] [CrossRef]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef]
- Mathivathana, M.K.; Murukarthick, J.; Karthikeyan, A.; Jang, W.; Dhasarathan, M.; Jagadeeshselvam, N.; Sudha, M.; Vanniarajan, C.; Karthikeyan, G.; Yang, T.-J.; et al. Detection of QTLs associated with Mungbean Yellow Mosaic Virus (MYMV) resistance using the interspecific cross of Vigna radiata × Vigna umbellata. J. Appl. Genet. 2019, 60, 255–268. [Google Scholar] [CrossRef]
- Noble, T.J.; Tao, Y.; Mace, E.S.; Williams, B.; Jordan, D.R.; Douglas, C.A.; Mundree, S.G. Characterization of linkage disequilibrium and population structure in a mungbean diversity panel. Front. Plant Sci. 2018, 8, 2102. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Jiao, K.; Li, X.; Guo, W.; Yuan, X.; Cui, X.; Chen, X. Genome re-sequencing of two accessions and fine mapping the locus of lobed leaflet margins in mungbean. Mol. Breed. 2016, 36, 1–2. [Google Scholar] [CrossRef]
- Jain, A.; Roorkiwal, M.; Pandey, M.K.; Varshney, R.K. Current status and prospects of genomic selection in legumes. In Genomic Selection for Crop Improvement; Springer: Cham, Switzerland, 2017; pp. 131–147. [Google Scholar]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing future crops: Genomics-Assisted breeding comes of age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; Declerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- Hartmann, A.; Czauderna, T.; Hoffmann, R.; Stein, N.; Schreiber, F. HTPheno: An image analysis pipeline for high-throughput plant phenotyping. BMC Bioinform. 2011, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Kim, K.; Yoon, H.-J.; Lee, S.; Kim, B.; Siddiqui, Z.S. Phenotyping of plants for drought and salt tolerance using infra-red thermography. Plant Breed. Biotechnol. 2015, 3, 299–307. [Google Scholar] [CrossRef]
- Pratap, A.; Gupta, S.; Nair, R.M.; Gupta, S.K.; Schafleitner, R.; Basu, P.S.; Singh, C.M.; Prajapati, U.; Gupta, A.K.; Nayyar, H.; et al. Using plant phenomics to exploit the gains of genomics. Agronomy 2019, 9, 126. [Google Scholar] [CrossRef]
- Omari, M.K.; Lee, J.; Faqeerzada, M.A.; Joshi, R.; Park, E.; Cho, B.-K. Digital image-based plant phenotyping: A review. Korean J. Agric. Sci. 2020, 47, 119–130. [Google Scholar]
- Tracy, S.R.; Nagel, K.A.; Postma, J.A.; Fassbender, H.; Wasson, A.; Watt, M. Crop improvement from phenotyping roots: Highlights reveal expanding opportunities. Trends Plant Sci. 2020, 25, 105–118. [Google Scholar] [CrossRef]
- Finkel, E. With ‘phenomics,’plant scientists hope to shift breeding into overdrive. Science 2009, 325, 380–381. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef]
- Varshney, R.K.; Hiremath, P.J.; Lekha, P.; Kashiwagi, J.; Balaji, J.; Deokar, A.A.; Vadez, V.; Xiao, Y.; Srinivasan, R.; Gaur, P.M.; et al. A comprehensive resource of drought- and salinity- responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.). BMC Genom. 2009, 10, 523. [Google Scholar] [CrossRef]
- Wang, A.; Hu, J.; Huang, X.; Li, X.; Zhou, G.; Yan, Z. Comparative transcriptome analysis reveals heat-responsive genes in chinese cabbage (Brassica rapa ssp. chinensis). Front. Plant Sci. 2016, 7, 939. [Google Scholar] [CrossRef]
- Wang, J.; Sun, H.; Sheng, J.; Jin, S.; Zhou, F.; Hu, Z.; Diao, Y. Transcriptome, physiological and biochemical analysis of Triarrhena sacchariflora in response to flooding stress. BMC Genet. 2019, 20, 88. [Google Scholar] [CrossRef]
