Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust
Abstract
:1. Introduction
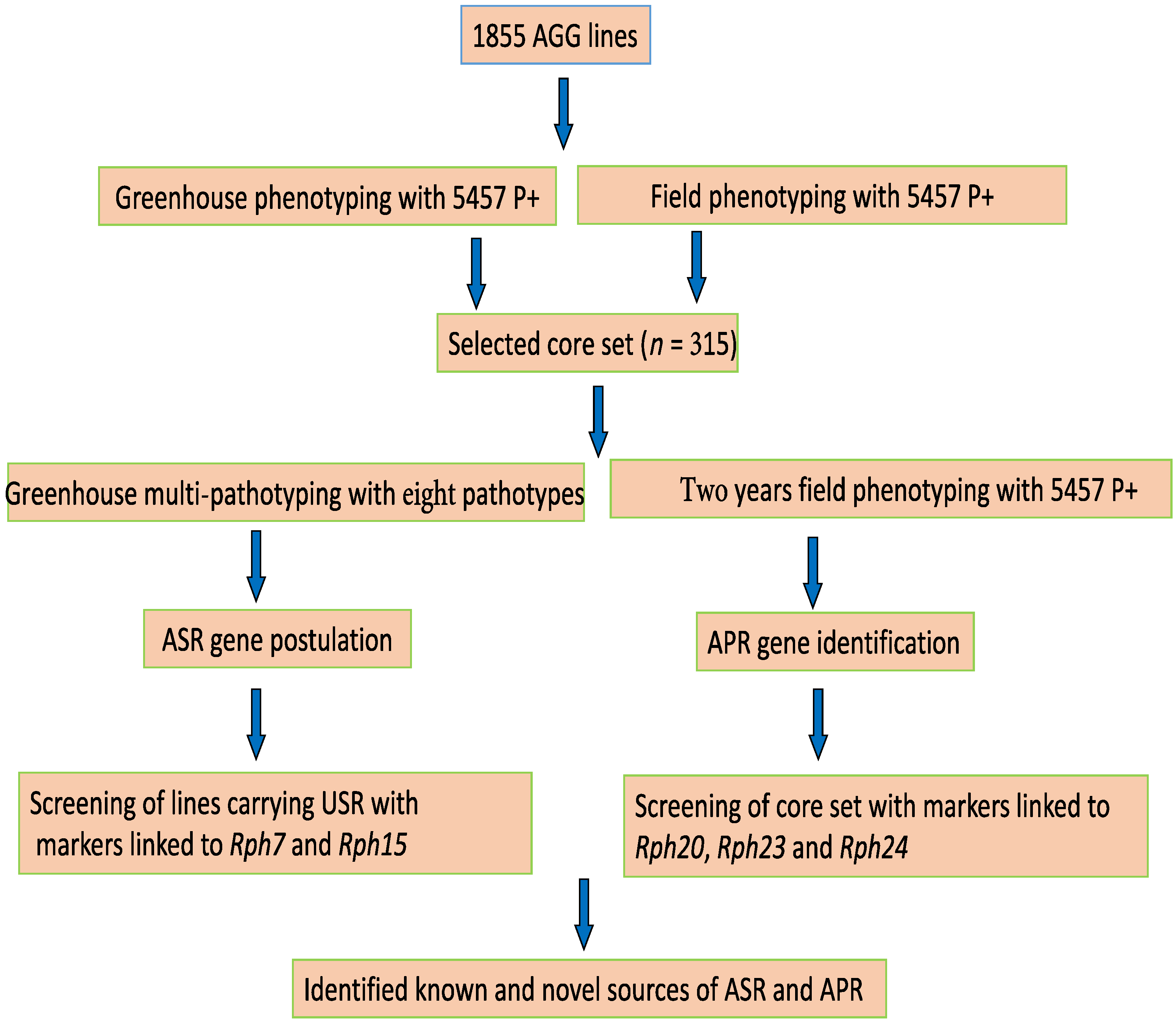
2. Materials and Methods
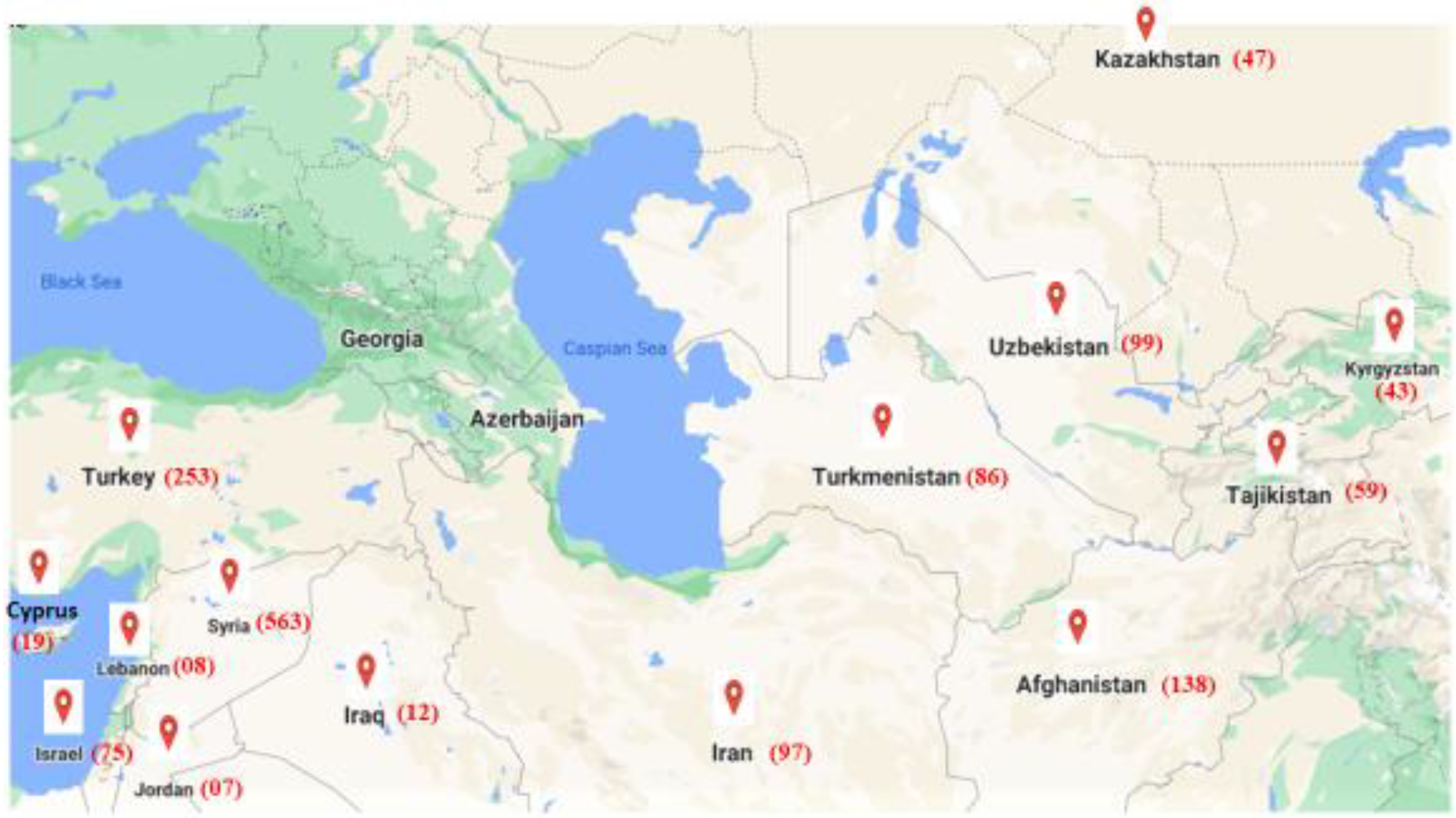
2.1. Plant Materials
2.2. Pathogen Isolates
2.3. Seedling Tests in the Greenhouse
