Abstract
Wheat is an essential constituent of cereal-based diets, and one of the most significant sources of calories. However, modern wheat varieties are low in proteins and minerals. Biofortification is a method for increasing the availability of essential elements in the edible portions of crops through agronomic or genetic and genomic interventions. Wheat biofortification, as a research topic, has become increasingly prevalent. Recent accomplishments in genomic biofortification could potentially be helpful for the development of biofortified wheat grains, as a sustainable solution to the issue of “hidden hunger”. Genomic interventions mainly include quantitative trait loci (QTL) mapping, marker-assisted selection (MAS), and genomic selection (GS). Developments in the identification of QTL and in the understanding of the physiological and molecular bases of the QTLs controlling the biofortification traits in wheat have revealed new horizons for the improvement of modern wheat varieties. Markers linked with the QTLs of desirable traits can be identified through QTL mapping, which can be employed for MAS. Besides MAS, a powerful tool, GS, also has great potential for crop improvement. We have compiled information from QTL mapping studies on wheat, carried out for the identification of the QTLs associated with biofortification traits, and have discussed the present status of MAS and different prospects of GS for wheat biofortification. Accelerated mapping studies, as well as MAS and GS schemes, are expected to improve wheat breeding efficiency further.
1. Introduction
Malnutrition impacts more than two billion individuals, and Asia and Africa are the most affected regions [1]. Biofortification is an approach for improving the levels of vital ingredients, like vitamins, minerals, and proteins, in the edible portions of crops through conventional breeding, as well as biotechnological and genomics approaches [2,3]. While minerals and vitamins are commonly provided as dietary supplements, they are out of reach for most people living in the developing world [4,5]. Biofortification is a one-time expense that provides a cost-effective, long-term, and sustainable method for combating concealed starvation [6]. Much of the population is dependent on cereals for their dietary requirements; therefore, the biofortification of cereals is essential [7]. Wheat is globally traded more than any other crop, and it is the second most-produced cereal, following maize [8]. Moreover, annual wheat production has almost tripled since the 1940s, and it is anticipated that this trend will continue, as the population continues to grow [9]. Biofortified wheat could help reduce starvation-related malnutrition, with which primarily low-income countries are faced [10].
A large variation in grain iron and zinc concentrations is found in the wild relatives, like Aegilops tasuchii, of wheat and continues to be exploited for the enhancement of modern elite cultivars [11]. Provitamin A is an additional essential nutrient, and it is promoted in wheat via breeding through biofortification. Some breeds of durum wheat are higher in their provitamin concentration and yellow pigment content material, due to the presence of carotenoids (xanthophyll and lutein), and this is an essential trait for improving the antioxidant content in wheat cultivars [12,13,14]. Similarly, the enhancement of the anthocyanin content in wheat is an important focus of wheat biofortification programs. Colored wheat (black, blue, and purple), due to the high concentration of phenolics, is utilized in several breeding programs, and certain varieties of it have already been released in several nations [3,15,16].
Several agronomic methods can lead to wheat biofortification [17]. However, augmenting mineral concentrations exclusively via agronomic practices, such as foliar sprays, is associated with high expenses for farmers [18]. This agronomic biofortification is a short-term solution to increasing micronutrient (especially Fe and Zn) availability. As these fertilizers are expensive, they cannot be accessed by resource-poor farmers of low-income countries. In contrast, genetic biofortification is considered a more sustainable solution for the long term. Next-generation sequencing is proving to be useful in the determination of precise information on cultivated crops. Moreover, developments in next-generation sequencing and statistical methods can aid in the identification of the regions within the wheat genome that is responsible for its higher mineral and other biofortification traits [19,20,21,22,23]. Recommended dietary allowances (RDAs) for important minerals, like iron, zinc, and selenium, are given in Table 1.

Table 1.
Recommended dietary allowances (RDAs) for iron, zinc, and selenium [24].
Moreover, this is a cost-effective approach and is known to have practical values. Biofortification by breeding is accomplished in crops when the genetic variability of a crop is readily accessible from its primary, secondary, or tertiary gene pool. Wheat has a large number of underexploited wild relatives that could contribute to the genetic improvement of wheat [25,26].
Qualitative traits are generally governed by a single major gene, whereas, traits, like yield, are quantitative in nature and are, therefore, considered to be governed by several genes [27,28,29]. These qualitative traits are easier to breed than quantitative traits through conventional breeding methods. Conventional breeding, mainly backcross breeding with participatory varietal selection (PVS), has played a significant role in wheat biofortification. Participatory varietal selection (PVS) is particularly used by farmers to identify varieties with better performance. Farmers evaluate multiple traits that are important to them and encourage them through breeding in order to achieve quicker varietal development. The genomic resources of wheat have provided important support for functional genomics and conservation biology (by conserving the important landraces) [30,31]. The wheat genome is difficult to interpret, because it has broadly dispersed repetitive sequences, heterozygosity, and polyploidy [32,33]. Nevertheless, developments in sequencing methodologies, the decreased sequencing price, and advancements in computational resources have permitted the spread of these resources [34,35]. Besides, the comparative genomics of plant species is proving to be an efficient method in the identification of novel genes with respect to the biofortification of modern wheat [36].
Various genomic approaches, such as quantitative trait loci (QTL) mapping, marker-assisted selection (MAS) and genomic selection, have been widely employed for the biofortification of wheat. There are several techniques for mapping QTL in an experimental cross [37]. The molecular basis of QTLs is challenging to dissect, even for model plants, like Arabidopsis and rice, because of the problems in precisely narrowing intervals to single genes [38]. The experimental design, type of plant population analyzed, and the level of polymorphisms between parental genomes also affect predictions of QTLs. Statistical methods for determining quantitative trait loci (QTL) require numerous molecular markers, with high-resolution genetic maps [39,40]. This approach is one of the major genomics methods that is geared toward the dissection of complex phenotypes [41]. Several QTL mapping studies have resulted in the identification of a number of stable and consistent QTLs, which can be helpful in elucidating the genetic basis of biofortification traits. Various mapping populations, such as doubled haploid lines (DH), recombinant inbred lines (RILs) or single seed descent lines (SSDs), F2-derived F3 (F2:3) or F4 (F2:4), the BC3F2:3 population, and BC5F2:F6 families have been used in mapping QTLs for biofortification traits (Table 2, Table 3 and Table 4). All of these populations have their own advantages and limitations [42]. The polyploidy level of wheat has allowed researchers to study some specific homozygous populations, such as recombinant inbred chromosome lines (RICLs) and recombinant substitution lines (RSLs) [43,44,45]. Disomic inter-varietal chromosome (DIC) substitution lines have been used in the development of recombinant inbred chromosome lines (RICLs) for chromosomes 6B and 5B, with the Triticum turgidum (L.) var. LDN background. Each of the lines was homozygous for a particular recombined or unrecombined chromosome, and near-isogenic to LDN durum for all of the other remaining chromosomes. These populations served as the best genetic stocks for the precise mapping of the GPC gene/QTLs [43,44] (Table 2). An important locus, Gpc-6B1, has also been fine mapped (1.5cM proximal) to Xcdo365 and (1.2 cM distal) to Xucw67 using recombinant substitution lines, developed from the cross LDN(DIC-6B) × LDN [45]. Mapped QTLs/genes can be introgressed into the elite lines using MAS. The closer the marker to the QTL/gene, the better will be the prediction.

Table 2.
List of the identified quantitative trait loci (QTL) for the grain protein content.

Table 3.
List of the identified QTL for the grain zinc (GZn), grain iron (GFe) and grain selenium (Se) contents.

Table 4.
List of the identified QTL for the yellow pigment content.
Marker aided selection has been used for the biofortification of the wheat, with several biofortification traits [91,92,93]. As we know, to conduct MAS on any trait in any crop, first, we need to identify marker–trait associations (MTAs), using either QTL mapping or association mapping, and then deploy these identified QTL-associated markers for the indirect selection of the concerned trait. However, there are limitations associated with the methods used for identifying MTAs [94], which partly makes MAS poorly suited to wheat improvement or wheat biofortification. The necessity for marker validation and escaping minor/small-effect QTLs in the detection, due to the stringent significance thresholds, are the major concerns. In 2001, a new method, now popularly known as genomic selection (GS) or genome-wide selection (GWS), was proposed to overcome the above-mentioned problems, as well as other problems, associated with QTL mapping and/or association mapping [95]. GS predicts the genetic value of selection candidates or individuals based on the genomic estimated breeding values (GEBVs), predicted from highly dense markers, located throughout the genome. Compared to MAS, GEBV captures the largest portion of the genetic variation for the particular trait under selection, owing to the inclusion of all markers, including both minor and major effects, in the determination of the GEBV [95].
As shown in various studies, this new approach has the potential for wheat biofortification [96,97,98]. GS has the potential to quickly improve complex traits with low heritability, like grain protein content, etc., as well as to significantly reduce the time and cost of wheat breeding. GS may support breeders in their selection decisions and may assist in efficiently combining biofortification traits with grain yield in newly developed wheat varieties. While recent research has shown the potential of GS for redesigning wheat breeding, researchers are still struggling to identify the best strategy for its successful implementation in wheat breeding. In 2016, Bassi et al. [99] suggested potential breeding schemes for GS in wheat. They further discussed the genotyping considerations and methods for effectively training the population design, and they also presented the components affecting the selection intensity and progress toward half- or full-sib recurrent schemes. In 2017, Crossa et al. [100] presented a detailed overview of the history, principles and basis of GS and GPs, along with the genetics and statistical complications associated with GP models, including G × E interaction. They analyzed the accuracy of GP models and methods using two cereal crops (maize and wheat) and a legume crop (chickpea), based on random cross-validation, and also showed the potential of GS in germplasm enhancement programs.
Therefore, in this review, we have compiled the QTLs identified for grain protein content, mineral elements, e.g., Fe, Zn and Se, and pigments, like anthocyanin. This review also discusses MAS and the various prospects of GS schemes associated with wheat, with an emphasis on improving the iron (Fe), zinc (Zn), selenium (Se), grain yield protein content (GYPC), anthocyanin content, etc. This review will be a useful and important resource for wheat breeders, providing information regarding the progress made so far in wheat biofortification.
2. Biofortification for the Grain Protein Content
Grain protein content (GPC) is among the important traits that contribute to the nutritional value, processing preference, quality of the end products (bread and pasta) and market value of both hexaploidy (Triticum aestivum L.) and durum (T. turgidum L. var. durum Desf.) wheat. The economic value of wheat grains relies on their GPC; therefore, an improvement in GPC and alteration in the composition of storage proteins in wheat grain have been a significant objective in wheat breeding programs, particularly for those working toward raising the nutritional quality [101]. Several efforts have been made by breeders to improve the GPC in wheat using conventional breeding methods, but the desired outputs could not be obtained. The reasons for this may include: (1) The significant influence of the environment on GPC; (2) negative correlation of GPC with grain yield; and (3) complex quantitative genetic control of GPC and its low heritability [102]. The QTLs for GPC have been identified and mapped on almost all of the chromosomes of both tetraploid and hexaploidy wheat.
To our knowledge, for GPC, a total of 325 main-effect QTLs have been reported so far using biparental populations. Among all the QTLs identified for GPC, the most critical QTL identified so far is Gpc-B1. This QTL was first detected in a wild accession (FA-15-3) of tetraploid wheat, Triticum turgidum var. dicoccoides [103]. Later on, the same accession was used to produce a complete set of chromosome substitution lines, with the background of modern durum wheat [104]. Using substitution lines, the Gpc-B1 gene was then mapped on chromosome arm 6BS, which explained 66% of the phenotypic variation of the GPC [45]. Gpc-B1 was cloned using a map-based cloning approach, and it was found that Gpc-B1 encodes a NAC transcription factor (NAM-B1), which accelerates the senescence and also affects the grain protein, zinc, and iron content in wheat [105]. The introgression of the functional GPC-B1 allele in the background of elite lines has resulted in the release of several varieties in different countries [106].
While major QTLs, with a large effect on GPCs, have been identified in various studies, most of the identified major QTLs were unfortunately found to be unstable across the environments (Table 2). Here, we listed the cross, population and its size, number of total QTLs, including all stable or unstable QTLs over the locations, which were consistent or inconsistent over the years, PVE range of QTLs, and chromosomes/chromosome arms, where QTLs are mapped (Table 2).
2.1. Epistatic Interactions for the Grain Protein Content
The development of efficient statistical and genomic tools has allowed geneticists to identify and map the QTLs involved in epistatic interactions for GPC [56,63,69,70,77]. Software, like QTLMapper [107], QTLNetwork [108], and IciMapping [109], have frequently been used to locate epistatic QTLs. In general, there are three different types of epistatic interactions. The first type comprises the interaction between two main-effect QTLs, in which each QTL has a noteworthy effect of its own (M-QTL × M-QTL or QQ epistatic interaction). The second type comprises the interaction between main-effect QTLs and epistatic QTLs (E-QTL), where the E-QTL does not have a noteworthy effect of its own, but it shows a significant effect when it interacts with another QTL (M-QTL × E-QTL). The third type comprises the interaction between two epistatic QTLs (E-QTL × E-QTL). In addition to these epistatic interactions, the interaction of QTLs with the environment is quite common, such as QTL × environment (QE) and QTL × QTL × environment (QQE) interactions [56]. Kulwal et al. [56] identified four QTLs involved in two digenic QQ epistatic interactions in one RIL population and six E-QTLs involved in three digenic QQ epistatic interactions in another RIL population of bread wheat. However, these interactions only accounted for 2.68% and 6.04% of the phenotypic variation in the first and second population, respectively. Patil et al. [63] identified one pair of epistatic interactions using a RIL population derived from a durum wheat cross between PDW 233 and Bhalegaon 4. Zhao et al. [69] identified two digenic epistatic interactions for GPC, both only involving E-QTL, using a population developed from two Chinese wheat cultivars. Conti et al. [70] identified five pairs of epistatic interactions for GPC, in addition to one QQE interaction using a RIL population, obtained from the cross between the UC Davis wheat breeding line, UC1113, and the Kofa variety. In another study, Xu et al. [77] identified two significant digenic interactions, involving four M-QTLs, for GPC using a RIL population developed from Chinese cultivars.
We believe that the QTLs involved in different types of interactions could contribute considerably to the total observed variation of a quantitative trait, and so they should not be ignored. The genetic control of a complex trait can be efficiently understood by dissecting these interactions. The above-mentioned different types of interactions, such as QQ, QE and QQE, observed for the GPC in wheat, make sense in light of the present knowledge on molecular genetics, where DNA and protein interactions, protein and protein interactions, and epigenetic variations have been revealed to be directed by changes in the environment. It has been evidenced by the characterization of cloned QTLs from higher plants that several QTLs do not directly contribute to the observed variation for the concerned trait, but may instead be either directly involved in interacting with a coding sequence (CDS) or else their gene products, e.g., mRNA or protein may be involved in such interactions [110,111]. It has been suggested that an interaction between a CDS and a regulatory sequence may be significant for the expression of a trait [112].
2.2. Different Prospects of GS for Wheat Biofortification with Protein
Phenotypic selection (PS), MAS, and GS prediction accuracy have been compared in wheat breeding programs. In one study, Heffner et al. [113] compared the above-mentioned three strategies for thirteen agronomic traits, including grain protein content, using a population of 374 winter wheat advanced cycle breeding lines. To evaluate the effects of model selection, the training population size, and the marker density in the presence of a G × E interaction, a cross-validation approach was employed. Heffner et al. observed 28% and 95% higher prediction accuracies with GS, compared to MAS and PS, respectively, providing empirical evidence of the advantage of multifamily GS in wheat breeding. In another study, they compared the phenotypic- and marker-based prediction accuracy of the genetic value of nine different grain quality traits, including flour protein content in two doubled haploid biparental soft winter wheat populations, ‘Cayuga’ × ‘Caledonia’ and ‘Foster’ × ‘KanQueen’ [96]. They reported a significantly greater prediction accuracy with GS, compared to MAS, for all nine traits. With the training population sizes of 96, 48, and 24, they achieved GS accuracy to PS accuracy average ratios of 0.66, 0.54, and 0.42, respectively.
The simultaneous improvement of GPC and grain yield has been a major challenge in wheat breeding, owing to a severe negative trade-off. Simultaneous GS, for grain yield and GPC, using various breeding strategies in the form of selection indices, may be a good approach to mitigating the negative trade-off between these two important traits [114]. Michel et al. compared two breeding strategies based on various “genomic selection indices, viz., (a) to select high-protein genotypes with acceptable yield potential, and (b) to develop high-yielding varieties, while maintaining protein content”. They achieved promising results using the second breeding strategy. In 2016, at CIMMYT, Battenfield et al. [115] developed and validated whole genome prediction models for phenotypes with an end-use quality in the CIMMYT bread wheat breeding program and also tested the accuracy of the developed model using forward prediction on breeding lines, tested in unbalanced yield trials from 2009 to 2015 at three different locations. They phenotyped a total of 5520 breeding lines for nine different traits, including grain and flour protein. They reported a substantial increase in prediction accuracy over time, as the data available to train the model increased. The forward prediction accuracies for the studied parameters ranged from 0.32 to 0.62, and the expected genetic gain was 1.4 to 2.7 times higher than PS for all traits. The predictive abilities of the genomic predictions (GPs) can be improved using genetically correlated traits in multi-trait models [97,98]. Michel et al. [97] phenotyped more than 400 genotyped wheat lines for protein content and baking quality traits, in multi-environment trials from 2009 to 2016, and applied GS to select the best individuals in terms of their good protein content and baking quality traits, as well as grain yield. They achieved an average prediction accuracy r = 0.39 across three independent validation populations, which could be increased to r = 0.47 by modelling the major QTLs, as fixed effects, as well as engaging multi-trait prediction models. They observed nearly twice the selection response, compared to the indirect selection, by protein content for the baking quality-related traits, using GS, which was applied 2–3 years earlier than direct phenotypic selection. Recently, in 2019, Kristensen et al. [98] studied multi-trait and trait-assisted genomic prediction models, dealing with two or four traits, including GPC, phenotyped in 1152 advanced winter wheat lines, and compared the GPs of these models with single-trait models. They observed increased predictive abilities for GPC with trait-assisted models. By including phenotypic data for GPC in the trait-associated models, they also reported increased predictive abilities for other traits, such as zeleny sedimentation, which may be advantageous for breeding programs for improving wheat quality. Overall, the above studies suggest that GS is a powerful strategy that can be used to facilitate an early generation selection for end-use quality in wheat, which can provide greater advantages in terms of both quality and yield in wheat breeding programs.
As already mentioned, an effect of the G × E interaction on the grain protein content in wheat is quite common [56,63,69,70]. Different GS models need to be designed to have better accuracy in the presence of different G × E interactions. The prediction accuracy of GS can be enhanced using multi-environment (multi-trait) models, which allow for the borrowing of information across environments (traits). An enhanced prediction accuracy has been reported in a multi-environment (multi-trait) analysis based on both pedigree and marker information, compared to those based on pedigree information only. Burgueno et al. [116] achieved better predictive accuracy than simple linear mixed models by using both marker and pedigree information in multi-environment (multi-trait) models in wheat. These types of the model may be promising for the improvement of the GPC in wheat. Furthermore, in 2015, at CIMMYT, a marker × environment interaction (M.E) genomic selection model was developed. The researchers analyzed three CIMMYT wheat data sets, where more than 1000 lines were genotyped using GBS and evaluated at CIMMYT’s research station under controlled environmental conditions using this M·E model, and proposed that the interaction model may provide information on the variants that have stable effects across environments and those which are responsible for G × E interactions [117]. It has also been shown that a sizeable proportion of the prediction accuracy is due to the pedigree or population structure [118]. When prediction equations were trained by predictions from unrelated populations, the prediction accuracy became negligible. However, an increase in the prediction accuracy was achieved when the genomic prediction included modeling GE, in which information from correlated environments was borrowed [118].
As already mentioned, GS is the better option for breeding wheat in terms of producing grain end-use quality traits, as breeding these traits is difficult, owing to the requirement for different assays, which require flour quantities that are only obtainable late in the breeding cycle. Efforts have been made to use near-infrared (NIR) or nuclear magnetic resonance (NMR), requiring a very small amount of flour and only a subset of accessions to obtain GPs of end-use quality in wheat [119]. They derived predictions for nineteen traits with an end-use quality in 398 accessions using a multi-trait approach and obtained increased prediction accuracies, ranging from 0 to 0.69, using NIR/NMR data, compared to the control. This study suggested that the NIR and NMR predictions of quality traits can successfully overcome the chief problem associated with the use of GS in order to hasten the improvement of the traits with a grain end-use quality in wheat.
3. Biofortification for the Grain Fe and Zn Content
The agronomical biofortification method involves fertilizing plants with Zinc fertilizers, which can increase the Zn content of grain. For instance, Zhang et al. [120] reported a 58% increase in whole grain Zn and a 76% increase in wheat flour Zn using a foliar application of 0.4% ZnSO4·7H2O. In another study, Zou et al. [121] increased grain Zn by 84% and 90%, using Zn as a foliar spray. However, in the case of iron, these agronomic approaches have been less effective [122], except if combined with increased nitrogen fertilizers [123], which may not be economically acceptable.
Backcross breeding, with the participatory varietal selection operated by collaboration among CIMMYT, (El Batán, Mexico), the Indian Institute of Wheat and Barley Research (Karnal, India) and Punjab Agricultural University (Ludhiana, India), resulted in a release of zinc biofortified wheat varieties. To date, four Zn biofortified varieties have been released: ‘Zinc Shakti’ (developed by transferring the genes from Ae. Squarrosa into the Indian variety, PBW343), ‘Zincol 2016′ (developed by transferring the genes from T. spelta into the Pakistani variety, NARC2011) and ‘WB02′ and ‘HPBW-01′ (developed by transferring the genes from Ae. squarrosa and T. dicoccon), with 40%, 25%, 20% and 20% increases in the zinc content in their grains, respectively [124]. These four varieties are currently being grown in India and Pakistan. Furthermore, human intervention trials to examine the effectiveness of consuming flour made from Zn biofortified wheat are presently being carried out in Pakistan [125]. While success in Zn biofortification has been achieved, no Fe-biofortified variety has been produced so far using conventional breeding methods.
3.1. QTLs for the Grain Fe and Zn Concentrations in Wheat
Marker-assisted breeding is a potential strategy for the development of Fe- and Zn-biofortified wheat. Knowledge of the genetic basis of Fe and Zn concentrations is required for the successful application of MAS. Genes acting through one or more steps, e.g., root uptake, translocation from root to shoot, storage and remobilization, may be reflected by the QTLs responsible for grain mineral concentrations. Various QTL mapping studies have allowed for the identification of many QTLs for both Fe and Zn (Table 3). We list the QTLs identified for both GZn and GFe concentrations in the same place, because many studies reported the QTLs for these two traits simultaneously, and some studies even colocalized the QTLs for these two traits (Table 3). Shi et al. [126] detected four and seven QTLs for the Zn concentration and Zn content, respectively. They suggested a possibility for improving both the grain Zn concentration and content simultaneously, because all four QTLs for the Zn concentration were co-located with the QTLs for the Zn content. The QTLs for the Zn concentration on chromosomes 4A and 4D and four QTLs for the grain Zn content on chromosome 2D, 3A and 4A were co-located with the QTLs for the P contents, indicating a possibility for simultaneously improving the grain Zn and P density in wheat.