- Liu, M.-S.; Kuo, T.C.-Y.; Ko, C.-Y.; Wu, D.-C.; Li, K.-Y.; Lin, W.-J.; Lin, C.-P.; Wang, Y.-W.; Schafleitner, R.; Lo, H.-F.; et al. Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean (Vigna radiata [L.] R. Wilczek). BMC Plant Biol. 2016, 16, 46. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, S.; Liu, Y.; Liu, X. Transcriptomic profiling reveals metabolic and regulatory pathways in the desiccation tolerance of mungbean (Vigna radiata [L.] R. Wilczek). Front. Plant Sci. 2016, 7, 1921. [Google Scholar] [CrossRef]
- Butsayawarapat, P.; Juntawong, P.; Khamsuk, O.; Somta, P. Comparative transcriptome analysis of waterlogging-sensitive and tolerant zombi pea (Vigna Vexillata) reveals energy conservation and root plasticity controlling waterlogging tolerance. Plants 2019, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Cloonan, N.; Grimmond, S.M. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008, 9, 234. [Google Scholar] [CrossRef]
- Chen, H.; Wang, L.; Wang, S.; Liu, C.; Blair, M.W.; Cheng, X. Transcriptome sequencing of mung bean (Vigna radiate L.) genes and the identification of EST-SSR markers. PLoS ONE 2015, 10, e0120273. [Google Scholar] [CrossRef]
- Shinwari, Z.K.; Jan, S.A.; Nakashima, K.; Yamaguchi-Shinozaki, K. Genetic engineering approaches to understanding drought tolerance in plants. Plant Biotechnol. Rep. 2020, 14, 151–152. [Google Scholar] [CrossRef]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cheng, K.; Wan, L.; Yan, L.; Jiang, H.; Liu, S.; Lei, Y.; Liao, B. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genom. 2015, 16, 1053. [Google Scholar] [CrossRef]
- Labbo, A.M.; Mehmood, M.; Akhtar, M.N.; Khan, M.J.; Tariq, A.; Sadiq, I. Genome-wide identification of AP2/ERF transcription factors in mungbean (Vigna radiata) and expression profiling of the VrDREB subfamily under drought stress. Crop Pasture Sci. 2018, 69, 1009–1019. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Srivastava, R.; Kumar, S.; Kobayashi, Y.; Kusunoki, K.; Tripathi, P.; Kobayashi, Y.; Koyama, H.; Sahoo, L. Comparative genome-wide analysis of WRKY transcription factors in two Asian legume crops: Adzuki bean and Mung bean. Sci. Rep. 2018, 8, 1–19. [Google Scholar]
- Wu, J.; Wang, L.; Wang, S. Comprehensive analysis and discovery of drought-related NAC transcription factors in common bean. BMC Plant Biol. 2016, 16, 193. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Cheng, H.-Y.; Yu, X.-W.; Shi, Q.-H.; Zhang, H.; Li, J.-G.; Ma, H. Characterization of a chickpea (Cicer arietinum L.) NAC family gene, CarNAC5, which is both developmentally-and stress-regulated. Plant Physiol. Biochem. 2009, 47, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Quach, T.N.; Guttikonda, S.K.; Aldrich, D.L.; Kumar, R.; Neelakandan, A.; Valliyodan, B.; Nguyen, H.T. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genom. 2009, 281, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-H.; Chen, F.-J.; Li, H.-Y.; Li, W.; Guo, J.-J. The soybean GmRACK1 gene plays a role in drought tolerance at vegetative stages. Russ. J. Plant Physiol. 2018, 65, 541–552. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant. 2021, 172, 847–868. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, J.; Li, X.; Wang, S.; Wu, J. Salt and drought stress and ABA responses related to bZIP genes from V. radiata and V. angularis. Gene 2018, 651, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, L.; Wang, L.; Wang, S.; Cheng, X. VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J. Plant Res. 2016, 129, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, N.; Gogoi, N.; Barthakur, S.; Basumatary, N. Morphophysiological responses in different mungbean genotypes under drought stress. Res. J. Recent Sci. 2018, 7, 1–5. [Google Scholar]