Disease Scoring
2.4. Adult Plant Tests in the Field
Disease Scoring
2.5. Marker Genotyping
2.5.1. DNA Extraction
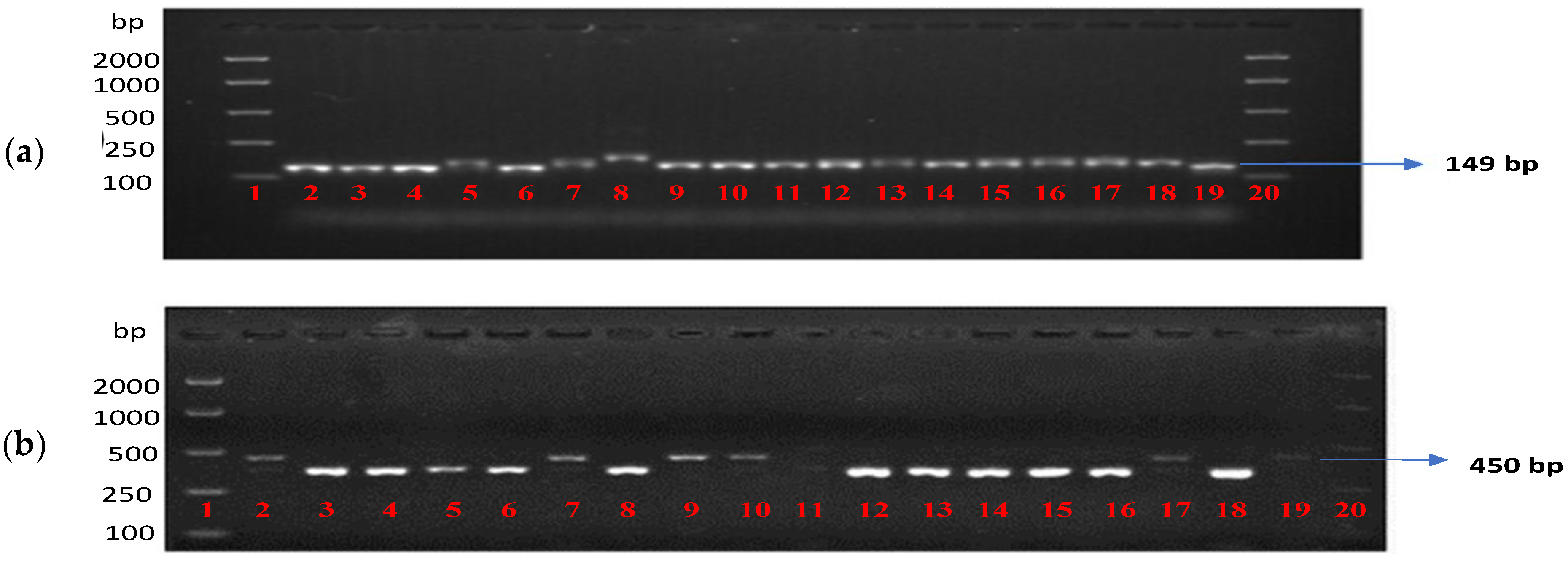
2.5.2. Genotyping with Markers for the APR Genes Rph20, Rph23 and Rph24
2.5.3. Genotyping with the Rph7 and Rph15 Markers
