A Pesticide Biopurification System: A Source of Biosurfactant-Producing Bacteria with Environmental Biotechnology Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Media
2.2. Biosurfactant-Producing Bacteria
2.3. Selection of Biosurfactant-Producing Bacteria
2.4. Biosurfactant Production by Selected Strains
2.5. Oil Displacement and Emulsification Assays
2.6. Biosurfactant Characterization
2.6.1. TLC
2.6.2. FTIR Spectroscopy
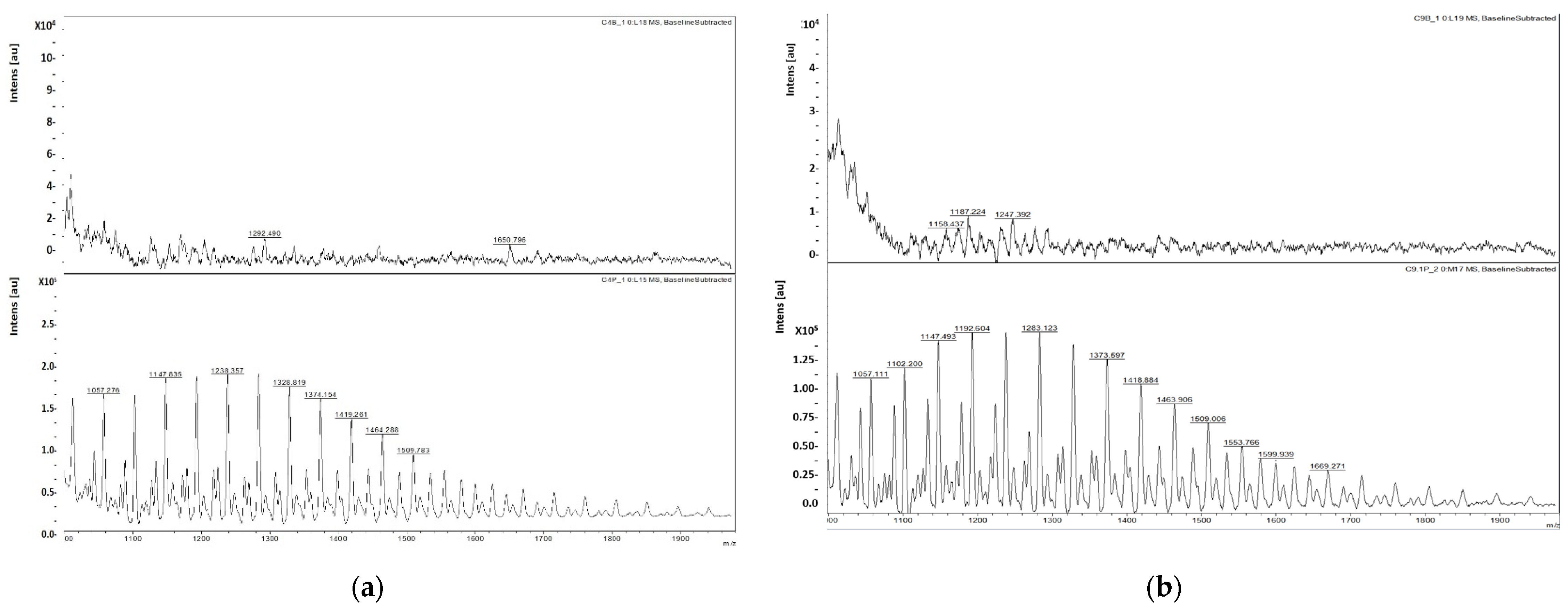
2.6.3. MALDI TOF-TOF Analysis
2.7. Biosurfactants Analyses
2.7.1. Drop-Collapse Test
2.7.2. Surface Tension Assay
2.7.3. Oil Displacement and Emulsification Analyses
2.8. Chlorpyrifos Degradation by Biosurfactant-Producing Bacteria
2.9. Pesticide Extraction and Analysis
2.10. Kinetic Calculations
2.11. Statistical Analyses
3. Results
3.1. Selection of Biosurfactant-Producing Bacteria
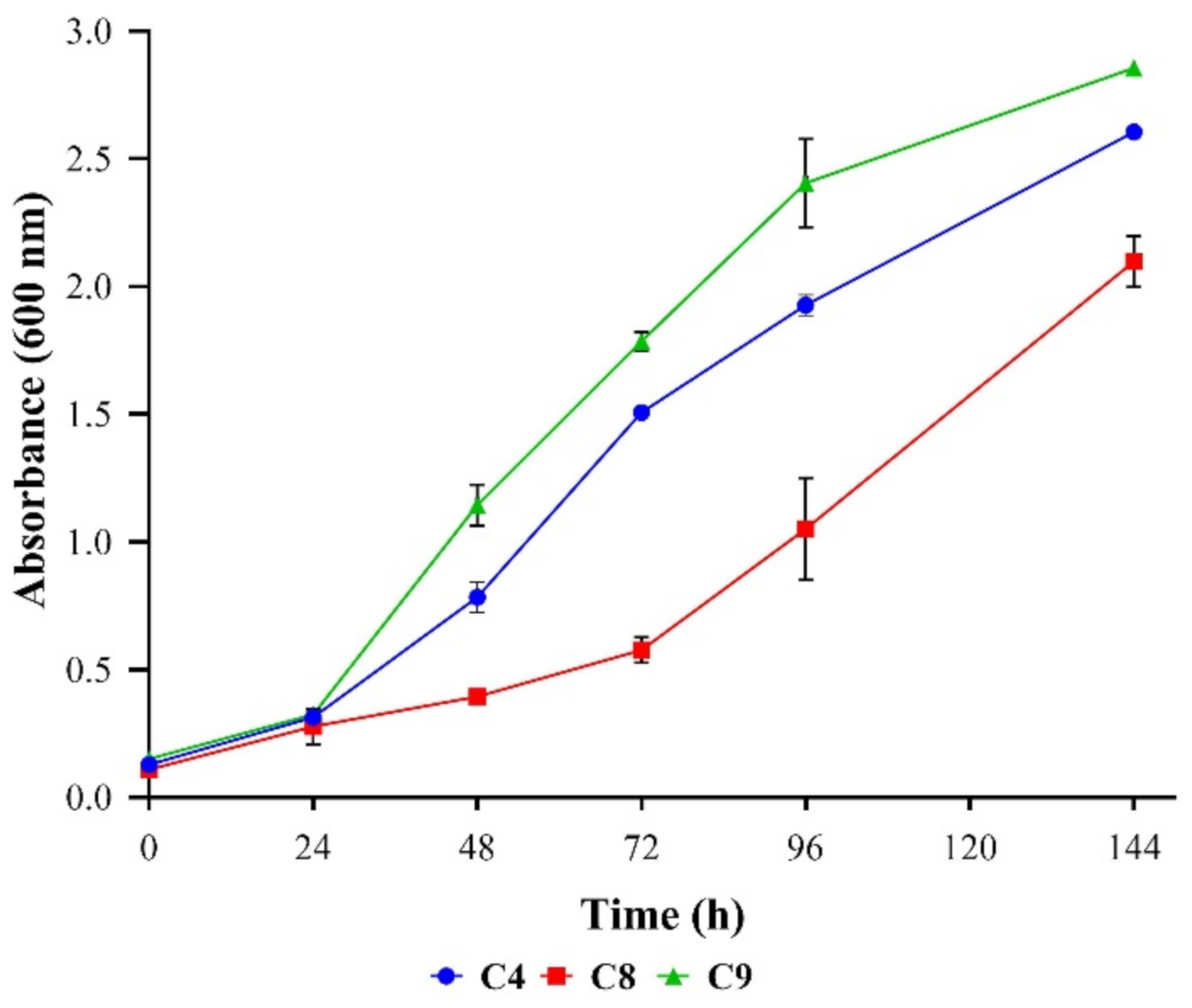
3.2. The Kinetics of Biosurfactant Production by the Selected Strains
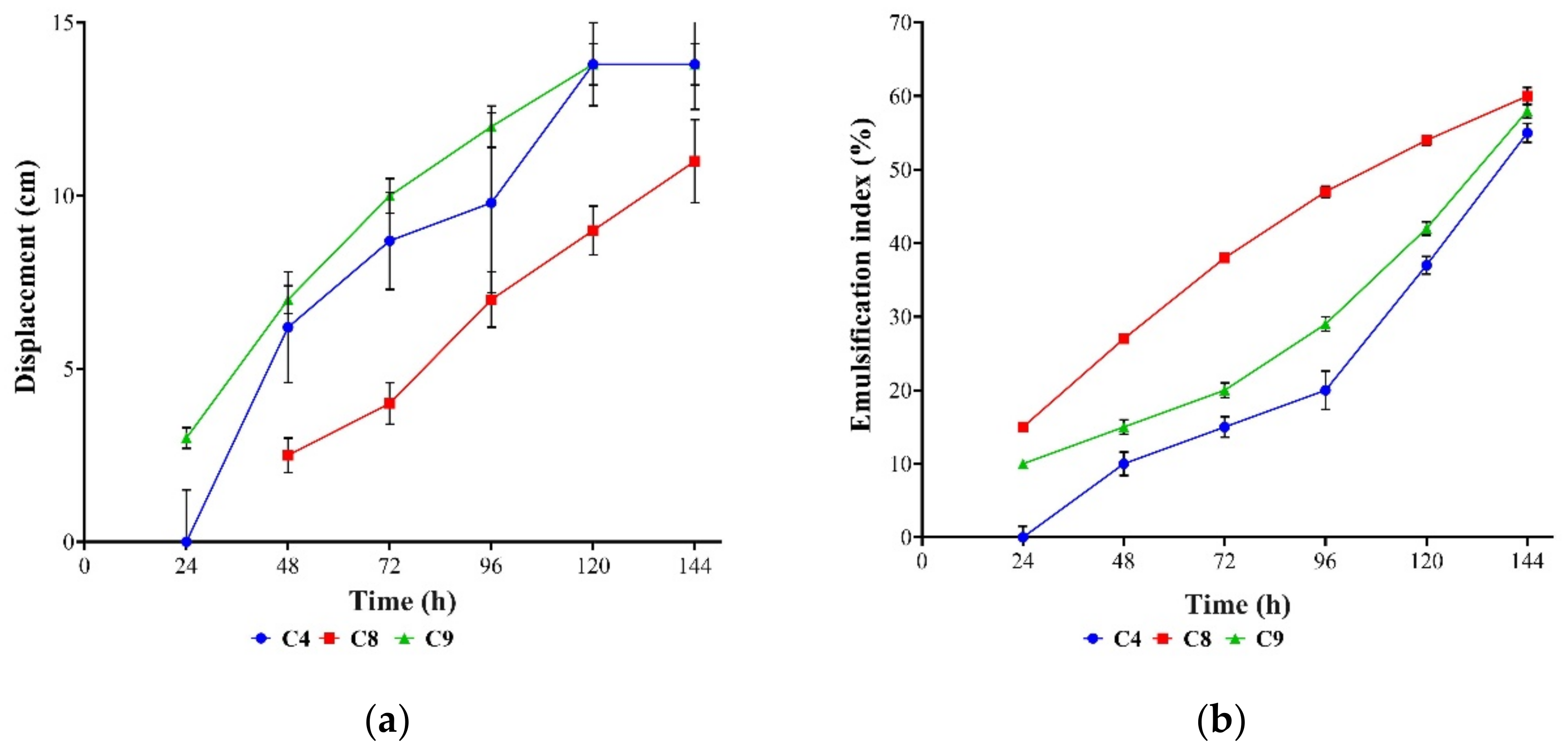
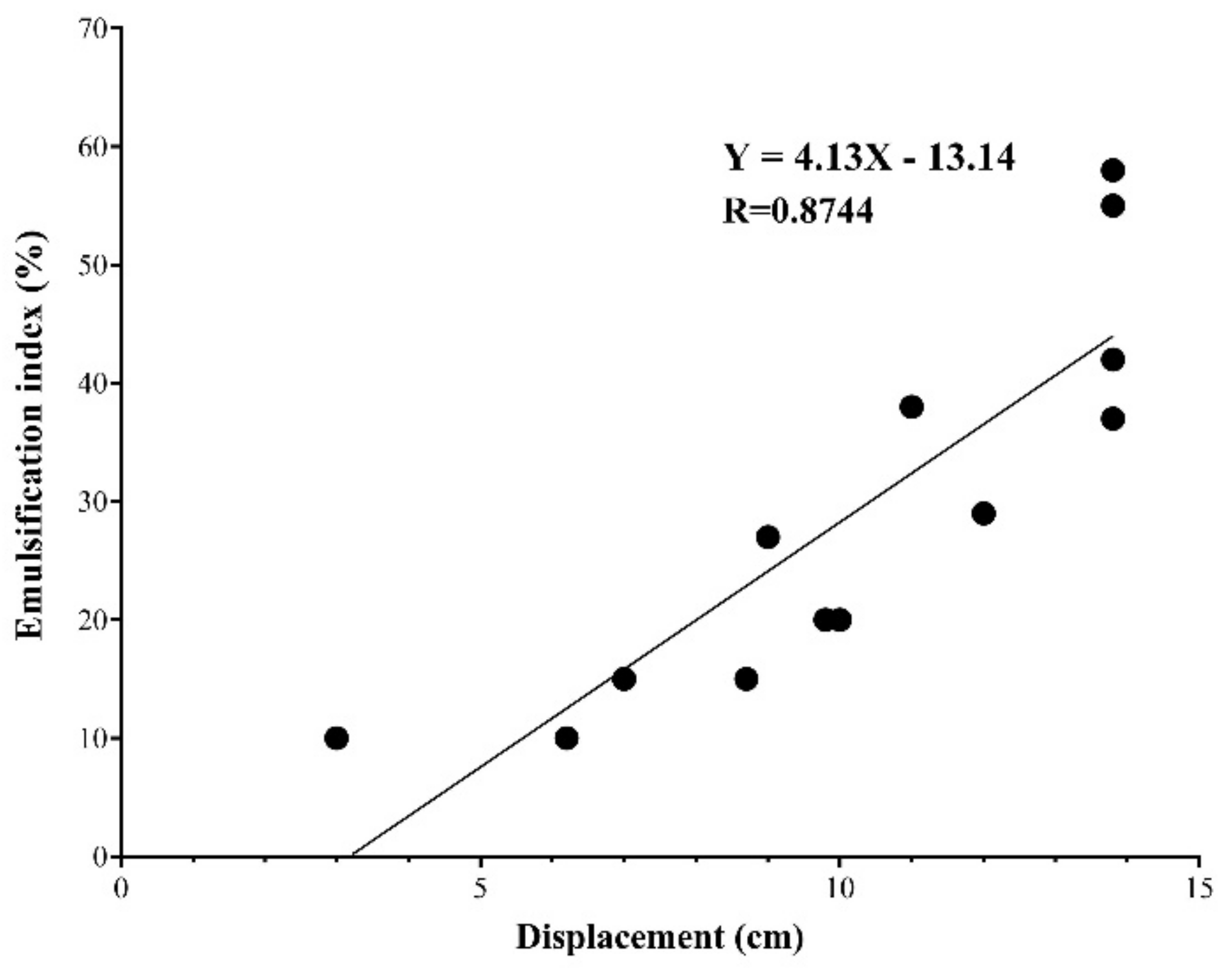
3.3. Oil Displacement and Emulsification Assays
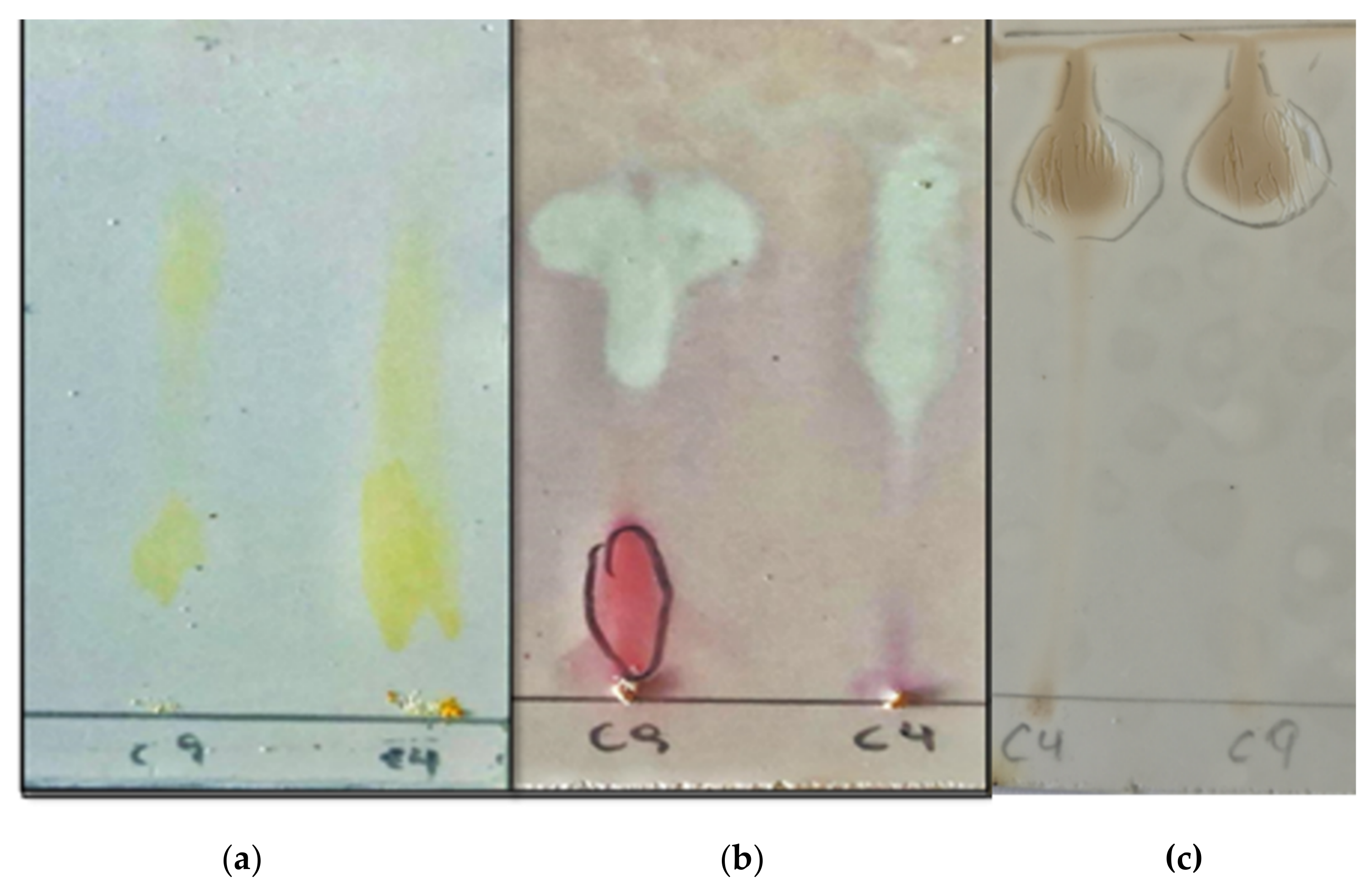
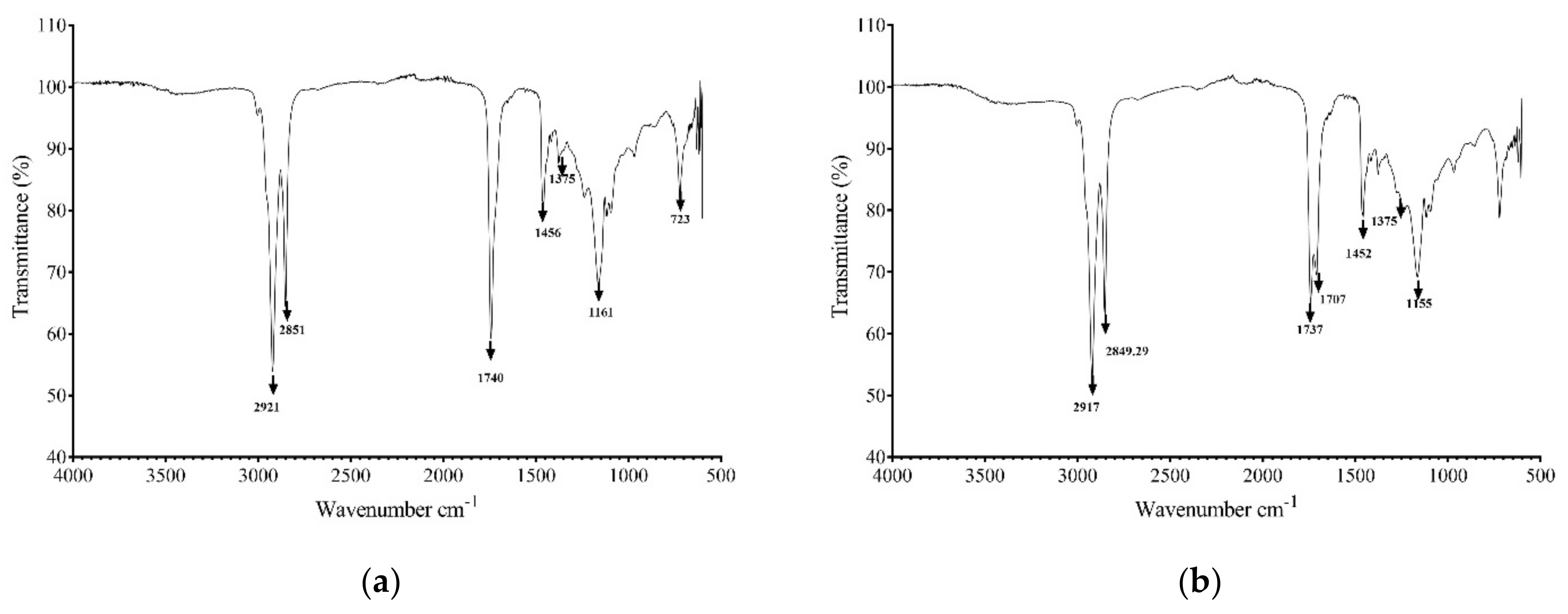
3.4. Biosurfactant Characterization
3.5. Degradation of Chlorpyrifos
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chaprão, M.J.; Ferreira, I.N.S.; Correa, P.F.; Rufino, R.D.; Luna, J.M.; Silva, E.J.; Sarubbo, L.A. Application of bacterial and yeast biosurfactants for enhanced removal and biodegradation of motor oil from contaminated sand. Electron. J. Biotechnol. 2015, 18, 471–479. [Google Scholar] [CrossRef]
- Lotfabad, T.B.; Shourian, M.; Roostaazad, R.; Najafabadi, A.R.; Adelzadeh, M.R.; Noghabi, K.A. An efficient biosurfactant-producing bacterium Pseudomonas aeruginosa MR01, isolated from oil excavation areas in south of Iran. Colloids Surf. B Biointerfaces 2009, 69, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Thavasi, R.; Jayalakshmi, S.; Banat, I.M. Application of biosurfactant produced from peanut oil cake by Lactobacillus delbrueckii in biodegradation of crude oil. Bioresour. Technol. 2011, 102, 3366–3372. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Saharan, B.S.; Chauhan, N.; Procha, S.; Lal, S. Isolation and functional characterization of novel biosurfactant produced by Enterococcus faecium. SpringerPlus 2015, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M. Biosurfactants production and possible uses in microbial enhanced oil recovery and oil pollution remediation: A review. Bioresour. Technol. 1995, 51, 1–12. [Google Scholar] [CrossRef]
- Odukkathil, G.; Vasudevan, N. Residues of endosulfan in surface and subsurface agricultural soil and its bioremediation. J. Environ. Manag. 2016, 165, 72–80. [Google Scholar] [CrossRef]
- Singh, P.; Saini, H.S.; Raj, M. Rhamnolipid mediated enhanced degradation of chlorpyrifos by bacterial consortium in soil-water system. Ecotoxicol. Environ. Saf. 2016, 134, 156–162. [Google Scholar] [CrossRef]
- Wattanaphon, H.T.; Kerdsin, A.; Thammacharoen, C.; Sangvanich, P.; Vangnai, A.S. A biosurfactant from Burkholderia cenocepacia BSP3 and its enhancement of pesticide solubilization. J. Appl. Microbiol. 2008, 105, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Mata-Sandoval, J.C.; Karns, J.; Torrents, A. Effect of nutritional and environmental conditions on the production and composition of rhamnolipids by P. aeruginosa UG2. Microbiol. Res. 2001, 155, 249–256. [Google Scholar] [CrossRef]
- García-Reyes, S.; Yáñez-Ocampo, G.; Wong-Villarreal, A.; Rajaretinam, R.K.; Thavasimuthu, C.; Patiño, R.; Ortiz-Hernández, M.L. Partial characterization of a biosurfactant extracted from Pseudomonas sp. B0406 that enhances the solubility of pesticides. Environ. Technol. 2018, 39, 2622–2631. [Google Scholar] [CrossRef]
- Singh, P.; Sharma, S.; Saini, H.; Chadha, B. Biosurfactant production by Pseudomonas sp. and its role in aqueous phase partitioning and biodegradation of chlorpyrifos. Lett. Appl. Microbiol. 2009, 49, 378–383. [Google Scholar] [CrossRef]
- Damalas, C.A.; Eleftherohorinos, I.G. Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 2011, 8, 1402–1419. [Google Scholar] [CrossRef]
- Ippolito, A.; Fait, G. Pesticides in surface waters: From edge-of-field to global modelling. Curr. Opin. Environ. Sustain. 2019, 36, 78–84. [Google Scholar] [CrossRef]
- Karanasios, E.; Karpouzas, D.G.; Tsiropoulos, N.G. Key parameters and practices controlling pesticide degradation efficiency of biobed substrates. J. Environ. Sci. Health Part B 2012, 47, 589–598. [Google Scholar] [CrossRef]
- Karas, P.A.; Perruchon, C.; Karanasios, E.; Papadopoulou, E.S.; Manthou, E.; Sitra, S.; Ehaliotis, C.; Karpouzas, D.G. Integrated biodepuration of pesticide-contaminated wastewaters from the fruit-packaging industry using biobeds: Bioaugmentation, risk assessment and optimized management. J. Hazard. Mater. 2016, 320, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Gennari, M.; Messina, C.; Abbate, C.; Baglieri, A.; Boursier, C. Solubility and adsorption behaviors of chlorpyriphos-methyl in the presence of surfactants. J. Environ. Sci. Health Part B 2009, 44, 235–240. [Google Scholar] [CrossRef]
- Sequinatto, L.; Reichert, J.M.; Dos Santos, D.R.; Reinert, D.J.; Copetti, A.C.C. Occurrence of agrochemicals in surface waters of shallow soils and steep slopes cropped to tobacco. Química Nova 2013, 36, 768–772. [Google Scholar] [CrossRef]
- Diez, M.C.; Leiva, B.; Gallardo, F. Novel insights in biopurification system for dissipation of a pesticide mixture in repeated applications. Environ. Sci. Pollut. Res. 2018, 25, 21440–21450. [Google Scholar] [CrossRef] [PubMed]
- Tortella, G.R.; Rubilar, O.; Cea, M.; Wulff, C.; Martinez, O.; Diez, M.C. Biostimulation of agricultural biobeds with npk fertilizer on chlorpyrifos degradation to avoid soil and water contamination. J. Soil Sci. Plant Nutr. 2010, 10, 464–475. [Google Scholar] [CrossRef]
- Briceño, G.; Lamilla, C.; Leiva, B.; Levio, M.; Donoso-Piñol, P.; Schalchli, H.; Gallardo, F.; Diez, M.C. Pesticide-tolerant bacteria isolated from a biopurification system to remove commonly used pesticides to protect water resources. PLoS ONE 2020, 15, e0234865. [Google Scholar] [CrossRef]
- Castillo, M.D.P.; Torstensson, L.; Stenström, J. Biobeds for environmental protection from pesticide use--a review. J. Agric. Food Chem. 2018, 56, 6206–6219. [Google Scholar] [CrossRef] [PubMed]
- Diez, M.C.; Elgueta, S.; Rubilar, O.; Tortella, G.R.; Schalchli, H.; Bornhardt, C.; Gallardo, F. Pesticide dissipation and microbial community changes in a biopurification system: Influence of the rhizosphere. Biodegradation 2017, 28, 395–412. [Google Scholar] [CrossRef]
- Fernández-Alberti, S.; Rubilar, O.; Tortella, G.R.; Diez, M.C. Chlorpyrifos degradation in a Biomix: Effect of pre-incubation and water holding capacity. J. Soil Sci. Plant Nutr. 2012, 12, 785–799. [Google Scholar] [CrossRef][Green Version]
- Sachdev, D.P.; Cameotra, S.S. Biosurfactants in agriculture. Appl. Microbiol. Biotechnol. 2013, 97, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Abouseoud, M.; Maachi, R.; Amrane, A.; Boudergua, S.; Nabi, A. Evaluation of different carbon and nitrogen sources in production of biosurfactant by Pseudomonas fluorescens. Desalination 2008, 223, 143–151. [Google Scholar] [CrossRef]
- Odukkathil, G.; Vasudevan, N. Enhanced biodegradation of endosulfan and its major metabolite endosulfate by a biosurfactant producing bacterium. J. Environ. Sci. Health Part B 2013, 48, 462–469. [Google Scholar] [CrossRef]
- Luna, J.M.; Rufino, R.D.; Sarubbo, L.A.; Campos-Takaki, G.M. Characterisation, surface properties and biological activity of a biosurfactant produced from industrial waste by Candida sphaerica UCP0995 for application in the petroleum industry. Colloids Surf. B Biointerfaces 2013, 102, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Rufino, R.D.; Luna, J.M.; Sarubbo, L.A.; Rodrigues, L.R.M.; Teixeira, J.A.C.; Campos-Takaki, G.M. Antimicrobial and anti-adhesive potential of a biosurfactant rufisan produced by Candida lipolytica UCP 0988. Colloids Surf. B Biointerfaces 2011, 84, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Rostás, M.; Blassmann, K. Insects had it first: Surfactants as a defence against predators. Proc. R. Soc. B Boil. Sci. 2009, 276, 633–638. [Google Scholar] [CrossRef]
- Awashti, N.; Kumar, A.; Makkar, R.C.S. A chlorinated pesticide in presence of biosurfactant. Environ. Sci. Health B 1999, 34, 793–803. [Google Scholar]
- Lotfabad, T.B.; Ebadipour, N.; Roostaazad, R.; Partovi, M.; Bahmaei, M. Two schemes for production of biosurfactant from Pseudomonas aeruginosa MR01: Applying residues from soybean oil industry and silica sol-gel immobilized cells. Colloids Surf. B Biointerfaces 2017, 152, 159–168. [Google Scholar] [CrossRef]
- Chakraborty, S.; Ghosh, M.; Chakraborti, S.; Jana, S.; Sen, K.K.; Kokare, C.; Zhang, L. Biosurfactant produced from Actinomycetes nocardiopsis A17: Characterization and its biological evaluation. Int. J. Biol. Macromol. 2015, 79, 405–412. [Google Scholar] [CrossRef]
- Song, B.; Springer, J. Determination of interfacial tension from the profile of a pendant drop using computer-aided image processing. J. Colloid Interface Sci. 1996, 184, 77–91. [Google Scholar] [CrossRef]
- Abbasi, H.; Sharafi, H.; Alidost, L.; Bodagh, A.; Zahiri, H.; Noghabi, K. Response surface optimization of biosurfactant produced by Pseudomonas aeruginosa MA01 isolated from spoiled apples. Prep. Biochem. Biotechnol. 2013, 43, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.M.; Mody, K.; Joshi, N.; Mishra, A.; Jha, B. Production and structural characterization of biosurfactant produced by an alkaliphilic bacterium, Klebsiella sp.: Evaluation of different carbon sources. Colloids Surf. B Biointerfaces 2013, 108, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Haloi, S.; Sarmah, S.; Gogoi, S.B.; Medhi, T. Characterization of Pseudomonas sp. TMB2 produced rhamnolipids for ex-situ microbial enhanced oil recovery. 3 Biotech 2020, 10, 120. [Google Scholar] [CrossRef]
- Pacheco, G.J.; Ciapina, E.M.P.; Gomes, E.D.B.; Pereira, N., Jr. Biosurfactant production by Rhodococcus rythropolis and its application to oil removal. Braz. J. Microbiol. 2010, 41, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Thavasi, R.; Nambaru, V.R.M.S.; Jayalakshmi, S.; Balasubramanian, T.; Banat, I.M. Biosurfactant production by Pseudomonas aeruginosa from renewable resources. Indian J. Microbiol. 2011, 51, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Rosen, M.J. Emulsification by surfactants. In Surfactants and Interfacial Phenomena; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2004; pp. 303–331. ISBN 9780471670568. [Google Scholar]
- Abouseoud, M.; Maachi, R.; Amrane, A. Biosurfactant production from olive oil by Pseudomonas fluorescens. Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 1, 340–347. [Google Scholar]
- Hassen, W.; Neifar, M.; Cherif, H.; Mahjoubi, M.; Souissi, Y.; Raddadi, N.; Fava, F.; Cherif, A. Assessment of genetic diversity and bioremediation potential of pseudomonads isolated from pesticide-contaminated artichoke farm soils. 3 Biotech 2018, 8, 263. [Google Scholar] [CrossRef] [PubMed]
- Leite, G.G.F.; Figueirôa, J.V.; Almeida, T.C.M.; Valões, J.L.; Marques, W.F.; Duarte, M.D.D.C.; Gorlach-Lira, K. Production of rhamnolipids and diesel oil degradation by bacteria isolated from soil contaminated by petroleum. Biotechnol. Prog. 2016, 32, 262–270. [Google Scholar] [CrossRef]
- Nurfarahin, A.H.; Mohamed, M.S.; Phang, L.Y. Culture medium development for microbial-derived surfactants production-an overview. Molecules 2018, 23, 1049. [Google Scholar] [CrossRef]
- Viramontes-Ramos, S.; Portillo-Ruiz, M.C.; Ballinas-Casarrubias, M.D.L.; Torres-Muñoz, J.V.; Rivera-Chavira, B.E.; Nevárez-Moorillón, G.V. Selection of biosurfactan/bioemulsifier-producing bacteria from hydrocarbon-contaminated soil. Braz. J. Microbiol. 2010, 41, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Belcher, R.W.; Huynh, K.V.; Hoang, T.V.; Crowley, D.E. Isolation of biosurfactant-producing bacteria from the Rancho La Brea Tar Pits. World J. Microbiol. Biotechnol. 2012, 28, 3261–3267. [Google Scholar] [CrossRef]
- Rocha, V.A.L.; de Castilho, L.V.A.; de Castro, R.P.V.; Teixeira, D.B.; Magalhães, A.V.; Gomez, J.G.C.; Freire, D.M.G. Comparison of mono-rhamnolipids and di-rhamnolipids on microbial enhanced oil recovery (MEOR) applications. Biotechnol. Prog. 2020, 36, e2981. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.K.F.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional Biomolecules of the 21st Century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Tiwary, B.N. Isolation and characterization of glycolipid biosurfactant produced by a Pseudomonas otitidis strain isolated from Chirimiri coal mines, India. Bioresour. Bioprocess. 2016, 3, 42. [Google Scholar] [CrossRef]