- Li, S.; Wang, R.; Jin, H.; Ding, Y.; Cai, C. Molecular characterization and expression profile analysis of heat shock transcription factors in mungbean. Front. Genet. 2019, 9, 736. [Google Scholar] [CrossRef]
- Czech, L.; Stöveken, N.; Bremer, E. EctD-mediated biotransformation of the chemical chaperone ectoine into hydroxyectoine and its mechanosensitive channel-independent excretion. Microb. Cell Fact. 2016, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Deokar, A.A.; Kondawar, V.; Jain, P.K.; Karuppayil, S.M.; Raju, N.L.; Vadez, V.; Varshney, R.K.; Srinivasan, R. Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and -susceptible genotypes of chickpea under terminal drought stress. BMC Plant Biol. 2011, 11, 70. [Google Scholar] [CrossRef]
- Hiremath, P.J.; Farmer, A.; Cannon, S.B.; Woodward, J.; Kudapa, H.; Tuteja, R.; Kumar, A.; Bhanuprakash, A.; Mulaosmanovic, B.; Gujaria, N.; et al. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnol. J. 2011, 9, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Gogoi, N.; Barthakur, S.; Baroowa, B.; Bharadwaj, N.; Alghamdi, S.S.; Siddique, K.H.M. Drought stress in grain legumes during reproduction and grain filling. J. Agron. Crop Sci. 2017, 203, 81–102. [Google Scholar] [CrossRef]
- Baloda, A.; Madanpotra, S.; Aiwal, P.K.J. Transformation of mungbean plants for salt and drought tolerance by introducing a gene for an osmoprotectant glycine betaine. J. Plant Stress Physiol. 2017, 3, 5–11. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Nayyar, H. Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed. 2020, 139, 1–27. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H.M. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 466. [Google Scholar] [CrossRef]
- Bohra, A.; Gandham, P.; Rathore, A.; Thakur, V.; Saxena, R.K.; Naik, S.J.S.; Varshney, R.K.; Singh, N.P. Identification of microRNAs and their gene targets in cytoplasmic male sterile and fertile maintainer lines of pigeonpea. Planta 2021, 253, 1–15. [Google Scholar] [CrossRef]
- Zhao, J.; He, Q.; Chen, G.; Wang, L.; Jin, B. Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 2016, 7, 1213. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, G.; Peng, X.; Sun, F.; Liu, S.; Xi, Y. Long non-coding RNAs of switchgrass (Panicum virgatum L.) in multiple dehydration stresses. BMC Plant Biol. 2018, 18, 79. [Google Scholar] [CrossRef]
- Ding, Z.; Tie, W.; Fu, L.; Yan, Y.; Liu, G.; Yan, W.; Li, Y.; Wu, C.; Zhang, J.; Hu, W. Strand-specific RNA-seq based identification and functional prediction of drought-responsive lncRNAs in cassava. BMC Genomics 2019, 20, 214. [Google Scholar] [CrossRef]
- Pang, J.; Zhang, X.; Ma, X.; Zhao, J. Spatio-temporal transcriptional dynamics of maize long non-coding RNAs responsive to drought stress. Genes 2019, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen, J.; Mysore, K.S.; Zhang, W.-H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef]
- Weidong, Q.I.; Hongping, C.; Zuozhen, Y.; Biaolin, H.U.; Xiangdong, L.; Bing, A.; Yuan, L.; Yu, H.; Jiankun, X.; Fantao, Z. Systematic characterization of long non-coding RNAs and their responses to drought stress in Dongxiang wild rice. Rice Sci. 2020, 27, 21–31. [Google Scholar] [CrossRef]
- Ben Amor, B.; Wirth, S.; Merchan, F.; Laporte, P.; d’Aubenton-Carafa, Y.; Hirsch, J.; Maizel, A.; Mallory, A.; Lucas, A.; Deragon, J.M.; et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009, 19, 57–69. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.J.; Jung, H.; Jeong, D.-H.; Ha, S.-H.; Do Choi, Y.; Kim, J.-K. Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics 2016, 17, 563. [Google Scholar] [CrossRef]
- Zhang, W.; Han, Z.; Guo, Q.; Liu, Y.; Zheng, Y.; Wu, F.; Jin, W. Identification of maize long non-coding RNAs responsive to drought stress. PLoS ONE 2014, 9, e98958. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Figueroa, B.E.; Gao, L.; Diop, N.N.; Wu, Z.; Ehlers, J.D.; Roberts, P.A.; Close, T.J.; Zhu, J.-K.; Liu, R. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 2011, 11, 127. [Google Scholar] [CrossRef]
- Wu, J.; Wang, L.; Wang, S. MicroRNAs associated with drought response in the pulse crop common bean (Phaseolus vulgaris L.). Gene 2017, 628, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Pal, A. Genome-wide characterization of microRNAs from mungbean (Vigna radiata L.). Biotechnol. J. Int. 2017, 17, 1–9. [Google Scholar] [CrossRef]
- Sharma, N.; Tripathi, A.; Sanan-Mishra, N. Profiling the expression domains of a rice-specific microRNA under stress. Front. Plant Sci. 2015, 6, 333. [Google Scholar] [CrossRef] [PubMed]
| Sl. N. | Key Traits | Stage of Tolerance | References |
|---|---|---|---|
| 1 | Early vigour, relative water content (RWC) | Seedling/vegetative stage | [51] |
| 2 | Seedling biomass, stress tolerance index | Seedling stage | [45] |
| 3 | Photosynthesis rate, stomatal conductance, transpiration rate, relative leaf water content (RLWC), leaf temperature | Vegetative/reproductive stage | [52] |
| 4 | Biomass, harvest index | Reproductive stage | [53] |
| 5 | RLWC | Vegetative stage | [54] |
| 6 | Root length | Reproductive stage | [26] |
| 7 | Number of floral buds, shoot dry weight, number of lateral roots, root length, number of root nodules, dry matter weight of root system | Seedling, vegetative/reproductive stage | [26] |
| 8 | Shoot length, root length, root shoot ratio, stem diameter, shoot weight, dead leaf percent, emergence percent, energy of emergence | Seedling stage | [49] |
| 9 | Early flowering, specific leaf area | Vegetative/reproductive stage | [55] |
| 10 | Yield components | Reproductive stage | [56] |
| 11 | Root length, shoot length, root volume, root diameter, root and shoot weight | Vegetative/reproductive stage | [57] |
| 12 | Shoot length, root length, number of roots, root diameter | Vegetative/reproductive stage | [58] |
| 13 | Survival rate of seedling, wilt index, RWC, stress index | Seedling stage | [59] |
| 14 | leaf area, RLWC | Seedling stage | [60] |
| 15 | Stomata size, net photosynthesis, osmotic stress injury, Biomass | -- | [61] |
| 16 | RWC, relative injury (RI), chlorophyll stability index (CSI), specific leaf area (SLA), chlorophyll content | Vegetative/reproductive stage | [56] |
| 17 | Germination percent, root length, shoot length | Seedling stage | [62] |
| 18 | SPAD chlorophyll meter reading and specific leaf area | Vegetative/reproductive stage | [63] |
| 19 | RWC, membrane stability index (MSI) | Vegetative/reproductive stage | [17] |
| 20 | Germination percentage, promptness index, radicle length, root length stress index, germination stress index, seed vigour | Seedling stage | [50] |
| 21 | Shoot length, biomass, leaf area index, RWC, stomatal conductance | Vegetative/reproductive stage | [64] |
| Sl. N. | Drought Tolerant Genotypes | Screening Method | References |
|---|---|---|---|
| 1 | V 1281, V 2013, V 3372, VC 2754, VC 2768A | -- | [79] |
| 2 | WGG 2, MGG 347, EC 396117, MGG 350 and Asha, LGG 450 | -- | [80] |
| 3 | VC 2917 | Removal of irrigation for 15 days | [59] |
| 4 | HUM 1, VMGG 67, VMGG 82, VMGG 83 and VMGG 90 | -- | [57] |
| 5 | ML 267, MGG 347 | Drought induced by PEG | [63] |
| 6 | K 851 | Drought induced by PEG | [60] |
| 7 | C. No. 35, OUM 14-1, OUM 49-2, Pusa 9072, OM 99-3, Banapur local B, Nipania munga, Kalamunga 1-A, TCR 20 | Drought induced by PEG | [81] |
| 8 | AU-M4 | Drought induced by PEG | [61] |
| 9 | SML-1411, SML 1136 | Drought induced by PEG | [62] |
| 10 | ML 267 | Drought induced by PEG | [63] |
| 11 | Pusa 1131 | Drought induced by PEG | [82] |
| 12 | Vigna sublobata, MCV-1, PLM-32, LGG-407, LGG-450, TM-96-2, and Sattya | Removal of irrigation for 15 days | [17] |
| 13 | COGG 1332, VGG 16069, VGG 17003, VGG 17004, VGG 17009, VGG 17019 and VGG 17045 | Drought induced by PEG | [50] |
| Sl. N. | Key Traits | Expression | References |
|---|---|---|---|
| 1 | H2O2 | Increased | [102] |
| 2 | Glutathione disulphide, GSH, ascorbate peroxidase and glutathione S-transferase activities | Increased | [103] |
| 3 | Superoxide dismutase (SOD), guaiacol peroxidase (GPOX), ROS | Increased | [82] |
| 4 | Cu/Zn superoxide dismutase, oxidoreductase and aldehyde reductase | Increased | [104] |
| 5 | Monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR) and catalase | Decreased | [105] |
| 6 | Phenols and ascorbic acid content, H2O2 | Increased | [60] |
| 7 | malondialdehyde (MDA), total soluble sugars (TSS) and proline contents | Increased | [64] |
| 8 | Proline content | Decreased | [17] |
| 9 | Soluble protein content | Decreased | [64] |
| 10 | Globulin seed storage protein | 12.8 kD band present in drought tolerant genotypes | [81] |
| Sl. N. | Candidate Gene(s) | Test Species | Stress Condition | References |
|---|---|---|---|---|
| 1 | VabZIP6, VabZIP34, VabZIP50, VrbZIP | V. angularis | Drought | [179] |
| 2 | VaWRKY61 | V. angularis | Osmotic stress | [173] |
| 3 | VaWRKY16, VaWRKY45, VrWRKY49 | V. angularis | Drought | [173] |
| 4 | VrDREB2A | V. radiata | Drought | [180] |
| 5 | VrbZIP17, VrbZIP27, VrbZIP31, VrbZIP50 | V. radiata | Drought | [179] |
| 6 | VrDREB5, VrDREB12, VrDREB13, VrDREB22, VrDREB30 | V. radiata | Drought | [171] |
| 7 | VrWRKY73 | V. radiata | Osmotic stress | [173] |
| 8 | VrSKP1 | V. radiata | Drought | [181] |
| 9 | VrHsfA6a, VrHsfA6b | V. radiata | Drought | [182] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, C.M.; Singh, P.; Tiwari, C.; Purwar, S.; Kumar, M.; Pratap, A.; Singh, S.; Chugh, V.; Mishra, A.K. Improving Drought Tolerance in Mungbean (Vigna radiata L. Wilczek): Morpho-Physiological, Biochemical and Molecular Perspectives. Agronomy 2021, 11, 1534. https://doi.org/10.3390/agronomy11081534
Singh CM, Singh P, Tiwari C, Purwar S, Kumar M, Pratap A, Singh S, Chugh V, Mishra AK. Improving Drought Tolerance in Mungbean (Vigna radiata L. Wilczek): Morpho-Physiological, Biochemical and Molecular Perspectives. Agronomy. 2021; 11(8):1534. https://doi.org/10.3390/agronomy11081534
Chicago/Turabian StyleSingh, Chandra Mohan, Poornima Singh, Chandrakant Tiwari, Shalini Purwar, Mukul Kumar, Aditya Pratap, Smita Singh, Vishal Chugh, and Awdhesh Kumar Mishra. 2021. "Improving Drought Tolerance in Mungbean (Vigna radiata L. Wilczek): Morpho-Physiological, Biochemical and Molecular Perspectives" Agronomy 11, no. 8: 1534. https://doi.org/10.3390/agronomy11081534
APA StyleSingh, C. M., Singh, P., Tiwari, C., Purwar, S., Kumar, M., Pratap, A., Singh, S., Chugh, V., & Mishra, A. K. (2021). Improving Drought Tolerance in Mungbean (Vigna radiata L. Wilczek): Morpho-Physiological, Biochemical and Molecular Perspectives. Agronomy, 11(8), 1534. https://doi.org/10.3390/agronomy11081534