3. Results
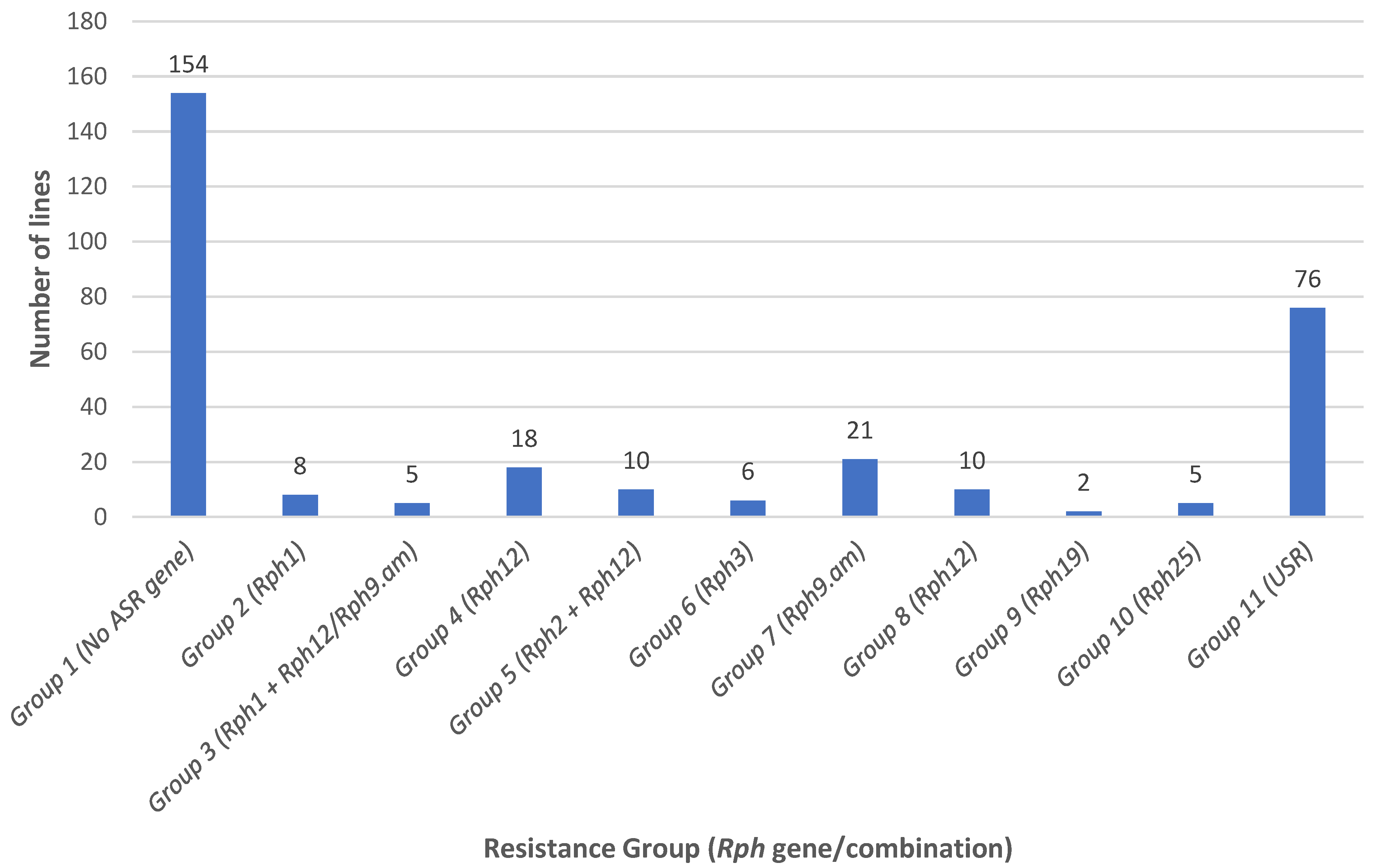
3.1. ASR Gene Postulation and Marker Analysis
3.2. Characterization of APR and Marker Analysis
3.2.1. Group A
3.2.2. Group B
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Von Bothmer, R. The Gene Pool of Barley and Preservation of Wild Species of Hordeum; IBPGR: Rome, Italy, 1992; Volume 5, pp. 32–35. [Google Scholar]
- FAOSTAT. Food and Agriculture Organization of the United Nations. Available online: www.fao.ogr/faostat (accessed on 10 May 2019).