- Angelini, R.; Babudri, F.; Lobasso, S.; Corcelli, A. MALDI-TOF/MS analysis of Archaebacterial lipids in lyophilized membranes dry-mixed with 9-aminoacridine. J. Lipid Res. 2010, 51, 2818–2825. [Google Scholar] [CrossRef]
- Diez, M.C.; Levio, M.; Briceño, G.; Rubilar, O.; Tortella, G.; Gallardo, F. Biochar as a partial replacement of peat in pesticide-degrading biomixtures formulated with different soil types. J. Biobased Mater. Bioenergy 2013, 7, 741–747. [Google Scholar] [CrossRef]
- Elgueta, S.; Correa, A.; Campo, M.; Gallardo, F.; Karpouzas, D.; Diez, M.C. Atrazine, chlorpyrifos, and iprodione effect on the biodiversity of bacteria, Actinomycetes, and fungi in a pilot biopurification system with a green cover. J. Environ. Sci. Health Part B 2017, 52, 651–657. [Google Scholar] [CrossRef]
- Tortella, G.R.; Rubilar, O.; Castillo, M.D.P.; Cea, M.; Mella-Herrera, R.; Diez, M.C. Chlorpyrifos degradation in a biomixture of biobed at different maturity stages. Chemosphere 2012, 88, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, C.; Peng, C.; Liu, H.; Hu, M.; Zhong, G. Biodegradation of Chlorpyrifos and Its Hydrolysis Product 3,5,6-Trichloro-2-Pyridinol by a New Fungal Strain Cladosporium cladosporioides Hu-01. PLoS ONE 2012, 7, e47205. [Google Scholar] [CrossRef] [PubMed]
- Chishti, Z.; Hussain, S.; Arshad, K.R.; Khalid, A.; Arshad, M. Microbial degradation of chlorpyrifos in liquid media and soil. J. Environ. Manag. 2013, 114, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Briceño, G.; Fuentes, M.S.; Palma, G.; Jorquera, M.A.; Amoroso, M.J.; Diez, M.C. Chlorpyrifos biodegradation and 3,5,6-trichloro-2-pyridinol production by Actinobacteria isolated from soil. Int. Biodeterior. Biodegrad. 2012, 73, 1–7. [Google Scholar] [CrossRef]

| N° | Bacteria | Strain 1 | CTAB | Emulsification (%) | Displacement (cm) | Drop-Collapse |
|---|---|---|---|---|---|---|
| 1 | P. rhodesiae | C4 | + | 55 | 14 ± 0.1 | + |
| 2 | A. deleyi | C7 | – | 48 | 12 ± 1.1 | + |
| 3 | R. jialingiae | C8 | + | 54 | 13 ± 1.5 | + |
| 4 | P. marginalis | C9 | + | 49 | 14 ± 1.2 | + |
| 5 | A. kerstersii | C10 | – | 53 | 1 ± 0.5 | – |
| Control (+) | – | 76 | 14 ± 0.0 | + | ||
| Control (–) | – | 0 | 0 ± 0.0 | – |
| CFS Addition (%) | Residual CHL (mg L−1) | Degradation (%) | Increment * (%) | TCP (mg L−1) |
|---|---|---|---|---|
| 0 (biotic control) | 15.22±0.13 | 39.3 c | 0 | 0.51± 0.01 |
| 2.5 | 13.37±0.02 | 46.7 a,b | 15.8 | 0.53 ±0.02 |
| 5.0 | 12.77±0.02 | 49.1 a | 19.9 | 0.56± 0.01 |
| 7.5 | 12.46±0.05 | 50.4 a | 22.0 | 0.82±0.06 |
| 10 | 12.15±0.04 | 51.6 a | 23.8 | 0.90±0.01 |
| 5 (abiotic control) | 24.92±0.07 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamilla, C.; Schalchli, H.; Briceño, G.; Leiva, B.; Donoso-Piñol, P.; Barrientos, L.; Rocha, V.A.L.; Freire, D.M.G.; Diez, M.C. A Pesticide Biopurification System: A Source of Biosurfactant-Producing Bacteria with Environmental Biotechnology Applications. Agronomy 2021, 11, 624. https://doi.org/10.3390/agronomy11040624
Lamilla C, Schalchli H, Briceño G, Leiva B, Donoso-Piñol P, Barrientos L, Rocha VAL, Freire DMG, Diez MC. A Pesticide Biopurification System: A Source of Biosurfactant-Producing Bacteria with Environmental Biotechnology Applications. Agronomy. 2021; 11(4):624. https://doi.org/10.3390/agronomy11040624
Chicago/Turabian StyleLamilla, Claudio, Heidi Schalchli, Gabriela Briceño, Bárbara Leiva, Pamela Donoso-Piñol, Leticia Barrientos, Vanessa A. L. Rocha, Denise M. G. Freire, and M. Cristina Diez. 2021. "A Pesticide Biopurification System: A Source of Biosurfactant-Producing Bacteria with Environmental Biotechnology Applications" Agronomy 11, no. 4: 624. https://doi.org/10.3390/agronomy11040624
APA StyleLamilla, C., Schalchli, H., Briceño, G., Leiva, B., Donoso-Piñol, P., Barrientos, L., Rocha, V. A. L., Freire, D. M. G., & Diez, M. C. (2021). A Pesticide Biopurification System: A Source of Biosurfactant-Producing Bacteria with Environmental Biotechnology Applications. Agronomy, 11(4), 624. https://doi.org/10.3390/agronomy11040624