The QTLs for grain zinc and iron have also been mapped in the populations derived from the crosses between T. boeoticum and T. monococcum [128], durum wheat and wild emmer [64], and synthetic hexaploid wheat and T. spelta [132,136,137]. Tiwari et al. [128] mapped 2 QTLs for grain Fe on chromosomes 2A and 7A, explaining 12.6% and 11.7% of the phenotypic variation, and 1 QTL for grain Zn on chromosome 7A, explaining 18.8% of the total phenotypic variation, using a RIL population derived from a cross between the T. boeoticum accession, ‘pau5088′, and T. monococcum accession, ‘pau1,4087′. Recently, in 2017, Crespo-Herrera et al. [136] identified several significant QTLs in a region, named nQGZn.cimmyt-7B_1P2, on chromosome 7B, explaining the largest proportion (32.7%) of the total phenotypic variance for GZn, and one QTL on chromosome 4A (QGFe.cimmyt-4A_P2), explaining the largest (21.14%) proportion of the phenotypic variance of the GFe in two RIL populations derived from T. spelta L. and synthetic hexaploid wheat crosses. In another study, Krishnappa et al. [137] mapped four QTLs, explaining 20% of the total phenotypic variation, and five QTLs, explaining 32% of the total phenotypic variation for GFe and GZn, respectively, using a RIL population derived from a cross between an Indian wheat variety, ‘WH542′, and a synthetic derivative. Further, they identified an association between GFe, GZn and GPC and a region in the interval of Xgwm359-Xwmc407 on chromosome 2A. The QTLs for GZn and GFe co-localized on chromosome 5A (Xgwm126-Xgwm595) and 7A (Xbarc49-Xwmc525). Furthermore, Xu et al. [77] also clearly indicated the role of epistasis in the expression of these traits in wheat grains. One QTL, located on chromosome 2A (Xgwm501-Xgwm156.2), showed an additive × additive epistatic interaction with the other QTL (Xwmc181-Xcfd267.1), located on the same chromosome 2A for GZn concentration, and one QTL on chromosome 2B (Xbarc1138.2-Xcfd238) showed the same additive × additive epistatic interaction with the other QTL (Xgwm617-Xcfa2114), located on chromosome 6A for GFe.
Several studies have identified and mapped QTLs for high GFe and GZn concentrations on different chromosomes, 1A, 1B, 1D, 2A, 2B, 3A, 3B, 3D, 4A, 4B, 4D,5A, 5B, 5D, 6A, 6B, 7A, 7B and 7D, found in different diploid, tetraploid and hexaploid wheat species (Table 3). Among these studies, some have reported a significant positive correlation between GZn and GFe across different environments, indicating the co-localization of QTL or pleiotropic effect in regulating the concentrations of both GZn and GFe in wheat grains. For instance, Tiwari et al. [128] showed the colocalization of QTLs for GZn and GFe on chromosome 7A between the flanking markers Xcfd31-Xcfa2049, and the closest markers, viz., wPt-9555 and Xcfa2019, were also mapped on the 7A chromosome, indicating an association with both GZn and GFe [64,137]. The QTLs for GZn and GFe were co-localized on other chromosomes, such as 2A [137], 2B [87], 4BS [134], 5A [77,137] and 6B [135]. The colocalizations or associations between QTLs, affecting different mineral nutrients, may be related to the physiological coupling of certain processes that regulate mineral accumulation in grains. Genomic regions regulating one or a few minerals should not be overlooked, as they might be involved in other mineral-specific mechanisms. Thus, the simultaneous mapping of QTLs for several minerals is advisable, as this will dissect their inter and intra relationships and provide insight into the functional basis of the genomic architecture, physiology and evolution of the system of mineral accumulation in the grain or any other part of wheat.
Furthermore, this colocalization of QTLs provides the opportunity to employ only one MAS program in elevating the concentrations of different minerals, for instance, utilizing both GZn and GFe simultaneously. A shift in the chromosomal locations of the QTLs may also be observed, owing to the contamination that may result in sampling and experimentation, in addition to disturbances caused by variations in the nutrition distribution and the availability of nutrients in the soil [61]. Thus, researchers need to be careful in planning the mapping of QTLs for minerals.
3.2. Breeding Strategies to Develop Zn- and Fe-Biofortified Wheat
Conventional breeding methods have been successfully employed at various research institutes, in collaboration with CIMMYT, to biofortify wheat grains using Zn [124,125]. However, the Fe content in wheat grain could not be increased using these methods. Several QTL mapping studies have been conducted, and many QTLs for GFe and GZn have been mapped on different chromosomes of wheat. Thus, now, efforts can be made to utilize the QTL information in marker-assisted backcrossing schemes to produce Zn- and Fe-biofortified wheat. The colocalization of the QTLs for GFe and GZn may further provide the opportunity to target them to simultaneously improve the concentration of both in wheat grains [137]. Moreover, in various QTL studies, some QTLs for these GFe and GZn are also co-localized with those of phosphorus [64,126], selenium [132], calcium, manganese and magnesium [64], or other agronomically essential traits, including the grain protein content [77] and thousand-grain weight [134]. Using MAS, these specific regions can be transferred to the elite wheat genotypes to simultaneously increase the contents of various minerals.
The genetic variation available for breeders is not limited. For instance, Gorafi et al. [141] assessed 47 synthetic wheat lines derived from the crosses between the tetraploid wheat cultivar, ‘Langdon’, and 47 Ae. tauschii lines, collected from different geographical areas. Grain Fe and Zn ranged from 22.2 to 78.5 (mg/kg) and 20.6 to 65.8 (mg/kg), respectively, in these synthetic wheat lines, which can be utilized as potential genetic resources for breeding wheat cultivars with high mineral content. In 2017, Magallanes-López et al. [142], reported a range from 25.7 to 40.5 mg/kg and from 24.8 to 48.8 mg/kg for zinc and iron, respectively, in 46 durum varieties. In a recent study, conducted in Iran, Amiri et al. [143] assessed five elite lines and 75 historical and modern cultivars (released or introduced from 1942 to 2012) and reported a wide range, with a mean of 72.30 ± 0.69 (mg/kg), 39.54 ± 0.51 (mg/kg), 528.92 ± 11.0 (g/ha) and 282.66 ± 5.7 (g/ha) for GFeC, GZnC, GFeY and GZnY, respectively. They grouped all assessed materials into two clusters; older genotypes and landraces with a high GFeC and GZnC were kept in one group, and the remaining genotypes and landraces with a low GFeC and GZnC, which can be utilized as parents in crossing programs, were kept in the other.
3.3. Different Prospects of GS in Wheat Biofortification with Fe and Zn
GS may also be a potential strategy for biofortifying wheat with grain zinc and iron, as indicated by Velu et al. [144] in 2016. They recorded genomic and phenotypic data from 330 diverse wheat lines (Harvest Plus Association Mapping panel) to determine the GPs for GZnC and GFeC and two agronomical traits. The estimated genomic predictions ranged from 0.331 to 0.694 for Zn and from 0.324 to 0.734 for Fe. In a study carried out using wheat landraces from Afghanistan, a moderate to high accuracy for major elemental contents (Mg, K, and P) and a low to moderate accuracy for minor elements (Mn, Fe, and Zn) were achieved with the GPs. This level of prediction accuracy is sufficient to consider selecting desirable individuals using marker information alone [145]. Alomari et al. [146] confirmed the usefulness of GPs in exploring the genetic base of GFeC for breeding programs that aim at the biofortification of bread wheat with iron. The GPs for the GFeC trait varied from low to moderate values.
4. Biofortification for the Grain Selenium Content
Selenium (Se), an essential mineral element, is incorporated in proteins to make seleno-proteins, which plays a critical role in human health. These seleno-proteins are necessary antioxidant enzymes that prevent cellular damage from free radicals, resulting in the prevention of chronic diseases, such as cancer and heart disease [147]. In staple crops, Se is present at a low concentration (<100 mg of Se kg−1), so the genetic resources with a high amount of Se, as well as the genes/QTLs controlling the Se concentration, need to be identified to breed the Se-rich varieties, either using the conventional or molecular breeding approach, such as MAS [139].
There are conflicting reports on the amount of genetic variability among the wheat cultivars for the Se density in grain. Some studies have found no evidence of genetic variability [148,149], while another found a higher Se density in wheat grains [150]. A higher density of Se could be due to a more efficient uptake system of the tested species. The Se density in wheat grain was about 16 µg kg−1, which is insufficient to meet the Se requirement for humans [149]. In 2005, Lyons et al. [151] surveyed 665 ancestral and wild relatives of wheat, wheat landrace accessions, populations, and commercial cultivars, grown in Mexico and Australia, for the Se concentration in grain. For the precise assessment of the genotypic variation in the Se density, field trials were also conducted on CIMMYT’s field in the Yaqui Valley, near Ciudad Obregon, Sonora, Mexico, under similar conditions. They found the grain Se concentrations to be within a range of 5–720 μg kg−1, but unfortunately, much of this variation was correlated with the spatial variation in soil selenium, and no significant genotypic variation in the grain Se density was observed among commercial bread or durum wheat varieties. However, a Se grain concentrations, that was 42% and 35% higher, was found in Aegilops tauschii and rye, respectively. On the other hand, Piergiovanni et al., in 1997 [150], found significant differences between emmer (T. dicoccon Schrank) and spelt (T. spelta L.) accessions and wheat cultivars with higher contents of Se, Li, Mg, P and Zn. For the Se content, they reported a range from 1.9 to 5.8 μg/100 g, with a mean of 3.9 μg/100 g, and from 1.8 to 3.5 μg/100 g, with a mean of 2.8 μg/100 g, for the spelt and emmer accessions, respectively.
QTLs for the Se Content in Wheat Grain
Knowledge of the underlying genetic mechanism of the Se content is a necessary step for the Se biofortification of wheat. QTL mapping aids in the understanding of the genetic basis, but unfortunately, only a small number of QTL mapping studies, in which the QTLs for the Se concentration in wheat grain are mapped, have been conducted [132,138,139,140]. In 2014, Pu et al. [132] identified a total of 39 QTLs for five micronutrient (Se, Fe, Zn, Cu and Mn) concentrations using two RIL populations derived from the crosses between SHW-L1 (synthetic hexaploid wheat) and Chuanmai 32 and, Chuanmai 42 and Chuannong 16. In the first population, they mapped four QTLs on chromosomes 3D, 4A, 5B, and 7D, explaining 6.4–28.5% of the genetic variance, while in the second population, they mapped only one QTL on chromosome 4D, revealing 35.1% of the genetic variance for the Se concentration in wheat grain. Wang et al. [138] mapped 16 QTLs (seven at the seedling stage and nine at the adult stage) for six Se content-related traits on eight chromosomes, viz., 1B, 2B, 4B, 5A, 5B, 5D, 6A, and 7D, using a RIL population derived from a cross between two Chinese winter wheat varieties (Tainong18 and Linmai6) under both field-grown and hydroponic conditions. Each mapped QTL explained between 7.37% and 20.22% of the total phenotypic variance for the Se content. Recently, in 2018, Pu et al. [139] documented a Se-rich synthetic wheat line for the first time and mapped a total of 24 QTLs for the Se component traits on chromosomes 1B, 3D, 5A, 6A, 6B, 6D and 7D. Notably, a QTL, located on chromosome 3D (marker interval: 214.00–218.00, Qse.sau-3D), explained the maximum amount (up to 28.38%) of genetic variation.
In another study, Yan et al. [140] mapped a total of 15 QTLs on chromosomes 1A, 1B, 2B, 3A, 4B, 5A, 6A, 7A, and 7B, explaining 1.4% to 18.6% of the phenotypic variation for GSeC (grain Se conc.) and GSeY (grain Se yield) using a RIL population derived from a cross between T. dicoccoides (accession G18-16) and Langdon (Durum wheat). These findings provided various main-effect QTLs and their linked markers, which can be utilized in the MAS program for the Se biofortification of wheat grain. QTLs regulating the selenium concentration in wheat grains have been mapped on chromosomes 1A, 1B, 2B, 3A, 3D, 5A, and 5B, as well as homeologous groups of 4 and 6 chromosomes (Table 3). Pu et al. [132] identified a QTL on chromosome 4D, which explained 35.1% of the total phenotypic variance for the Se content in common wheat. The details are provided in Table 3.
5. Biofortification for the Grain Yellow Pigment Content (GYPC)
GYPC is responsible for the yellowness of wheat flour, which is regarded as good for human health because of the antioxidant properties of the carotenoids involved in this pigmentation. The yellow pigment in the wheat grain is predominantly associated with carotenoid compounds (carotenes and xanthophyll), which affect the quality and nutritional value of wheat grain products [152].
5.1. QTLs Identified for GYPC in Wheat
GYPC is a complex quantitative trait, and several efforts have been made to identify the genetic regions associated with it. To our knowledge, in the last two decades, several QTL mapping studies (mostly on durum wheat crosses) have been conducted to map more than eighty QTLs associated with GYPC in wheat (Table 4). Interestingly, in each mapping study, at least one QTL was mapped on the chromosome 7 of both T. turgidum L. var durum and T. aestivum, explaining a significant part of the phenotypic variation [153,154,155,156,157,158,159,160,161,162,163,164,165]. Parker et al. [153] identified two regions on chromosomes 3A and 7A, explaining 13% and 60% of the genetic variation, respectively. They further fine-mapped the region on chromosome 7A and identified seven additional AFLP markers within the target region. Using a RIL population derived from a cross between T. turgidum L. var. durum cultivar and T. dicoccoides (acc.600545), Elouafi et al. [154] mapped three QTLs on the chromosomal group 7 (one on 7AL and two on 7BL telomeres), explaining 62% of the total phenotypic variation. All three QTLs were consistent and showed a strong genetic effect and a weak QTL × E interaction. Pozniak et al. [155] located phytoene synthase 1 (Psy1) and phytoene synthase 1 (Psy2) genes in group 7 and 5 chromosomes, respectively. They also identified four QTLs underlying the phenotypic variation in the endosperm color on chromosomes 2A, 4B, 6B, and 7B. The Psy1-1 allelic locus of the Psy gene was co-segregated with the 7B QTL, demonstrating the role of this gene in the determination of the endosperm color. This became the first report of mapping Psy genes and supported the role of Psy1-1 in the increased levels of endosperm color in durum wheat. In another study, Zhang et al. [157] also mapped the Psy-A1 gene on chromosome 7A, with a PVE value of 33.9%, and, interestingly, also detected a QTL region on 1RS (1B.1R translocation), explaining 31.9% of the phenotypic variance.
In 2008, Patil et al. [156] identified four different stable QTLs on chromosome 1A, 3B, 5B, and 7B, explaining 5–8.75% of the phenotypic variation, and one significantly robust QTL on the distal part of chromosome 7AL, explaining 55.22% of the variation in the yellow pigment content. Markers (ISSR and AFLP) linked with the strongest QTL were also converted into SCAR markers to facilitate MAS. Blanco et al. [159] detected clusters of QTLs for the carotenoid compounds in durum wheat. In 2012, Roncallo et al. [161] reported an overlapping of the main QTLs affecting the flour yellow color (Fb*) and GYPC on the chromosomal arms, 4AL, 6AL, 7AS, 7AL, 7BS and 7BL. One QTL on 7BL also included the Psy-B1 locus. A novel minor QTL regulating Fb* (located on 7AS) showed an epistatic effect on YPC. One QTL on chromosome 4AL, with a strong effect on Fb*, was also involved in two digenic epistatic interactions. Colasuonno et al. [163] mapped a total of seven QTLs on different chromosome regions (1B, 2A, 2B, 5A, 5B, 7A and 7B) using a dense map, consisting of 5670 loci, comprising 5019 SNPs, 467 DArT, 182 SSR markers and eight genes. They also identified two candidate genes involved in the carotenoid biosynthesis pathway [aldehyde oxidase (AO1) and diphosphomevalonate decarboxylase (DMAPD)] by scanning the genome for QTLs and predicting the SNP homology against the annotated proteins in wheat and Brachypodium genomes. Recently, in 2016, in a common wheat population, Zhai et al. [165] mapped sixteen QTLs on chromosome 1B.1R, 2A, 2B, 2D, 5A, 5B, 6B, 7A and 7B, explaining 5.7 to 30.8% of the phenotypic variance for the trait. The QTLs for YPC have been mapped on different chromosomes—1A, 1B, 3A, 3B, 7A, and 7B, as well as homologous groups of 2, 4, 5 and 6 chromosomes (Table 4). The QTLs mapped on the homoeologous group 7 chromosomes explained up to 55.22% of the total phenotypic variation in durum wheat [156] and up to 77% of that in common wheat [164] in various biparental populations (Table 4), whereas, the QTLs on chromosomes 5B contributed up to 16.2% [161] and up to 14.8% [165] of the phenotypic variation in durum and common wheat, respectively. Zhai et al. [165] also identified a QTL on 1BL.1RS (H20), explaining 30.8% of the total phenotypic variance. Markers listed in Table 4 would facilitate the selection of improved durum and common varieties.
Details on the QTLs identified for YPC are provided in Table 4. The range of the PVE values, observed across environments, are also shown.
5.2. Allelic Variation and Marker-Assisted Selection
The allelic variation for GYPC at the Psy1-A1, Psy1-B1/Psy1-S1 and Psy1-D1 loci in wheat have been studied in great detail [160,167]. In 2009, Wang et al. [168] identified 27 alleles associated with the GYPC variation in wheat. While several gene/allele-specific markers have been developed, with the potential use in improving the yellow pigment in durum and common wheat [156,166,169], studies on the marker-assisted development of advanced wheat breeding lines, with an improved GYPC, are lacking [170]. In one study, a short terminal 7EL segment (from Lophopyrum ponticum), translocated to 7A, including the Lr19 and Y genes, has been transferred to durum wheat by marker-assisted backcrossing [171]. Patil et al. [91] identified eight alleles of Psy-A1 SSR, showing a significant association with GYPC, and seven alleles for Psy-B1 SSR, with no association with GYPC in 222 wheat accessions. Further, they introgressed the Psy-A1 SSR allele from PDW233 to durum wheat cultivars, MACS 3125 and HI 8498, and achieved an 89% to 98% gain in GYPC over recurrent parents.
5.3. GS for Yellow Pigment Improvement
Recently in 2019, a study was carried out involving a genetic dissection of agronomic and quality traits of interest, including whole grain protein and yellow color, in a panel of 179 durum wheat lines using GS. GS provided greater prediction accuracies for traits, such as GPC, sedimentation and gluten indexes, and can be utilized in durum wheat breeding programs [172]. GS showed promising results, which supported its use in durum wheat biofortification programs.
6. Biofortification for the Grain Phytic Acid Content
Phytic acid (PA) is primarily the storage form of phosphorus in cereal and legume seeds and accounts for 70 to 80% of the total P in grains [173,174]. Phytases are found in the aleurone layers of cereal grains and are activated due to the moisture. Hence, they are inactive in dry cereals, due to the lack of moisture needed for their activation. Because of the strong ability of phytic acid to make insoluble complexes of multi-charged metal ions, the consumption of great quantities of food containing high phytic acid levels could produce a deficit in the absorption of these dietary minerals [175]. Cook et al. [176] reported that, because of the high phytate content of cereal porridges, the iron absorption of native iron and fortification iron might be deficient. Barbro et al. [177] reported that, when the phytate is absent, the zinc and magnesium absorption rate from our food could be increased by 20% and 60%, respectively.
Genetic Variation and Breeding Strategies
Breeding opportunities for crops with a low level of phytic acid depend on the extent of the variation of phytic acid in the germplasm. Ram et al. [178] observed 3.4-fold variations in phytase levels among varieties and 5.9-fold variations among synthetic hexaploids, while investigating a total of four hundred wheat genotypes, including released varieties in India, advanced lines and synthetic hexaploids. On the other hand, they observed a lower variability (1.6-fold in varieties and 2.2-fold in synthetic hexaploids) in the phytate levels. Shitre et al. [179] observed a range of phytic acid, from 4.97 mg/g to 15.02 mg/g (mean of 9.58 mg/g), in 100 advanced breeding lines. This genetic variation of the trait can be utilized for the potential breeding of low phytic acid in wheat. Low phytic acid can also be achieved by manipulating the genes involved either in its biosynthesis or its transport in the vacuoles. Genes involved in the late stages of the phytic acid biosynthesis pathway are known to be in crops, such as maize, soybeans and barley, but none could be reported in wheat. For the first time, Bhati et al. [180] identified six in-silico wheat genes that might be involved in the biosynthesis of inositol phosphates.
Furthermore, they identified a homolog of Zmlpa-1, an ABCC subclass transporter protein (TaMRP3) that are putatively involved in phytic acid transportation. Bhati et al. [181] revealed the involvement of the ABCC13 transporter in wheat grain development, phytic acid accumulation and lateral root formation. Naidoo et al. [182] successfully introgressed the lpa1-1 gene into tropical and subtropical adapted maize germplasm using marker-assisted selection. They significantly increased the efficiency of detection of the homozygous recessive (99.58%) and heterozygous (99.59%) genotypes using SNP, as well as improved the recovery of the recurrent parent (92.15%) in the BC2F1 generation using AFLP markers in a maize backcross breeding program. Similar efforts can be made to generate low-phytate wheat.
7. Biofortification for the Anthocyanin Content
Grains of common wheat cultivars are amber in color all over the world, and anthocyanin-biofortified wheat is quite uncommon. However, researchers have started to focus on the anthocyanin content in wheat because of their vital importance for human health. Anthocyanins act as antioxidants and prevent cardiovascular diseases [183], diabetes [184], cancer [185], obesity [186], etc.
7.1. Genetic Basis of Purple-Colored Wheat Grains
The genetic base of the purple pericarp in wheat has been explored in some detail. Some researchers found it to be monogenic [187], while others reported a digenic inheritance of the pericarp color [164,165]. Dobrovolskaya et al. [187] attempted to find crosses between purple-grained hexaploidy wheat, ‘Purple Feed’-Pp1Pp1/Pp2Pp2, and ‘Purple’–Pp1Pp1/Pp3Pp3 with non-purple-grained cultivars, ‘Novosibirskaya 67′ (‘N67′) and ‘Saratovskaya 29′ (‘S29′), to map the genes, Pp1, Pp2, and Pp3. Both Pp2 and Pp3, showing a mono-factorial (dominant) inheritance, were mapped in the centromeric region of chromosome 2A. Therefore, these two genes were suggested as different alleles at the same locus and then designated as Pp3a and Pp3b. A mutual interaction (9:7) of two dominant genes, Pp1 and Pp3, was further reported in the crosses between purple-grained wheat and ‘S29′. Khlestkina et al. [188] mapped the Pp1 gene on the short arm of chromosome 7B in an F2 population from the durum wheat (Triticum durum) cross, TRI 15744/TRI 2719, whereas, in common wheat, it was reportedly located on 7BL. The mapping suggested that the Pp3 gene of T. durum is allelic to T. aestivum Pp3. Tereshchenko et al. [189] suggested that the Pp genes on T. durum chromosome 7B and T. aestivum chromosome 7D are orthologous and designated them as Pp-B1 and Pp-D1, respectively. Overall, it can be concluded that the purple pericarp trait in T. aestivum is controlled by genes on chromosomes 7D and 2A; still, the underlying molecular mechanisms by which they regulate the pericarp color is unknown.
A marker-assisted backcross strategy was followed to produce a set of bread wheat near-isogenic lines (NILs), carrying various combinations of Pp alleles, such as Pp3 (chr. 2A), Pp-A1 (7A) and Pp-D1 (7D) [93]. Based on a qRT-PCR-based study, the authors suggested that the Pp genes up-regulate the transcription of anthocyanin synthesis-related structural genes [Chi (chalcone-flavanone isomerase) and F3h (flavanone 3-hydroxylase)] in contrasting ways.
In 2016, Liu et al. [190] compared the transcriptomes of purple and white pericarps in common wheat and reported significant differential expression of a total of 23,642 unigenes (9945 up-regulated and 13,697 down-regulated). The analysis found that three unigenes of the MYB gene on the long arm of chromosome 7B and three unigenes of MYC on the long arm of chromosome 2A are candidate genes for the purple grain trait. Further, they also observed a higher expression level of TaMYC1 in purple wheat grains, compared to white (amber) wheat grains. Shoeva et al. [191] isolated the TaMYC1 gene-encoding MYC-like transcription factor from the bread wheat genome, which was co-located with the Pp3 gene regulating the purple pericarp color. Later on, in 2017, Zong et al. [192] provided additional strong evidence to prove that TaMYC1 is a synonym of Pp3 and found differences between dominant and recessive Pp3 alleles (TaMYC1p and TaMYC1w). In another study, Li et al. [193] isolated an MYB transcription factor gene, TaMYB3, localized on chromosome 4BL, which induces anthocyanin synthesis in pericarp cells. Recently, Jiang et al. [194] characterized two transcription factors: TaPpm1 (purple pericarp-MYB 1) and TaPpb1 (purple pericarp-bHLH 1) in wheat, and it was demonstrated that the interaction of TaPpm1 and TaPpb1 co-regulates the synthesis of anthocyanin in pericarps.
7.2. Development of Anthocyanin-Biofortified Purple Wheat
Burešová et al. [195] confirmed different chromatin introgressions, carrying genomic regions associated with the production of blue-aleurone, from Thinopyrum ponticum and Triticum monococcum in the background of blue-aleurone wheat elite lines. They analyzed a total of 26 blue aleurone wheat lines and reported an introgression (ranging from a ditelosomic addition to a disomic substitution, which is a substitution of entire chromosome arms and various translocations) from Th. Ponticum in 17 lines and presumably from T. monococcum in nine lines. This study supports the hypothesis that the introgressions activated the blue aleurone trait present in blue aleurone wheat lines, but they inactivated it in common wheat germplasm.
Anthocyanin-biofortified wheat has attracted the attention of many researchers across the world, but the developed lines exhibit a low yield, due to linkage drag [196]. In India, Garg et al. [92] developed colored wheat lines, viz., blue, purple and black, with a reasonable yield potential and regional adaptation. They transferred genes for blue, purple and wheat pericarps from blue aleurone wheat, “TA3972”, purple wheat, “TA3851”, and black wheat to the high yielding, disease-resistant and locally adapted Indian wheat cultivars, viz., PBW550, PBW621 and HD2967. These developed colored wheat lines exhibited higher anthocyanin contents and antioxidant activities (black > purple > blue > white), compared to amber wheat cultivars [16].
8. Conclusions and Future Prospects
Micronutrient deficiency, micronutrient malnutrition or hidden hunger is an increasingly severe global challenge for humankind [2]. As wheat is the major staple food crop in temperate countries and is frequently consumed in developing countries, it has become necessary to biofortify wheat with micronutrients, especially iron and zinc, in order to fulfil the requirements for better human health [87].
In general, there are two major biofortification approaches: First, agronomic biofortification; and second, genetic biofortification (including conventional breeding and molecular breeding). The existence of substantial genetic variability in biofortification traits presents superior possibilities for raising the availability of minerals and proteins in wheat grains. Biofortification traits are complex quantitative traits. Hence, their improvement via traditional breeding approaches is very complicated. In this post-genomic and computational systems biology era, research specifically focused on QTL mapping has helped to dissect the molecular basis of the biofortification of wheat. Several major-effect and stable QTLs for biofortification traits, such as GPC, minerals and pigments, have been identified. These can be successfully transferred into the elite lines using MAS. Four wheat varieties with a high zinc content have been released in India and Pakistan, and several biofortified lines are in the advanced stages of evaluation for release in other countries as well. In parallel, GS, a genome scale method has great potential for wheat biofortification. It allows for the significant benefits of a drastically reduced cost and increased throughput of genotyping assays, in combination with advances in high throughput phenotyping. In this review, information on the QTL mapping studies carried out on wheat, in which the QTLs associated with various biofortification traits are mapped, has been summarized, and the progress made so far in wheat biofortification breeding using MAS and GS has been discussed. A summary of the significant QTLs, with a PVE value > 15% for biofortification traits, is presented in Figure 1. Moreover, the biofortification of all relevant QTLs together will require detailed knowledge of the traits and their coexistence. Nonetheless, this also appears to be a promising strategy for the future. Additionally, the combination of functional and genetic proof, in conjunction with genome sequencing, will hopefully deliver additional insights into the emerging biofortification genomics. We anticipate that producing nutrient-rich genotypes of wheat through the development of wheat biofortification breeding will be helpful for addressing the expanding problem of hidden hunger.
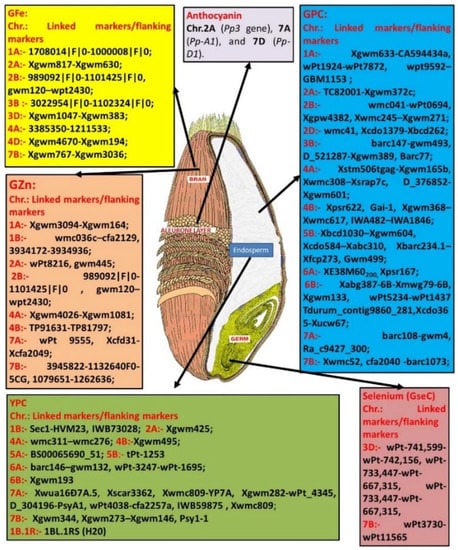
Figure 1.
Illustration of the locations of various biofortification traits in wheat grain and the closest markers/marker intervals on the chromosomes associated with the major stable QTLs (PVE > 15%).
While the biofortified varieties of wheat have been developed as a proof of the concept of biofortification, further systemic studies are needed to improve the bioavailability of these elevated protein, mineral and pigment concentrations. Research coordination between breeders and nutrition specialists are needed to decide upon the target level of micronutrients and protein, their retention after storage, processing and cooking, as well as the probable levels of consumption by humans. Adequate information programs are required to increase public awareness of the adoption of biofortified varieties by farmers and acceptance by consumers, particularly if there are noticeable changes in the grain phenotype, such as pigmented wheat (purple and black wheat, etc.). The high price of biofortified grains or products in the market may inspire the farmers to grow more biofortified crops.
Coordination and collaboration among various specialists from different disciplines, namely, plant breeders, biochemists, biotechnologists, agronomists, seed technologists and postharvest technologists, across various private and public organizations may further speed up the development of biofortified wheat varieties in a more active way [197]. Genetic engineering technology may also be a potential tool for the development of biofortified crops, but regulatory issues need to be dealt with first. Generally, nutritional traits, such as minerals and pigments, do not affect the grain yield. This need to become public knowledge, so that the wheat varieties developed in future possess at least a set level of one or more desirable nutritional traits. The high yielding biofortified wheat crop has great implications for nutritional security. The father of the Green Revolution (in India), Prof. M.S. Swaminathan, has said that ‘we need to move from food security to nutrition security, where not only calories and proteins, but also micronutrients, are taken care of. All that needs to be done is to bring agriculture, health and nutrition together in a triangular relationship, which can only be achieved through partnerships’ [198].
Author Contributions
P.K. conceived of and designed the project. P.K. supervised the study. D.K.S., P.D. and P.K. wrote the paper. P.K. corrected the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors are thankful to the anonymous reviewers for their careful reading of the manuscript and for providing insightful suggestions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Grew, R. Food in Global History; Routledge: New York, NY, USA, 2018. [Google Scholar]
- Bouis, H.E.; Saltzman, A. Improving nutrition through biofortification: A review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Secur. 2017, 12, 49–58. [Google Scholar] [CrossRef]
- Garg, M.; Sharma, N.; Sharma, S.; Kapoor, P.; Kumar, A.; Chunduri, V.; Arora, P. Biofortified Crops Generated by Breeding, Agronomy, and Transgenic Approaches Are Improving Lives of Millions of People around the World. Front. Nutr. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Hossein-Nezhad, A.; Holick, M.F. Vitamin D for Health: A Global Perspective. Mayo Clin. Proc. 2013, 88, 720–755. [Google Scholar] [CrossRef] [PubMed]
- Ward, E. Addressing nutritional gaps with multivitamin and mineral supplements. Nutr. J. 2014, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- De Valença, A.W.; Bake, A.; Brouwer, I.D.; Giller, K.E. Agronomic biofortification of crops to fight hidden hunger in sub-Saharan Africa. Glob. Food Secur. 2017, 12, 8–14. [Google Scholar] [CrossRef]
- Cassman, K.G. Ecological intensification of cereal production systems: Yield potential, soil quality, and precision agriculture. Proc. Natl. Acad. Sci. USA 1999, 96, 5952–5959. [Google Scholar] [CrossRef]
- Curtis, T.; Halford, N.G. Food security: The challenge of increasing wheat yield and the importance of not compromising food safety. Ann. Appl. Biol. 2014, 164, 354–372. [Google Scholar] [CrossRef]
- Iizumi, T.; Furuya, J.; Shen, Z.; Kim, W.; Okada, M.; Fujimori, S.; Hasegawa, T.; Nishimori, M. Responses of crop yield growth to global temperature and socioeconomic changes. Sci. Rep. 2017, 7, 7800. [Google Scholar] [CrossRef]
- Shewry, P.R.; Hey, S.J. The contribution of wheat to human diet and health. Food Energy Secur. 2015, 4, 178–202. [Google Scholar] [CrossRef]
- Arora, S.; Cheema, J.; Poland, J.; Uauy, C.; Chhuneja, P. Genome-Wide Association Mapping of Grain Micronutrients Concentration in Aegilops tauschii. Front. Plant Sci. 2019, 10, 54. [Google Scholar] [CrossRef]
- Fambrini, M.; Pugliesi, C. Carotenoids in Crops: Roles, Regulation of the Pathway, Breeding to Improve the Content. In Beta Carotene Dietary Sources, Cancer and Cognition; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2009; pp. 1–57. [Google Scholar]
- Mellado-Ortega, E.; Hornero-Méndez, D. Lutein esterification in wheat flour increases the carotenoid retention and is induced by storage temperatures. Foods 2017, 6, 111. [Google Scholar] [CrossRef] [PubMed]
- Young, A.J.; Lowe, G.L. Carotenoids—Antioxidant Properties. Antioxidants 2018, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Jin, L.; Zhang, G.; Lu, Y.; Shen, Y.; Bao, J. Association mapping of grain color, phenolic content, flavonoid content and antioxidant capacity in dehulled rice. Theor. Appl. Genet. 2011, 122, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Chunduri, V.; Kumar, A.; Kumar, R.; Khare, P.; Kondepudi, K.K.; Bishnoi, M.; Garg, M. Anthocyanin bio-fortified colored wheat: Nutritional and functional characterization. PLoS ONE 2018, 13, e0194367. [Google Scholar] [CrossRef]
- Cakmak, I.; Kutman, U. Agronomic biofortification of cereals with zinc: A review: Agronomic zinc biofortification. Eur. J. Soil Sci. 2017, 69, 172–180. [Google Scholar] [CrossRef]
- Goulding, K.; Jarvis, S.; Whitmore, A. Optimizing nutrient management for farm systems. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2008, 363, 667–680. [Google Scholar] [CrossRef]
- Deschamps, S.; Llaca, V.; May, G.D. Genotyping-by-Sequencing in Plants. Biology 2012, 1, 460–483. [Google Scholar] [CrossRef]
- Hashmi, U.; Shafqat, S.; Khan, F.; Majid, M.; Hussain, H.; Kazi, A.G.; John, R.; Ahmad, P. Plant exomics: Concepts, applications and methodologies in crop improvement. Plant Signal Behav. 2014, 10, e976152. [Google Scholar] [CrossRef]
- Li, C.; Lin, F.; An, D.; Wang, W.; Huang, R. Genome Sequencing and Assembly by Long Reads in Plants. Genes 2017, 9, 6. [Google Scholar] [CrossRef]
- Klimenko, I.; Razgulayeva, N.; Gau, M.; Okumura, K.; Nakaya, A.; Tabata, S.; Kozlov, N.N.; Isobe, S. Mapping candidate QTLs related to plant persistency in red clover. Theor. Appl. Genet. 2010, 120, 1253–1263. [Google Scholar] [CrossRef]
- Zhang, S.; Meng, L.; Wang, J.; Zhang, L. Background controlled QTL mapping in pure-line genetic populations derived from four-way crosses. Heredity 2017, 119, 256–264. [Google Scholar] [CrossRef]
- Institute of Medicine, Food and Nutrition Board. Dietary Reference Intakes for Vitamin A, Vitamin K, Arsenic, Boron, Chromium, Copper, Iodine, Iron, Manganese, Molybdenum, Nickel, Silicon, Vanadium, and Zinc; National Academy Press: Washington, DC, USA, 2001. [Google Scholar]
- Ahmadi, J.; Pour-Aboughadareh, A.; Ourang, S.F.; Mehrabi, A.A.; Siddique, K.H.M. Wild relatives of wheat: Aegilops–Triticum accessions disclose differential antioxidative and physiological responses to water stress. Acta Physiol. Plant. 2018, 40, 1–14. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and Future Use of Wild Relatives in Crop Breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Breseghello, F.; Coelho, A.S.G. Traditional and modern plant breeding methods with examples in rice (Oryza sativa L.). J. Agric. Food Chem. 2013, 61, 8277–8286. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, F.; Xu, J.; Li, Z.; Xu, S. Mapping quantitative trait loci in selected breeding populations: A segregation distortion approach. Heredity 2015, 115, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef]
- Khan, S.; Nabi, G.; Ullah, M.W.; Yousaf, M.; Manan, S.; Siddique, R.; Hou, H. Overview on the Role of Advance Genomics in Conservation Biology of Endangered Species. Int. J. Genom. 2016, 2016, 3460416. [Google Scholar] [CrossRef]
- Wambugu, P.W.; Ndjiondjop, M.N.; Henry, R.J. Role of genomics in promoting the utilization of Plant Genetic Resources in genebanks. Brief. Funct. Genom. 2018, 17, 198–206. [Google Scholar] [CrossRef]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar]
- International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 7191. [Google Scholar]
- Muir, P.; Li, S.; Lou, S.; Wang, D.; Spakowicz, D.J.; Salichos, L.; Zhang, J.; Weinstock, G.M.; Isaacs, F.; Rozowsky, J.; et al. The real cost of sequencing: Scaling computation to keep pace with data generation. Genome Biol. 2016, 17, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chiodini, R.; Badr, A.; Zhang, G. The impact of next-generation sequencing on genomics. J. Genet. Genom. 2011, 38, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Baenziger, P.S.; Waters, B.M.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3237. [Google Scholar] [CrossRef] [PubMed]
- Miles, C.M.; Wayne, M. Quantitative trait locus (QTL) analysis. Nat. Educ. 2008, 1, 208. [Google Scholar]
- Wang, B.; Liu, H.; Liu, Z.; Dong, X.; Guo, J.; Li, W.; Chen, J.; Gao, C.; Zhu, Y.; Zheng, X.; et al. Identification of minor effect QTLs for plant architecture related traits using super high density genotyping and large recombinant inbred population in maize (Zea mays). BMC Plant Biol. 2018, 18, 17. [Google Scholar] [CrossRef]
- Ishikawa, A. A Strategy for Identifying Quantitative Trait Genes Using Gene Expression Analysis and Causal Analysis. Genes 2017, 8, 347. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Guo, N.; Zhang, Y.; Bu, Y.; Zhao, J.; Xing, H. Combining QTL-seq and linkage mapping to fine map a wild soybean allele characteristic of greater plant height. BMC Genom. 2018, 19, 226. [Google Scholar] [CrossRef]
- Civelek, M.; Lusis, A.J. Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 2014, 15, 34–48. [Google Scholar] [CrossRef]
- Schneider, K. Mapping populations and principles of genetic mapping. In The Handbook of Plant Genome Mapping Genetic and Physical Mapping; Meksem, K., Kahl, G., Eds.; Wiley-VCH: Weinheim, Germany, 2005. [Google Scholar]
- Joppa, L.R.; Du, C.; Hart, G.E.; Hareland, G.A. Mapping gene (s) for grain protein in tetraploid wheat (Triticum turgidum L.) using a population of recombinant inbred chromosome lines. Crop Sci. 1997, 37, 1586–1589. [Google Scholar] [CrossRef]
- Gonzalez-Hernandez, J.L.; Elias, E.M.; Kianian, S.F. Mapping genes for grain protein concentration and grain yield on chromosome 5B of Triticum turgidum (L.) var. Dicoccoides Euphytica 2004, 139, 217–225. [Google Scholar] [CrossRef]
- Olmos, S.; Distelfeld, A.; Chicaiza, O.; Schlatter, A.R.; Fahima, T.; Echenique, V.; Dubcovsky, J. Precise mapping of a locus affecting grain protein content in durum wheat. Theor. Appl. Genet. 2003, 107, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.; Giovanni, C.D.; Laddomada, B.; Sciancalepore, A.; Simeone, R.; Devos, K.M.; Gale, M.D. Quantitative trait loci influencing grain protein content in tetraploid wheats. Plant Breed. 1996, 115, 310–316. [Google Scholar] [CrossRef]
- Prasad, M.; Varshney, R.K.; Kumar, A.; Balyan, H.S.; Sharma, P.C.; Edwards, K.J.; Dhaliwal, H.S.; Roy, J.K.; Gupta, P.K. A microsatellite marker associated with a QTL for grain protein content on chromosome arm 2DL of bread wheat. Theor. Appl. Genet. 1999, 99, 341–345. [Google Scholar] [CrossRef]
- Perretant, M.R.; Cadalen, T.; Charmet, G.; Sourdille, P.; Nicolas, P.; Boeuf, C.; Tixier, M.H.; Branlard, G.; Bernard, S. QTL analysis of bread-making quality in wheat using a doubled haploid population. Theor. Appl. Genet. 2000, 100, 1167–1175. [Google Scholar] [CrossRef]
- Dholakia, B.B.; Ammiraju, J.S.S.; Santra, D.K.; Singh, H.; Katti, M.V.; Lagu, M.D.; Tamhankar, S.A.; Rao, V.S.; Gupta, V.S.; Dhaliwal, H.S.; et al. Molecular marker analysis of protein content using PCR-based markers in wheat. Biochem. Genet. 2001, 39, 325–338. [Google Scholar] [CrossRef]
- Singh, H.; Prasad, M.; Varshney, R.K.; Roy, J.K.; Balyan, H.S.; Dhaliwal, H.S.; Gupta, P.K. STMS markers for grain protein content and their validation using near-isogenic lines in bread wheat. Plant Breed. 2001, 120, 273–278. [Google Scholar] [CrossRef]
- Blanco, A.; Pasqualone, A.; Troccoli, A.; Di Fonzo, N.; Simeone, R. Detection of grain protein content QTLs across environments in tetraploid wheats. Plant Mol. Biol. 2002, 48, 615–623. [Google Scholar] [CrossRef]
- Börner, A.; Schumann, E.; Fürste, A.; Cöster, H.; Leithold, B.; Röder, M.; Weber, W. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef]
- Groos, C.; Robert, N.; Bervas, E.; Charmet, G. Genetic analysis of grain protein-content, grain yield and thousand-kernel weight in bread wheat. Theor. Appl. Genet. 2003. 106, 1032–1040. [CrossRef]
- Prasad, M.; Kumar, N.; Kulwal, P.; Röder, M.; Balyan, H.; Dhaliwal, H.; Gupta, P. QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor. Appl. Genet. 2003, 106, 659–667. [Google Scholar] [CrossRef]
- Groos, C.; Bervas, E.; Charmet, G. Genetic analysis of grain protein content, grain hardness and dough rheology in a hard× hard bread wheat progeny. J. Cereal Sci. 2004, 40, 93–100. [Google Scholar] [CrossRef]
- Kulwal, P.; Kumar, N.; Kumar, A.; Gupta, R.K.; Balyan, H.S.; Gupta, P.K. Gene networks in hexaploid wheat: Interacting quantitative trait loci for grain protein content. Funct. Integr. Genom. 2005, 5, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Q.; Cloutier, S.; Lycar, L.; Radovanovic, N.; Humphreys, D.G.; Noll, J.S.; Somers, D.J.; Brown, P.D. Molecular detection of QTLs for agronomic and quality traits in a doubled haploid population derived from two Canadian wheats (Triticum aestivum L.). Theor. Appl. Genet. 2006, 113, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.C.; Andreescu, C.; Breseghello, F.; Finney, P.L.; Gualberto, D.G.; Bergman, C.J.; Pena, R.J.; Perretant, M.R.; Leroy, P.; Qualset, C.O.; et al. Quantitative trait locus analysis of wheat quality traits. Euphytica 2006, 149, 145–159. [Google Scholar] [CrossRef]
- Laperche, A.; Brancourt-Hulmel, M.; Heumez, E.; Gardet, O.; Hanocq, E.; Devienne-Barret, F.; Le Gouis, J. Using genotype × nitrogen interaction variables to evaluate the QTL involved in wheat tolerance to nitrogen constraints. Theor. Appl. Genet. 2007, 115, 399–415. [Google Scholar] [CrossRef]
- Sun, H.; Lü, J.; Fan, Y.; Zhao, Y.; Kong, F.; Li, R.; Wang, H.; Li, S. Quantitative trait loci (QTLs) for quality traits related to protein and starch in wheat. Prog. Nat. Sci. 2008, 18, 825–831. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Zhou, R.; Branlard, G.; Jia, J. Detection of QTLs for bread-making quality in wheat using a recombinant inbred line population. Plant Breed. 2009, 128, 235–243. [Google Scholar] [CrossRef]
- Mann, G.; Diffey, S.; Cullis, B.; Azanza, F.; Martin, D.; Kelly, A.; McIntyre, L.; Schmidt, A.; Ma, W.; Nath, Z.; et al. Genetic control of wheat quality: Interactions between chromosomal regions determining protein content and composition, dough rheology, and sponge and dough baking properties. Theor. Appl. Genet. 2009, 118, 1519–1537. [Google Scholar] [CrossRef]
- Patil, R.M.; Oak, M.D.; Tamhankar, S.A.; Rao, V.S. Molecular mapping of QTLs for gluten strength as measured by sedimentation volume and mixograph in durum wheat (Triticum turgidum L. ssp durum). J. Cereal Sci. 2009, 49, 378–386. [Google Scholar] [CrossRef]
- Peleg, Z.; Cakmak, I.; Ozturk, L.; Yazici, A.; Jun, Y.; Budak, H.; Korol, A.B.; Fahima, T.; Saranga, Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat × wild emmer wheat RIL population. Theor. Appl. Genet. 2009, 119, 353–369. [Google Scholar] [CrossRef]
- Raman, R.; Allen, H.; Diffey, S.; Raman, H.; Martin, P.; McKelvie, K. Localisation of quantitative trait loci for quality attributes in a doubled haploid population of wheat (Triticum aestivum L.). Genome 2009, 52, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Suprayogi, Y.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Knox, R.E.; Singh, A.K. Identification and validation of quantitative trait loci for grain protein concentration in adapted Canadian durum wheat populations. Theor. Appl. Genet. 2009, 119, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Marza, F.; Ma, H.; Carver, B.F.; Bai, G. Mapping quantitative trait loci for quality factors in an inter-class cross of US and Chinese wheat. Theor. Appl. Genet. 2010, 120, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Tsilo, T.J.; Hareland, G.A.; Simsek, S.; Chao, S.; Anderson, J.A. Genome mapping of kernel characteristics in hard red spring wheat breeding lines. Theor. Appl. Genet. 2010, 121, 717–730. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, K.P.; Liu, B.; Deng, Z.Y.; Qu, H.L.; Tian, J.C. A comparison of grain protein content QTLs and flour protein content QTLs across environments in cultivated wheat. Euphytica 2010, 174, 325–335. [Google Scholar] [CrossRef]
- Conti, V.; Roncallo, P.F.; Beaufort, V.; Cervigni, G.L.; Miranda, R.; Jensen, C.A.; Echenique, V.C. Mapping of main and epistatic effect QTLs associated to grain protein and gluten strength using a RIL population of durum wheat. J. Appl. Genet. 2011, 52, 287–298. [Google Scholar] [CrossRef]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between grain protein content and grain yield components through quantitative trait locus analyses in a recombinant inbred line population derived from two elite durum wheat cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Golabadi, M.; Arzani, A.; Mirmohammadi Maibody, S. Identification of microsatellite markers associated with grain protein content in durum wheat grown under drought stress at terminal growth stages. Cereal Res. Commun. 2012, 40, 215–224. [Google Scholar] [CrossRef]
- Li, J.; Cui, F.; Ding, A.M.; Zhao, C.H.; Wang, X.Q.; Wang, L.; Bao, Y.G.; Qi, X.L.; Li, X.F.; Gao, J.R.; et al. QTL detection of seven quality traits in wheat using two related recombinant inbred line populations. Euphytica 2012, 183, 207–226. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, R.; Wang, J.; Liao, X.; Branlard, G.; Jia, J. Novel and favorable QTL allele clusters for end-use quality revealed by introgression lines derived from synthetic wheat. Mol. Breed. 2012, 29, 627–643. [Google Scholar] [CrossRef]
- Simons, K.; Anderson, J.A.; Mergoum, M.; Faris, J.D.; Klindworth, D.L.; Xu, S.S.; Sneller, C.; Ohm, J.B.; Hareland, G.A.; Edwards, M.C.; et al. Genetic mapping analysis of bread-making quality traits in spring wheat. Crop Sci. 2012, 52, 2182–2197. [Google Scholar] [CrossRef]
- Wang, L.I.N.; Cui, F.A.; Wang, J.; Jun, L.I.; Ding, A.; Zhao, C.; Li, X.; Feng, D.; Gao, J.; Wang, H. Conditional QTL mapping of protein content in wheat with respect to grain yield and its components. J. Genet. 2012, 91, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; An, D.; Liu, D.; Zhang, A.; Xu, H.; Li, B. Molecular mapping of QTLs for grain zinc, iron and protein concentration of wheat across two environments. Field Crops Res. 2012, 138, 57–62. [Google Scholar] [CrossRef]
- El-Feki, W.M.; Byrne, P.F.; Reid, S.D.; Lapitan, N.L.; Haley, S.D. Quantitative trait locus mapping for end-use quality traits in hard winter wheat under contrasting soil moisture levels. Crop Sci. 2013, 53, 1953–1967. [Google Scholar] [CrossRef]
- Heo, H.; Sherman, J. Identification of QTL for grain protein content and grain hardness from winter wheat for genetic improvement of spring wheat. Plant Breed. Biotechnol. 2013, 1, 347–353. [Google Scholar] [CrossRef]
- Deng, Z.; Hu, S.; Chen, F.; Li, W.; Chen, J.; Sun, C.; Zhang, Y.; Wang, S.; Song, X.; Tian, J. Genetic dissection of interaction between wheat protein and starch using three mapping populations. Mol. Breed. 2015, 35, 12. [Google Scholar] [CrossRef]
- Maphosa, L.; Langridge, P.; Taylor, H.; Emebiri, L.C.; Mather, D.E. Genetic control of grain protein, dough rheology traits and loaf traits in a bread wheat population grown in three environments. J. Cereal Sci. 2015, 64, 147–152. [Google Scholar] [CrossRef]
- Moore, C.M.; Richards, R.A.; Rebetzke, G.J. Phenotypic variation and QTL analysis for oil content and protein concentration in bread wheat (Triticum aestivum L.). Euphytica 2015, 204, 371–382. [Google Scholar] [CrossRef]
- Echeverry-Solarte, M.; Kumar, A.; Kianian, S.; Simsek, S.; Alamri, M.S.; Mantovani, E.E.; McClean, P.E.; Deckard, E.L.; Elias, E.; Schatz, B.; et al. New QTL alleles for quality-related traits in spring wheat revealed by RIL population derived from supernumerary × non-supernumerary spikelet genotypes. Theor. Appl. Genet. 2015, 128, 893–912. [Google Scholar] [CrossRef]
- Li, C.; Bai, G.; Chao, S.; Carver, B.; Wang, Z. Single nucleotide polymorphisms linked to quantitative trait loci for grain quality traits in wheat. Crop J. 2016, 4, 1–11. [Google Scholar] [CrossRef]
- Mahjourimajd, S.; Taylor, J.; Rengel, Z.; Khabaz-Saberi, H.; Kuchel, H.; Okamoto, M.; Langridge, P. The genetic control of grain protein content under variable nitrogen supply in an Australian wheat mapping population. PLoS ONE 2016, 11, e0159371. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, Y.; Ito, M.; Tabiki, T.; Nagasawa, K.; Hatta, K.; Nishio, Z. Mapping of a major QTL associated with protein content on chromosome 2B in hard red winter wheat (Triticum aestivum L.). Breed. Sci. 2016, 66, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, C.; Wallwork, H.; Arun, B.; Mishra, V.K.; Velu, G.; Stangoulis, J.; Kumar, U.; Joshi, A.K. Molecular mapping of quantitative trait loci for zinc, iron and protein content in the grains of hexaploid wheat. Euphytica 2016, 207, 563–570. [Google Scholar] [CrossRef]
- Sun, X.; Wu, K.; Zhao, Y.; Qian, Z.; Kong, F.; Guo, Y.; Wang, Y.; Li, S. Molecular genetic analysis of grain protein content and flour whiteness degree using RILs in common wheat. J. Genet. 2016, 95, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Marcotuli, I.; Gadaleta, A.; Mangini, G.; Signorile, A.; Zacheo, S.; Blanco, A.; Simeone, R.; Colasuonno, P. Development of a high-density SNP-based linkage map and detection of QTL for β-glucans, protein content, grain yield per spike and heading time in durum wheat. Int. J. Mol. Sci. 2017, 18, 1329. [Google Scholar] [CrossRef]
- Fatiukha, A.; Lupo, I.; Lidzbarsky, G.; Klymiuk, V.; Korol, A.B.; Pozniak, C.; Fahima, T.; Krugman, T. Grain Protein Content QTLs Identified in a Durum × Wild Emmer Wheat Mapping Population Tested in Five Environments. bioRxiv 2019. [Google Scholar] [CrossRef]
- Patil, R.; Oak, M.; Deshpande, A.; Tamhankar, S. Development of a robust marker for Psy-1 homoeologs and its application in improvement of yellow pigment content in durum wheat. Mol. Breed. 2018, 38, 136. [Google Scholar] [CrossRef]
- Garg, M.; Chawla, M.; Chunduri, V.; Kumar, R.; Sharma, S.; Sharma, N.K.; Kaur, N.; Kumar, A.; Mundey, J.K.; Saini, M.K.; et al. Transfer of grain colors to elite wheat cultivars and their characterization. J. Cereal Sci. 2016, 71, 138–144. [Google Scholar] [CrossRef]
- Gordeeva, E.I.; Shoeva, O.Y.; Khlestkina, E.K. Marker-assisted development of bread wheat near-isogenic lines carrying various combinations of purple pericarp (Pp) alleles. Euphytica 2015, 203, 469–476. [Google Scholar] [CrossRef]
- Gupta, P.K.; Kulwal, P.L.; Jaiswal, V. Association mapping in crop plants: Opportunities and challenges. Adv. Genet. 2014, 85, 109–147. [Google Scholar]
- Meuwissen, T.H.E.; Hayes, B.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [PubMed]
- Heffner, E.L.; Jannink, J.L.; Iwata, H.; Souza, E.; Sorrells, M.E. Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci. 2011, 51, 2597–2606. [Google Scholar] [CrossRef]
- Michel, S.; Kummer, C.; Gallee, M.; Hellinger, J.; Ametz, C.; Akgöl, B.; Epure, D.; Löschenberger, F.; Buerstmayr, H. Improving the baking quality of bread wheat by genomic selection in early generations. Theor. Appl. Genet. 2018, 131, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, P.S.; ed Jahoor, A.; Andersen, J.R.; ad Orabi, J.; Janss, L.; Jensen, J. Multi-Trait and Trait-Assisted Genomic Prediction of Winter Wheat Quality Traits Using Advanced Lines from Four Breeding Cycles. Crop Breed. Genet. Genom. 2019, 1. [Google Scholar] [CrossRef]
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 2016, 242, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef]
- Balyan, H.S.; Gupta, P.K.; Kumar, S.; Dhariwal, R.; Jaiswal, V.; Tyagi, S.; Agarwal, P.; Gahlaut, V.; Kumari, S. Genetic improvement of grain protein content and other health-related constituents of wheat grain. Plant Breed. 2013, 132, 446–457. [Google Scholar] [CrossRef]
- Avivi, L. High protein content in wild tetraploid Triticum dicoccoides Korn. In Proceedings of the 5th International Wheat Genetics Symposium, New Delhi, India, 23–28 February 1978; Ramanujam, S., Ed.; Indian Society of Genetics and Plant Breeding (ISGPB): New Delhi, India, 1978; pp. 372–380. [Google Scholar]
- Joppa, L.R.; Cantrell, R.G. Chromosomal location of genes for grain protein content of wild tetraploid wheat. Crop Sci. 1990, 30, 1059–1064. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Tabbita, F.; Pearce, S.; Barneix, A.J. Breeding for increased grain protein and micronutrient content in wheat: Ten years of the GPC-B1 gene. J. Cereal Sci. 2017, 73, 183–191. [Google Scholar] [CrossRef]
- Wang, D.L.; Zhu, J.; Li, Z.K.L.; Paterson, A.H. Mapping QTLs with epistatic effects and QTL × environment interactions by mixed linear model approaches. Theor. Appl. Genet. 1999, 99, 1255–1264. [Google Scholar] [CrossRef]
- Yang, J.; Hu, C.; Hu, H.; Yu, R.; Xia, Z.; Ye, X.; Zhu, J. QTL Network: Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 2008, 24, 721–723. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Paran, I.; Zamir, D. Quantitative traits in plants: Beyond the QTL. Trends Genet. 2003, 19, 303–306. [Google Scholar] [CrossRef]
- Morgante, M.; Salamini, F. From plant genomics to practice. Curr. Opin. Biotechnol. 2013, 14, 214–219. [Google Scholar] [CrossRef]
- Guillaumie, S.; Charmet, G.; Linossier, L.; Torney, V.; Robert, N.; Ravel, C. Colocation between a gene encoding the bZipfactor SPA and an eQTL for a high-molecular-weight glutenin subunit in wheat (Triticum aestivum). Genome 2004, 47, 705–713. [Google Scholar] [CrossRef]
- Heffner, E.L.; Jannink, J.L.; Sorrells, M.E. Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Genome 2011, 4, 65–75. [Google Scholar] [CrossRef]
- Michel, S.; Löschenberger, F.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Simultaneous selection for grain yield and protein content in genomics-assisted wheat breeding. Theor. Appl. Genet. 2019, 132, 1745–1760. [Google Scholar] [CrossRef]
- Battenfield, S.D.; Guzmán, C.; Gaynor, R.C.; Singh, R.P.; Peña, R.J.; Dreisigacker, S.; Fritz, A.K.; Poland, J.A. Genomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding program. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef]
- Burgueño, J.; de los Campos, G.; Weigel, K.; Crossa, J. Genomic prediction of breeding values when modeling genotype× environment interaction using pedigree and dense molecular markers. Crop Sci. 2012, 52, 707–719. [Google Scholar] [CrossRef]
- Lopez-Cruz, M.; Crossa, J.; Bonnett, D.; Dreisigacker, S.; Poland, J.; Jannink, J.L.; Singh, R.P.; Autrique, E.; de los Campos, G. Increased prediction accuracy in wheat breeding trials using a marker× environment interaction genomic selection model. G3 Genes Genomes Genet. 2015, 5, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Perez, P.; Hickey, J.; Burgueno, J.; Ornella, L.; Cerón-Rojas, J.; Zhang, X.; Dreisigacker, S.; Babu, R.; Li, Y.; et al. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 2014, 112, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Panozzo, J.; Walker, C.K.; Choy, A.L.; Kant, S.; Wong, D.; Tibbits, J.; Daetwyler, H.D.; Rochfort, S.; Hayden, M.J.; et al. Accelerating wheat breeding for end-use quality with multi-trait genomic predictions incorporating near infrared and nuclear magnetic resonance-derived phenotypes. Theor. Appl. Genet. 2017, 130, 2505–2519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Sun, Y.X.; Ye, Y.L. Zinc biofortification of wheat through fertilizer applications in different locations of China. Field Crops Res. 2012, 125, 1–7. [Google Scholar] [CrossRef]
- Zou, C.Q.; Zhang, Y.Q.; Rashid, A.; Ram, H.; Savasli, E.; Arisoy, R.Z.; Ortiz-Monasterio, I.; Simunji, S.; Wang, Z.H.; Sohu, V.; et al. Biofortification of wheat with zinc through zinc fertilization in seven countries. Plant Soil 2012, 361, 119–130. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, R.; Rezaul, K.M.; Zhang, F.; Zou, C. Iron and zinc concentrations in grain and flour of winter wheat as affected by foliar application. J. Agric. Food Chem. 2010, 58, 12268–12274. [Google Scholar] [CrossRef]
- Aciksoz, S.B.; Yazici, A.; Ozturk, L.; Cakmak, I. Biofortification of wheat with iron through soil and foliar application of nitrogen and iron fertilizers. Plant Soil 2011, 349, 215–225. [Google Scholar] [CrossRef]
- Singh, R.; Govindan, V.; Andersson, M.S. Zinc-Biofortified Wheat: Harnessing Genetic Diversity for Improved Nutritional Quality. Sci. Br. Biofortif. Ser. 2017, 1, 1–4. [Google Scholar]
- Lowe, N.M.; Khan, M.J.; Broadley, M.R.; Zia, M.H.; McArdle, H.J.; Joy, E.J.; Ohly, H.; Shahzad, B.; Ullah, U.; Kabana, G.; et al. Examining the effectiveness of consuming flour made from agronomically biofortified wheat (Zincol-2016/NR-421) for improving Zn status in women in a low-resource setting in Pakistan: Study protocol for a randomised, double-blind, controlled cross-over trial (BiZiFED). BMJ Open 2018, 8, e021364. [Google Scholar]
- Shi, R.; Li, H.; Tong, Y.; Jing, R.; Zhang, F.; Zou, C. Identification of quantitative trait locus of zinc and phosphorus density in wheat (Triticum aestivum L.) grain. Plant Soil 2008, 306, 95–104. [Google Scholar] [CrossRef]
- Genc, Y.; Verbyla, A.P.; Torun, A.A.; Cakmak, I.; Willsmore, K.; Wallwork, H.; McDonald, G.K. Quantitative trait loci analysis of zinc efficiency and grain zinc concentration in wheat using whole genome average interval mapping. Plant Soil 2009, 314, 49. [Google Scholar] [CrossRef]
- Tiwari, V.K.; Rawat, N.; Chhuneja, P.; Neelam, K.; Aggarwal, R.; Randhawa, G.S.; Dhaliwal, H.S.; Keller, B.; Singh, K. Mapping of quantitative trait loci for grain iron and zinc concentration in diploid A genome wheat. J. Hered. 2009, 100, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.L.; Tong, Y.P.; Jing, R.L.; Zhang, F.S.; Zou, C.Q. Characterization of quantitative trait loci for grain minerals in hexaploid wheat (Triticum aestivum L.). J. Integr. Agric. 2013, 12, 1512–1521. [Google Scholar] [CrossRef]
- Roshanzamir, H.; Kordenaeej, A.; Bostani, A. Mapping QTLs related to Zn and Fe concentrations in bread wheat (Triticum aestivum) grain using microsatellite markers. Iran. J. Genet. Plant Breed. 2013, 2, 10–17. [Google Scholar]
- Hao, Y.; Velu, G.; Peña, R.J.; Singh, S.; Singh, R.P. Genetic loci associated with high grain zinc concentration and pleiotropic effect on kernel weight in wheat (Triticum aestivum L.). Mol. Breed. 2014, 34, 1893–1902. [Google Scholar] [CrossRef]
- Crespo-Herrera, L.A.; Govindan, V.; Stangoulis, J.; Hao, Y.; Singh, R.P. QTL mapping of grain Zn and Fe concentrations in two hexaploid wheat RIL populations with ample transgressive segregation. Front. Plant Sci. 2017, 8, 1800. [Google Scholar] [CrossRef]
- Srinivasa, J.; Arun, B.; Mishra, V.K.; Singh, G.P.; Velu, G.; Babu, R.; Vasistha, N.K.; Joshi, A.K. Zinc and iron concentration QTL mapped in a Triticum spelta × T. aestivum cross. Theor. Appl. Genet. 2014, 127, 1643–1651. [Google Scholar] [CrossRef]
- Velu, G.; Tutus, Y.; Gomez-Becerra, H.F.; Hao, Y.; Demir, L.; Kara, R.; Crespo-Herrera, L.A.; Orhan, S.; Yazici, A.; Singh, R.P.; et al. QTL mapping for grain zinc and iron concentrations and zinc efficiency in a tetraploid and hexaploid wheat mapping populations. Plant Soil 2017, 411, 81–99. [Google Scholar] [CrossRef]
- Pu, Z.E.; Ma, Y.U.; He, Q.Y.; Chen, G.Y.; Wang, J.R.; Liu, Y.X.; Jiang, Q.T.; Wei, L.I.; Dai, S.F.; Wei, Y.M.; et al. Quantitative trait loci associated with micronutrient concentrations in two recombinant inbred wheat lines. J. Integr. Agric. 2014, 13, 2322–2329. [Google Scholar] [CrossRef]
- Krishnappa, G.; Singh, A.M.; Chaudhary, S.; Ahlawat, A.K.; Singh, S.K.; Shukla, R.B.; Jaiswal, J.P.; Singh, G.P.; Solanki, I.S. Molecular mapping of the grain iron and zinc concentration, protein content and thousand kernel weight in wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0174972. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Herrera, L.A.; Velu, G.; Singh, R.P. Quantitative trait loci mapping reveals pleiotropic effect for grain iron and zinc concentrations in wheat. Ann. Appl. Biol. 2016, 169, 27–35. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Liu, Q.; Tian, X.; Shi, Y.; Zhang, X. QTL mapping of selenium content using a RIL population in wheat. PLoS ONE 2017, 12, e0184351. [Google Scholar] [CrossRef]
- Pu, Z.; Pei, Y.; Yang, J.; Ma, J.; Li, W.; Liu, D.; Wang, J.; Wei, Y.; Zheng, Y. A QTL located on chromosome 3D enhances the selenium concentration of wheat grain by improving phytoavailability and root structure. Plant Soil 2018, 425, 287–296. [Google Scholar] [CrossRef]
- Yan, J.; Xue, W.T.; Yang, R.Z.; Qin, H.B.; Zhao, G.; Tzion, F.; Cheng, J.P. Quantitative trait loci conferring grain selenium nutrient in durum wheat× wild emmer wheat RIL population. Czech J. Genet. Plant Breed. 2018, 54, 52–58. [Google Scholar] [CrossRef]
- Gorafi, Y.S.; Ishii, T.; Kim, J.S.; Elbashir, A.A.E.; Tsujimoto, H. Genetic variation and association mapping of grain iron and zinc contents in synthetic hexaploid wheat germplasm. Plant Genet. Resour. 2018, 16, 9–17. [Google Scholar] [CrossRef]
- Magallanes-López, A.M.; Hernandez-Espinosa, N.; Velu, G.; Posadas-Romano, G.; Ordoñez-Villegas, V.M.G.; Crossa, J.; Ammar, K.; Guzmán, C. Variability in iron, zinc and phytic acid content in a worldwide collection of commercial durum wheat cultivars and the effect of reduced irrigation on these traits. Food Chem. 2017, 237, 499–505. [Google Scholar] [CrossRef]
- Amiri, R.; Bahraminejad, S.; Cheghamirza, K. Estimating genetic variation and genetic parameters for grain iron, zinc and protein concentrations in bread wheat genotypes grown in Iran. J. Cereal Sci. 2018, 80, 16–23. [Google Scholar] [CrossRef]
- Velu, G.; Crossa, J.; Singh, R.P.; Hao, Y.; Dreisigacker, S.; Perez-Rodriguez, P.; Joshi, A.K.; Chatrath, R.; Gupta, V.; Balasubramaniam, A.; et al. Genomic prediction for grain zinc and iron concentrations in spring wheat. Theor. Appl. Genet. 2016, 129, 1595–1605. [Google Scholar] [CrossRef]
- Manickavelu, A.; Hattori, T.; Yamaoka, S.; Yoshimura, K.; Kondou, Y.; Onogi, A.; Matsui, M.; Iwata, H.; Ban, T. Genetic nature of elemental contents in wheat grains and its genomic prediction: Toward the effective use of wheat landraces from Afghanistan. PLoS ONE 2017, 12, 0169416. [Google Scholar] [CrossRef]
- Alomari, D.; Eggert, K.; von Wirén, N.; Polley, A.; Plieske, J.; Ganal, M.; Liu, F.; Pillen, K.; Röder, M. Whole-genome association mapping and genomic prediction for iron concentration in wheat grains. Int. J. Mol. Sci. 2019, 20, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.X.; Xing, H.J.; Liu, C.P.; Zhang, Z.W.; Xu, S.W. Effect of Selenium Deficiency on Nitric Oxide and Heat Shock Proteins in Chicken Erythrocytes. Biol. Trace Elem. Res. 2016, 171, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Noble, R.M.; Barry, G.A. Survey of selenium concentrations in wheat, sorghum and soybean grains, prepared poultry feeds and feed ingredients from Queensland. Qld. J. Agric. Anim. Sci. 1982, 39, 1–8. [Google Scholar]
- Tveitnes, S.; Singh, B.R.; Ruud, L. Selenium concentration in spring wheat as influenced by basal application and top dressing of selenium-enriched fertilizers. Fertil. Res. 1995, 45, 163–167. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Rizzi, R.; Pannacciulli, E.; Gatta, C.D. Mineral composition in hulled wheat grains: A comparison between emmer (Triticum dicoccon Schrank) and spelt (T. spelta L.) accessions. Int. J. Food Sci. Nutr. 1997, 48, 381–386. [Google Scholar] [CrossRef]
- Lyons, G.; Ortiz-Monasterio, I.; Stangoulis, J.; Graham, R. Selenium concentration in wheat grain: Is there sufficient genotypic variation to use in breeding? Plant Soil 2005, 269, 369–380. [Google Scholar] [CrossRef]
- Yeum, K.J.; Russell, R.M. Carotenoid bioavailability and bioconversion. Annu. Rev. Nutr. 2002, 22, 483–504. [Google Scholar] [CrossRef]
- Parker, G.D.; Chalmers, K.J.; Rathjen, A.J.; Langridge, P. Mapping loci associated with flour colour in wheat (Triticum aestivum L.). Theor. Appl. Genet. 1998, 97, 238–245. [Google Scholar] [CrossRef]
- Elouafi, I.; Nachit, M.M.; Martin, L.M. