The Microflora of Maize Grains as a Biological Barrier against the Late Wilt Causal Agent, Magnaporthiopsis maydis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Origin and Growth of the Maize Pathogen, M. maydis
2.2. Preparation of Maize Grains
2.3. Isolation and Identification of Endophytes from Maize Grains
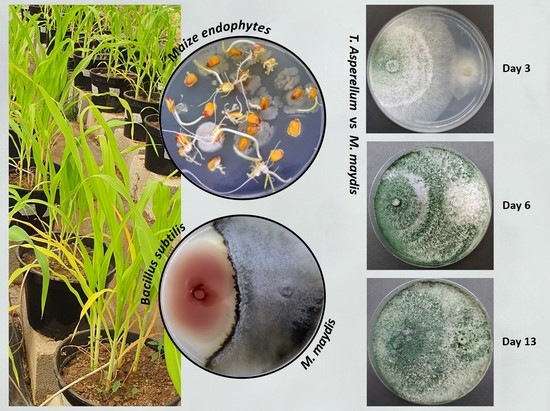
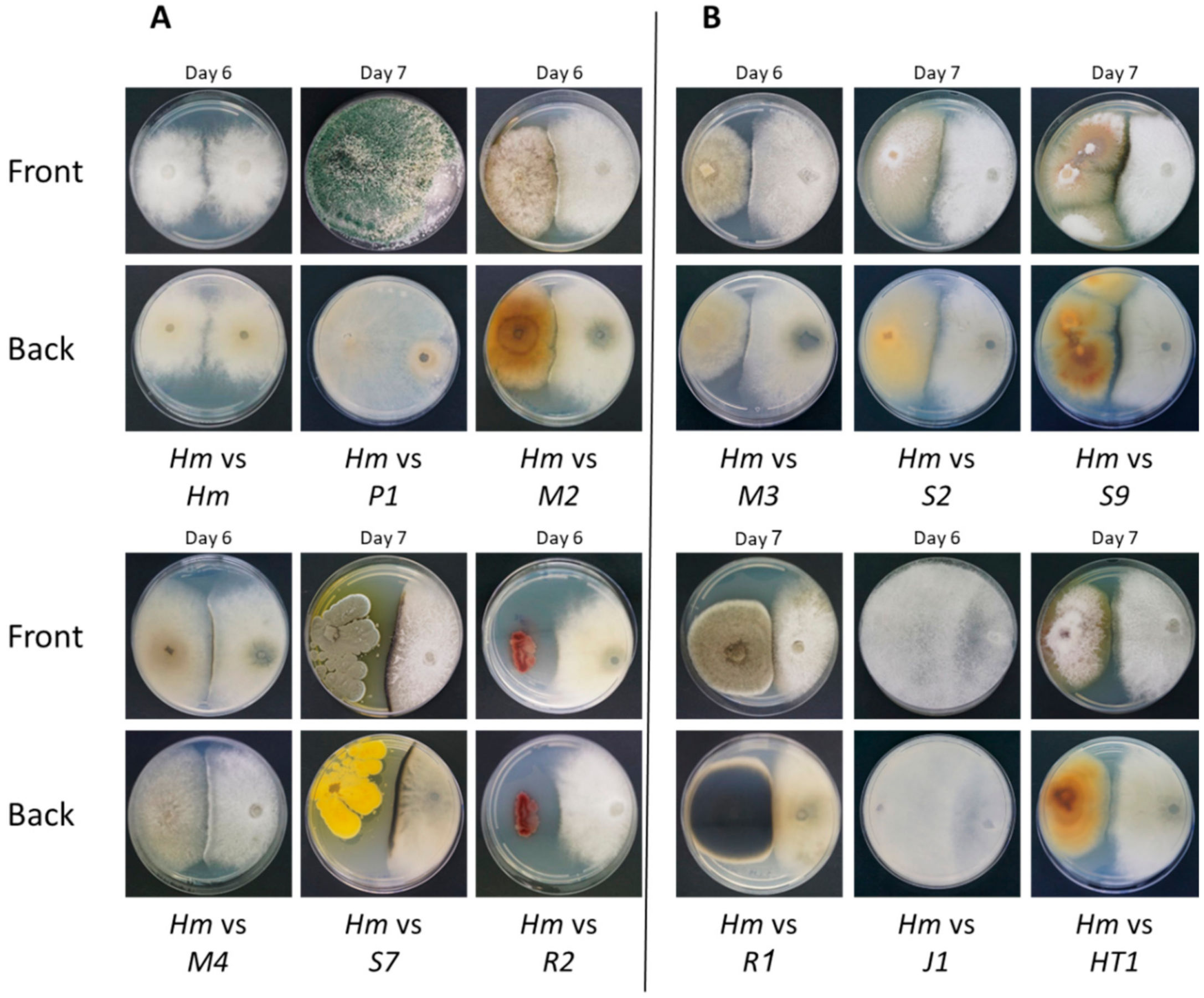
2.4. Plate Confrontation Assay
2.5. Pathogenicity Assay in Sprouts
2.6. Molecular Diagnosis
2.6.1. Molecular Identification of the Endophytes
2.6.2. qPCR Diagnosis of M. maydis DNA in the Maize Plants
2.7. Statistical Analysis
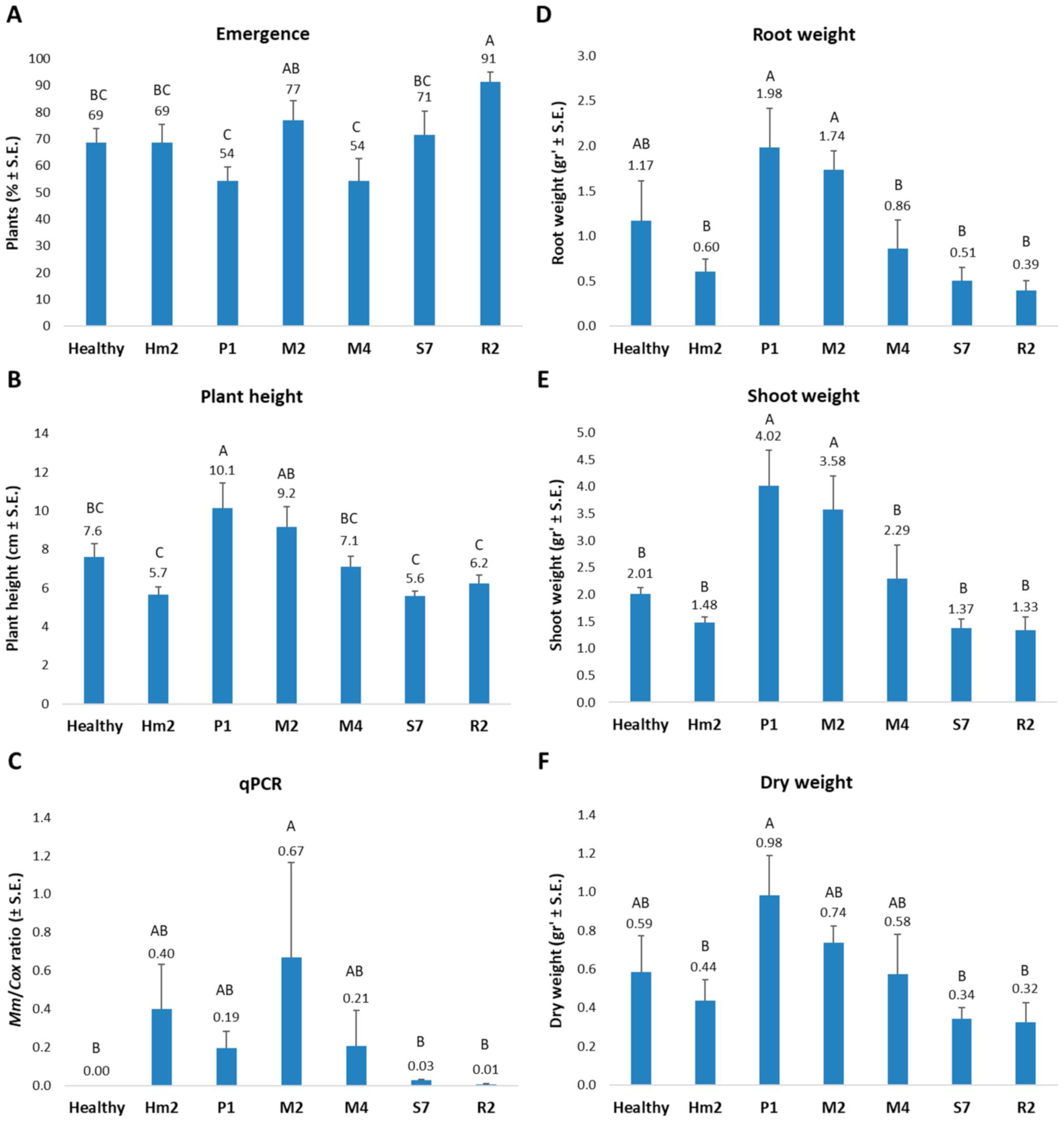
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klaubauf, S.; Tharreau, D.; Fournier, E.; Groenewald, J.Z.; Crous, P.W.; de Vries, R.P.; Lebrun, M.H. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Stud. Mycol. 2014, 79, 85–120. [Google Scholar] [CrossRef]
- Gams, W. Phialophora and some similar morphologically little-differentiated anamorphs of divergent ascomycetes. Stud. Mycol. 2000, 187–200. [Google Scholar]
- Payak, M.M.; Lal, S.; Lilaramani, J.; Renfro, B.L. Cephalosporium maydis—A new threat to maize in India. Indian Phytopathol. 1970, 23, 562–569. [Google Scholar]
- Pecsi, S.; Nemeth, L. Appearance of Cephalosporium maydis Samra Sabet and Hingorani in Hungary. Meded. Fac. Landbouwkd. Toegep. Biol. Wet. Univ. Gent 1998, 63, 873–877. [Google Scholar]
- Bergstrom, G.; Leslie, F.J.; Huber, D.; Lipps, P.; Warren, H.; Esker, P.; Grau, C.; Botratynski, T.; Bulluck, R.; Floyd, J.; et al. Recovery Plan for Late Wilt of Corn Caused by Harpophora maydis syn. Cephalosporium maydis; National Plant Disease Recovery System (NPDRS): Washington, DC, USA, 2008; p. 24.
- Molinero-Ruiz, M.L.; Melero-Vara, J.M.; Mateos, A. Cephalosporium maydis, the cause of late wilt in maize, a pathogen new to Portugal and Spain. Plant Dis. 2011, 94, 379. [Google Scholar] [CrossRef] [Green Version]
- Drori, R.; Sharon, A.; Goldberg, D.; Rabinovitz, O.; Levy, M.; Degani, O. Molecular diagnosis for Harpophora maydis, the cause of maize late wilt in Israel. Phytopathol. Mediterr. 2013, 52, 16–29. [Google Scholar]
- Subedi, S. A review on important maize diseases and their management in Nepal. J. Maize Res. Dev. 2015, 1, 28–52. [Google Scholar] [CrossRef]
- El-Shabrawy, E.-S.; Shehata, H. Controlling maize late-wilt and enhancing plant salinity tolerance by some rhizobacterial strains. Egypt. J. Phytopathol. 2018, 46, 235–255. [Google Scholar] [CrossRef]
- Degani, O.; Dor, S.; Movshowitz, D.; Fraidman, E.; Rabinovitz, O.; Graph, S. Effective chemical protection against the maize late wilt causal agent, Harpophora maydis, in the field. PLoS ONE 2018, 13, e0208353. [Google Scholar] [CrossRef]
- Degani, O.; Movshowitz, D.; Dor, S.; Meerson, A.; Goldblat, Y.; Rabinovitz, O. Evaluating Azoxystrobin Seed Coating against Maize Late Wilt Disease Using a Sensitive qPCR-Based Method. Plant Dis. 2019, 103, 238–248. [Google Scholar] [CrossRef] [Green Version]
- Degani, O.; Dor, S.; Movshovitz, D.; Rabinovitz, O. Methods for Studying Magnaporthiopsis maydis, the Maize Late Wilt Causal Agent. Agronomy 2019, 9, 181. [Google Scholar] [CrossRef] [Green Version]
- Degani, O.; Cernica, G. Diagnosis and Control of Harpophora maydis, the Cause of Late Wilt in Maize. Adv. Microbiol. 2014, 4, 94–105. [Google Scholar] [CrossRef] [Green Version]
- Sabet, K.A.; Samra, A.S.; Mansour, I.M. Saprophytic behaviour of Cephalosporium maydis and C. acremonium. Ann. Appl. Biol. 1970, 66, 265–271. [Google Scholar] [CrossRef]
- Michail, S.H.; Abou-Elseoud, M.S.; Nour Eldin, M.S. Seed health testing of corn for Cephalosporium maydis. Acta Phytopathol. Entomol. Hung. 1999, 34, 35–42. [Google Scholar]
- Sahab, A.F.; Osman, A.R.; Soleman, N.K.; Mikhail, M.S. Studies on root-rot of lupin in Egypt and its control. Egypt. J. Phytopathol. 1985, 17, 23–35. [Google Scholar]
- Dor, S.; Degani, O. Uncovering the Host Range for Maize Pathogen Magnaporthiopsis maydis. Plants 2019, 8, 259. [Google Scholar] [CrossRef] [Green Version]
- Degani, O.; Dor, S.; Abraham, D.; Cohen, R. Interactions between Magnaporthiopsis maydis and Macrophomina phaseolina, the Causes of Wilt Diseases in Maize and Cotton. Microorganisms 2020, 8, 249. [Google Scholar] [CrossRef] [Green Version]
- Degani, O.; Dor, S.; Chen, A.; Orlov-Levin, V.; Stolov-Yosef, A.; Regev, D.; Rabinovitz, O. Molecular Tracking and Remote Sensing to Evaluate New Chemical Treatments against the Maize Late Wilt Disease Causal Agent, Magnaporthiopsis maydis. J. Fungi 2020, 6, 54. [Google Scholar] [CrossRef]
- Degani, O.; Weinberg, T.; Graph, S. Chemical control of maize late wilt in the field. Phytoparasitica 2014, 42, 559–570. [Google Scholar] [CrossRef]
- Ons, L.; Bylemans, D.; Thevissen, K.; Cammue, B.P.A. Combining Biocontrol Agents with Chemical Fungicides for Integrated Plant Fungal Disease Control. Microorganisms 2020, 8, 1930. [Google Scholar] [CrossRef]
- El-Assiuty, E.M.; El-Shafey, H.A.; Ismael, A.-S.; Fahmy, Z.M. Pathogenic variation in Cephalosporium maydis. Phytopathology 1988, 88, S25. [Google Scholar]
- Ortiz-Bustos, C.M.; Testi, L.; García-Carneros, A.B.; Molinero-Ruiz, L. Geographic distribution and aggressiveness of Harpophora maydis in the Iberian peninsula, and thermal detection of maize late wilt. Eur. J. Plant Pathol. 2015, 144, 383–397. [Google Scholar] [CrossRef]
- Zeller, K.A.; Ismael, A.M.; El-Assiuty, E.M.; Fahmy, Z.M.; Bekheet, F.M.; Leslie, J.F. Relative Competitiveness and Virulence of Four Clonal Lineages of Cephalosporium maydis from Egypt Toward Greenhouse-Grown Maize. Plant Dis. 2002, 86, 373–378. [Google Scholar] [CrossRef] [Green Version]
- Kamara, M.M.; Ghazy, N.A.; Mansour, E.; Elsharkawy, M.M.; Kheir, A.M.S.; Ibrahim, K.M. Molecular Genetic Diversity and Line × Tester Analysis for Resistance to Late Wilt Disease and Grain Yield in Maize. Agronomy 2021, 11, 898. [Google Scholar] [CrossRef]
- El-Shafey, H.A.; El-Shorbagy, F.A.; Khalil, I.I.; El-Assiuty, E.M. Additional sources of resistance to the late-wilt disease of maize caused by Cephalosporium maydis. Agric. Res. Rev. 1988, 66, 221–230. [Google Scholar]
- Sunitha, N.; Gangappa, E.; Gowda, R.V.; Ramesh, S.; Biradar, S.; Swamy, D.; Hemareddy, H. Assessment of Impact of Late Wilt Caused by Harpophora maydis (Samra, Sabet and Hing) on Grain Yield and its Attributing Traits in Maize (Zea mays L.). Mysore J. Agric. Sci. 2020, 54, 30–36. [Google Scholar]
- Elshahawy, I.E.; El-Sayed, A.E.-K.B. Maximizing the efficacy of Trichoderma to control Cephalosporium maydis, causing maize late wilt disease, using freshwater microalgae extracts. Egypt. J. Biol. Pest Control 2018, 28, 48. [Google Scholar] [CrossRef] [Green Version]
- El-Moghazy, S.; Shalaby, M.E.; Mehesen, A.A.; Elbagory, M. Fungicidal effect of some promising agents in controlling maize late wilt disease and their potentials in developing yield productivity. Environ. Biodivers. Soil Secur. 2017, 1, 129–143. [Google Scholar] [CrossRef]
- Doleib, N.M.; Farfour, S.A.; Al-shakankery, F.M.; Ammar, M.; Hamouda, R.A. Antifungal activates of cyanobacteria and some marine algae against Cephalosporium maydis, the cause of Maize Late Wilt disease In Vitro. Biosci. Res. 2021, 18, 536–543. [Google Scholar]
- El-Mehalowy, A.A.; Hassanein, N.M.; Khater, H.M.; Daram El-Din, E.A.; Youssef, Y.A. Influence of maize root colonization by rhizosphere actinomycetes and yeast fungi on plant growth and on the biological control of late wilt disease. Int. J. Agric. Biol. 2004, 6, 599–605. [Google Scholar]
- Ghazy, N.; El-Nahrawy, S. Siderophore production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and their efficacy in controlling Cephalosporium maydis in maize plant. Arch. Microbiol. 2020, 1–15. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Harman, G.E. Overview of Mechanisms and Uses of Trichoderma spp. Phytopathology 2006, 96, 190–194. [Google Scholar] [CrossRef] [Green Version]
- Shalaby, M.; El-Moghazy, S.; Mehesen, A.A. Biological control of maize late wilt disease caused by Cephalosporiummaydis. J. Agric. Res. 2009, 35, 1–19. [Google Scholar]
- El-Gazzar, N.; El-Bakery, A.M.; Ata, A.A. Influence of some bioagents and chitosan nanoparticles on controlling maize late wilt and improving plants characteristics. Egypt. J. Phytopathol. 2018, 46, 243–264. [Google Scholar] [CrossRef]
- Degani, O.; Dor, S. Trichoderma Biological Control to Protect Sensitive Maize Hybrids against Late Wilt Disease in the Field. J. Fungi 2021, 7, 315. [Google Scholar] [CrossRef]
- Wallace, J.G.; May, G. Endophytes: The other maize genome. In The Maize Genome; Springer: Cham, Switzerland, 2018; pp. 213–246. [Google Scholar] [CrossRef]
- Hawkes, C.V.; Connor, E.W. Translating phytobiomes from theory to practice: Ecological and evolutionary considerations. Phytobiomes 2017, 1, 57–69. [Google Scholar] [CrossRef]
- Orole, O.; Adejumo, T. Activity of fungal endophytes against four maize wilt pathogens. Afr. J. Microbiol. Res. 2009, 3, 969–973. [Google Scholar]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White, J.F., Jr. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2015, 172, 79–87. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Saleh, A.A.; Leslie, J.F. Cephalosporium maydis is a distinct species in the Gaeumannomyces-Harpophora species complex. Mycologia 2004, 96, 1294–1305. [Google Scholar] [CrossRef]
- Zeller, K.A.; Jurgenson, J.E.; El-Assiuty, E.M.; Leslie, J.F. Isozyme and amplified fragment length polymorphisms from Cephalosporium maydis in Egypt. Phytoparasitica 2000, 28, 121–130. [Google Scholar] [CrossRef]
- Weller, S.; Elphinstone, J.; Smith, N.; Boonham, N.; Stead, D. Detection of Ralstonia solanacearumstrains with a quantitative, multiplex, Real-Time, fluorogenic PCR (TaqMan) assay. Appl. Environ. Microbiol. 2000, 66, 2853–2858. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N., Jr. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Hartung, J.S.; Levy, L. Quantitative Real-Time PCR for detection and identification of Candidatus liberibacter species associated with Citrus huanglongbing. J. Microbiol. Methods 2006, 66, 104–115. [Google Scholar] [CrossRef]
- Degani, O.; Regev, D.; Dor, S.; Rabinovitz, O. Soil Bioassay for Detecting Magnaporthiopsis maydis Infestation Using a Hyper Susceptible Maize Hybrid. J. Fungi 2020, 6, 107. [Google Scholar] [CrossRef]
- Ortiz-Bustos, C.; López-Bernal, A.; Testi, L.; Molinero-Ruiz, L.J.P.P. Environmental and irrigation conditions can mask the effect of Magnaporthiopsis maydis on growth and productivity of maize. Plant Pathol. 2019, 68, 1555–1564. [Google Scholar] [CrossRef]
- Galal, O.A.; Aboulila, A.A.; Motawei, A.; Galal, A. Cytology. Biochemical and Molecular Diversity and Their Re-Lationship To Late Wilt Disease Resistance In Yel-Low Maize Inbred Lines. Egypt. J. Genet. Cytol. 2019, 47, 2. [Google Scholar]
- Massi, F.; Torriani, S.F.; Borghi, L.; Toffolatti, S.L. Fungicide Resistance Evolution and Detection in Plant Pathogens: Plasmopara viticola as a Case Study. Microorganisms 2021, 9, 119. [Google Scholar] [CrossRef]
- Patanita, M.; Campos, M.D.; Félix, M.D.R.; Carvalho, M.; Brito, I. Effect of Tillage System and Cover Crop on Maize Mycorrhization and Presence of Magnaporthiopsis maydis. Biology 2020, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Tej, R.; Rodríguez-Mallol, C.; Rodríguez-Arcos, R.; Karray-Bouraoui, N.; Molinero-Ruiz, L. Inhibitory effect of Lycium europaeum extracts on phytopathogenic soil-borne fungi and the reduction of late wilt in maize. Eur. J. Plant Pathol. 2018, 152, 249–265. [Google Scholar] [CrossRef] [Green Version]
- Degani, O.; Drori, R.; Goldblat, Y. Plant growth hormones suppress the development of Harpophora maydis, the cause of late wilt in maize. Physiol. Mol. Biol. Plants 2015, 21, 137–149. [Google Scholar] [CrossRef] [Green Version]
- Ashour, A.; Sabet, K.; El-Shabrawy, E.-S.; Alhanshoul, A. Control of maize late wilt and enhancing plant growth parameters using rhizobacteria and organic compounds. Egypt. J. Phytopathol. 2013, 41, 187–207. [Google Scholar] [CrossRef]
- Hamza, A.M.; El-Kot, G.; El-Moghazy, S. Non-traditional methods for controlling maize late wilt disease caused by Cephalosporium maydis. Egypt. J. Biol. Pest Control 2013, 23, 87–93. [Google Scholar]
- Gao, F.-K.; Dai, C.-C.; Liu, X.-Z. Mechanisms of fungal endophytes in plant protection against pathogens. Afr. J. Microbiol. Res. 2010, 4, 1346–1351. [Google Scholar]
- Khan, S.A.; Hamayun, M.; Yoon, H.; Kim, H.-Y.; Suh, S.-J.; Hwang, S.-K.; Kim, J.-M.; Lee, I.-J.; Choo, Y.-S.; Yoon, U.-H.; et al. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008, 8, 231. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Wu, H.-J.; Qiao, J.; Gao, X.; Borriss, R. Novel routes for improving biocontrol activity of Bacillus based bioinoculants. Front. Microbiol. 2015, 6, 1395. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Yu, S.; Yang, Y.; Zhang, J.; Zhao, D.; Pan, Y.; Fan, S.; Yang, Z.; Zhu, J. Antifungal Effects of Volatiles Produced by Bacillus subtilis Against Alternaria solani in Potato. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Francl, L.J. The disease triangle: A plant pathological paradigm revisited. Plant Health Instr. 2001, 10. [Google Scholar] [CrossRef]
- Rodriguez, R.; White, J., Jr.; Arnold, A.E.; Redman, A.R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]

| Cultivar | Type | Seed Company | Supply Company | Degree of LWD Sensitivity |
|---|---|---|---|---|
| Simon | Fodder | Semillas Fitó, Barcelona, Spain | Tarsis Inc., Petach Tikva, Israel | Highly resistant |
| Hatai | Fodder | Semillas Fitó, Barcelona, Spain | Tarsis Inc., Petach Tikva, Israel | Highly resistant |
| Megaton | Sweet | Zeraim Gedera-Syngenta, Kibbutz, Revadim, Israel | Hazera Seeds Ltd., Berurim MP Shikmim, Israel | Hypersensitive |
| Prelude | Sweet | SRS Snowy River Seeds, Australia | Green 2000 Ltd., Israel | Sensitive |
| Jubilee | Sweet | Pop Vriend Seeds B.V., Andijk, The Netherlands | Eden Seeds, Reut, Israel | Sensitive |
| Royalty | Sweet | Pop Vriend Seeds B.V., Andijk, The Netherlands | Eden Seeds, Reut, Israel | Resistant |
| Pairs | Primer | Sequence | Uses | Amplification | References |
|---|---|---|---|---|---|
| Pair 1 | ITS1 | 5′-TCCGTAGGTGAACCTGCGG-3′ | PCR target gene | Target region for the identification of fungi species | [49] |
| ITS4 | 5′-TCCTCCGCTTATTGATATGC-3′ | ||||
| Pair 2 | A200a-for | 5′-CCGACGCCTAAAATACAGGA-3′ | qPCR target gene | 200 bp M. maydis species-specific fragment | [7] |
| A200a-rev | 5′-GGGCTTTTTAGGGCCTTTTT-3′ | ||||
| Pair 3 | COX-F | 5′-GTATGCCACGTCGCATTCCAGA-3′ | qPCR control | Cytochrome c oxidase (Cox) gene product | [47,50] |
| COX-R | 5′-CAACTACGGATATATAAGRRCCRRAACTG-3′ |
| Species | Designation | Systematic Classification | Maize Cultivar | NCBI Accession | NCBI Score | Confrontation Assay Winner | Tested in Sprouts |
|---|---|---|---|---|---|---|---|
| Magnaporthiopsis maydis | Hm-2 | Fungi | Zea mays | ||||
| Trichoderma asperellum | P1 | Fungi | Prelude cv. | MF871569 | 99.66% | P1 | + |
| Chaetomium subaffine | M2 | Fungi | Megaton cv. | HM365247 | 100.00% | Antagonism | + |
| Chaetomium cochliodes | M4 | Fungi | Megaton cv. | MT520580 | 97.53% | Antagonism | + |
| Penicillium citrinum | S7 | Fungi | Simon cv. | MN046972 | 99.57% | Antagonism | + |
| Bacillus subtilis | R2 | Bacteria | Royalty cv. | MT415782 | 99.06% | Antagonism | + |
| Chaetomium cochliodes | M3 | Fungi | Megaton cv. | MN534819 | 99.80% | Antagonism | − |
| Fusarium proliferatum | S2 | Fungi | Simon cv. | MT563410 | 99.44% | Antagonism | − |
| Fusarium proliferatum | S9 | Fungi | Simon cv. | EU272509 | 99.25% | Antagonism | − |
| Alternaria alternata | R1 | Fungi | Royalty cv. | MK174979 | 99.46% | Antagonism | − |
| Rhizopus oryzae | J1 | Fungi | Jubilee cv. | MT603963 | 100.00% | J1 | − |
| Fusarium proliferatum | HT1 | Fungi | Hatai cv. | EU272509 | 98.89% | Antagonism | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degani, O.; Regev, D.; Dor, S. The Microflora of Maize Grains as a Biological Barrier against the Late Wilt Causal Agent, Magnaporthiopsis maydis. Agronomy 2021, 11, 965. https://doi.org/10.3390/agronomy11050965
Degani O, Regev D, Dor S. The Microflora of Maize Grains as a Biological Barrier against the Late Wilt Causal Agent, Magnaporthiopsis maydis. Agronomy. 2021; 11(5):965. https://doi.org/10.3390/agronomy11050965
Chicago/Turabian StyleDegani, Ofir, Danielle Regev, and Shlomit Dor. 2021. "The Microflora of Maize Grains as a Biological Barrier against the Late Wilt Causal Agent, Magnaporthiopsis maydis" Agronomy 11, no. 5: 965. https://doi.org/10.3390/agronomy11050965
APA StyleDegani, O., Regev, D., & Dor, S. (2021). The Microflora of Maize Grains as a Biological Barrier against the Late Wilt Causal Agent, Magnaporthiopsis maydis. Agronomy, 11(5), 965. https://doi.org/10.3390/agronomy11050965