- Clifford, B.C. Barley leaf rust. In The Cereal Rusts II; Roelfs, A.P., Bushnell, W.R., Eds.; Academic Press: New York, NY, USA, 1985; pp. 173–205. [Google Scholar]
- Park, R.F.; Golegaonkar, P.G.; Derevnina, L.; Sandhu, K.S.; Karaoglu, H.; Elmansour, H.M.; Singh, D. Leaf rust of cultivated barley pathology and control. Annu. Rev. Phytopathol. 2015, 53, 565–589. [Google Scholar] [PubMed]
- Cotterill, P.J.; Rees, R.G.; Platz, G.J.; Dill-Macky, R. Effects of leaf rust on selected Australian barleys. Aust. J. Exp. Agric. 1992, 32, 747–751. [Google Scholar] [CrossRef]
- Van Der Plank, J.E. Disease Resistance in Plants; Academic Press: New York, NY, USA, 1968; 210p. [Google Scholar]
- Parlevliet, J.E. Race–specific aspects of polygenic resistance of barley to leaf rust, Puccinia hordei. Neth. J. Plant Pathol. 1978, 84, 121–126. [Google Scholar] [CrossRef]
- Gupta, S.; Vassos, E.; Sznajder, B.; Fox, R.; Khoo, K.H.; Loughman, R.; Chalmers, K.J.; Mather, D.E. A locus on barley chromosome 5H affects adult plant resistance to powdery mildew. Mol. Breed. 2018, 38, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavanagh, P.J.; Singh, D.; Bansal, U.K.; Park, R.F. Inheritance and characterization of the new and rare gene Rph25 conferring seedling resistance in Hordeum vulgare against Puccinia hordei. Plant Breed. 2017, 136, 908–912. [Google Scholar] [CrossRef]
- Yu, X.; Kong, H.Y.; Meiyalaghan, V.; Casonato, S.; Chng, S.; Jones, E.E.; Johnston, P.A. Genetic mapping of a barley leaf rust resistance gene Rph26 introgressed from Hordeum bulbosum. Theor. Appl. Gen. 2018, 131, 2567–2580. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, C.T.; Singh, D.; Dracatos, P.M.; Park, R.F. Inheritance and characterization of Rph27: A third race–specific resistance gene in the barley cultivar quinn. Phytopathology 2020, 110, 1067–1073. [Google Scholar] [CrossRef]
- Mehnaz, M.; Dracatos, P.; Pham, A.; March, T.; Maurer, A.; Pillen, K.; Forrest, K.; Kulkarni, T.; Pourkheirandish, M.; Park, R.F.; et al. Discovery and fine mapping of Rph28: A new gene conferring resistance to Puccinia hordei from wild barley. Theor. Appl. Gen. 2021, 134, 2167–2179. [Google Scholar] [CrossRef] [PubMed]
- Hickey, L.T.; Lawson, W.; Platz, G.J.; Dieters, M.; Arief, V.N.; German, S.; Fletcher, S.; Park, R.F.; Singh, D.; Pereyra, S.; et al. Mapping Rph20: A gene conferring adult plant resistance to Puccinia hordei in barley. Theor. Appl. Gen. 2011, 123, 55–68. [Google Scholar] [CrossRef]
- Singh, D.; Dracatos, P.M.; Derevnina, L.; Zhou, M.; Park, R.F. Rph23: A new designated additive adult plant resistance gene to leaf rust in barley on chromosome 7H. Plant Breed. 2015, 134, 62–69. [Google Scholar] [CrossRef]
- Ziems, L.A.; Hickey, L.T.; Platz, G.J.; Franckowiak, J.D.; Dracatos, P.M.; Singh, D.; Park, R.F. Characterization of Rph24: A gene conferring adult plant resistance to Puccinia hordei in barley. Phytopathology 2017, 107, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Singh, D.; Dracatos, P.M.; Loughman, R.; Park, R.F. Genetic mapping of resistance to Puccinia hordei in three barley doubled-haploid populations. Euphytica 2017, 213, 1–16. [Google Scholar] [CrossRef]
- Gilmour, J. Octal notation for designating physiologic races of plant pathogens. Nature 1973, 242, 620. [Google Scholar] [CrossRef]
- Park, R.F.; Poulsen, D.; Barr, A.R.; Cakir, M.; Moody, D.B.; Raman, H.; Read, B.J. Mapping genes for resistance to Puccinia hordei in barley. Aust. J. Agric. Res. 2003, 54, 1323–1333. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, R.A.; Wellings, C.R.; Park, R.F. Wheat Rusts: An Atlas of Resistance Genes; CSIRO: Canberra, Australia, 1995. [Google Scholar]
- McNeal, F.M.; Konzak, C.F.; Smith, E.P.; Tate, W.S.; Russell, T.S. A system for recording and processing cereal research data. US Agric. Res. Serv. 1971, 42, 34–121. [Google Scholar]
- Fulton, T.M.; Chunwongse, J.; Tanksley, S.D. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Bio. Rep. 1995, 13, 207–209. [Google Scholar] [CrossRef]
- Dracatos, P.M.; Park, R.F.; Singh, D. Validating molecular markers for barley leaf rust resistance genes Rph20 and Rph24. Plant Dis. 2021, 105, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Jost, M.; Clark, B.; Martin, M.; Matny, O.; Steffenson, B.J.; Franckowiak, J.D.; Mascher, M.; Singh, D.; Perovic, D.; et al. BED-domain-containing immune receptor from wild barley confers widely effective resistance to leaf rust. Plant Biotech. J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Park, R.F. Pathogenic specialization and pathotype distribution of Puccinia hordei in Australia, 1992 to 2001. Plant Dis. 2003, 87, 1311–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrell, P.L.; Clegg, M.T. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 2007, 104, 3289–3294. [Google Scholar] [CrossRef] [Green Version]
- Golegaonkar, P.G.; Singh, D.; Park, R.F. Evaluation of seedling and adult plant resistance to Puccinia hordei in barley. Euphytica 2009, 166, 183–197. [Google Scholar] [CrossRef]
- Derevnina, L.; Singh, D.; Park, R.F. Identification and characterization of seedling and adult plant resistance to Puccinia hordei in Chinese barley germplasm. Plant Breed. 2013, 132, 571–579. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Forrest, K.L.; Kong, S.; Bansal, U.K.; Singh, D.; Hayden, M.J.; Park, R.F. Inheritance and molecular mapping of a gene conferring seedling resistance against Puccinia hordei in the barley cultivar Ricardo. Theor. Appl. Gen. 2012, 125, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Elmansour, H.; Singh, D.; Dracatos, P.M.; Park, R.F. Identification and characterization of seedling and adult plant resistance to Puccinia hordei in selected African barley germplasm. Euphytica 2017, 213, 119. [Google Scholar] [CrossRef]
- Singh, D.; Ziems, L.A.; Dracatos, P.M.; Pourkheirandish, M.; Tshewang, S.; Czembor, P.; German, S.; Fowler, R.A.; Snyman, L.; Platz, G.J.; et al. Genome-wide association studies provide insights on genetic architecture of resistance to leaf rust in a worldwide barley collection. Mol. Breed. 2018, 38, 43. [Google Scholar] [CrossRef]
- Dracatos, P.M.; Singh, D.; Bansal, U.; Park, R.F. Identification of new sources of adult plant resistance to Puccinia hordei in international barley (Hordeum vulgare L.) germplasm. Eur. J. Plant Pathol. 2015, 141, 463–476. [Google Scholar] [CrossRef]
- Singh, D.; Dracatos, P.; Mehnaz, M.; Park, R.F. Australian barley cultivar pedigree and leaf rust seedling and adult plant resistance genotype information. Cereal Rust Report PBI. 2020, 17, 1–9. [Google Scholar]


| Host Response | Abbreviation | Infection Type | Disease Symptoms/Description |
|---|---|---|---|
| Resistant | R | 1 | No uredinia or flecking |
| Resistant or moderately resistant | RMR | 2 | No uredinia but flecking may be present |
| Moderately resistant | MR | 3 | Traces of uredinia without sporulation |
| Moderately resistant or moderately susceptible | MRMS | 4 | Small uredinia with restricted sporulation |
| Moderately susceptible | MS | 5 | Small or medium-sized uredinia with moderate sporulation |
| Moderately susceptible or susceptible | MSS | 6 | Medium-sized uredinia with heavy sporulation |
| Susceptible | S | 7 | Large-sized uredinia with abundant sporulation |
| Susceptible or very susceptible | SVS | 8 | Large-sized coalesced uredinia with abundant sporulation |
| Very susceptible | VS | 9 | Large-sized coalesced uredinia with abundant sporulation and lesions |
| Markers | Gene | Forward Primer Sequence 5′–3′ | Reverse Primer Sequence 5′–3′ | Reference |
|---|---|---|---|---|
| bPb-0837 | Rph20 | GACACTTCGTGCCAGTTTG | CCTCCCTCCCTCTTCTCAAC | [13] |
| Ebmac0603 | Rph23 | ACCGAAACTAAATGAACTACTTCG | TGCAAACTGTGCTATTAAGGG | [14] |
| sun43-44 | Rph24 | CTAGACACCACCACCACACC | ATACCAGAGTTTGCGTCCGG | [22] |
| Unknown | Rph7 | GAGATAAAAGCATTACCAAAGGCTCAT | GCGCGCGCAACAGCAAACGGC | Unpublished |
| Unknown | Rph15 | TGAAGAAGCTGGAAGGTCACC | AGCCAAAAACCCTTCTGGCT | [23] |
| Groups | Rph Genes | Pathotypes | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 200 P− | 220 P+ | 253 P− | 5652 P+ | 5610 P+ | 5453 P− | 5457 P− | 5457 P+ | ||
| 1 | No R genes | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ |
| 2 | Rph1 | ;N to ;+CN | ;N to ;N=C | ;1CN to 3+ | ;CN to ;=C | ;N to ;1=CN | 3+ | 3+ | 23- to 3+ |
| 3 | Rph1 + Rph9.am | ;N to ;1+CN | ;1-CN to ;12CN | ;1-CN to ;12C | ;1-CN to 23-CN | ;N to ;1-CN | 33-C to 3+ | 23C to 3+ | 23-C to 3+ |
| 4 | Rph2 | ;1+N to ;12C | ;N to ;12C | 3+ | 3 to 3+ | ;1 to ;12CN | 33+ to 3+ | 3+ | 3+ |
| 5 | Rph2 + Rph12 | ;N to ;12C | ;1 to ;12C | ;1C to ;12+C | 3+ | ;1N to ;12CN | 3+ | 3+ | 3+ |
| 6 | Rph3 | 0; to ;1+CN | 0; to ;1-CN | 0; to ;1+CN | 0; to ;1-CN | ;C to ;1+CN | ;C to ;1CN | 3+C to 3+ | 3+C to 3+ |
| 7 | Rph9.am | 3+ | 3+ | ;1C to ;12C | 3+ | 3+ | 3C to 3+ | 3C to 3+ | 3+ |
| 8 | Rph12 | ;1C to ;12+C | ;1C to ;12+C | ;1C to 12+C | 3 to 3+ | 33+C to 3+ | 3- to 3+ | 3 to 3+ | 3 to 3+ |
| 9 | Rph19 | ;1 to ;12C | 3+ | ;1+CN to ;12 | 3+ | 3+ | ;1+C to ;12CN | ;12 to;12+ | 3+ |
| 10 | Rph25 | 33+ to 3+ | ;1-CN to ;12+C | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ |
| 11 | USR # | 0; to 2-C | 0; to 3-C | ;C to 2C | 0; to 23-C | ;N to 2C | ; to 3-C | 0; to 23C | ;N to 2+3+C |
| 1 Gus | No R genes | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ |
| 2 Sudan | Rph1 | ;N | ;+N | 3+ | ;+N | ;-N | 3+ | 3+ | 3+ |
| 3 Peruvian | Rph2 | ;1-N | ;1-N | 3- | 3+ | ;1+CN | 3+ | 3+C | 3+C |
| 4 Estate | Rph3 | ;C | 0; | 0; | 0;C | ;-CN | ;1- | 3+C | 3+C |
| 5 Cantala | Rph9.am | 3+ | 3+ | 12-C | 3+ | 3+ | 3c | 3+c | 3+c |
| 6 Triumph | Rph12 | ;+N | ;+N | ;1CN | 3+ | 3 | 3+ | 3+ | 3+ |
| 7 Prior | Rph19 | ;1 | 33+ | ;1-CN | 3+ | 33+ | 12- | 12- | 3+ |
| 8 Fong Tien | Rph25 | 3+ | 12=CN | 3+ | 3+ | 3+ | 3+ | 3+ | 3+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehnaz, M.; Dracatos, P.M.; Park, R.F.; Singh, D. Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust. Agronomy 2021, 11, 2146. https://doi.org/10.3390/agronomy11112146
Mehnaz M, Dracatos PM, Park RF, Singh D. Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust. Agronomy. 2021; 11(11):2146. https://doi.org/10.3390/agronomy11112146
Chicago/Turabian StyleMehnaz, Mehnaz, Peter M. Dracatos, Robert F. Park, and Davinder Singh. 2021. "Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust" Agronomy 11, no. 11: 2146. https://doi.org/10.3390/agronomy11112146
APA StyleMehnaz, M., Dracatos, P. M., Park, R. F., & Singh, D. (2021). Mining Middle Eastern and Central Asian Barley Germplasm to Understand Diversity for Resistance to Puccinia hordei, Causal Agent of Leaf Rust. Agronomy, 11(11), 2146. https://doi.org/10.3390/agronomy11112146







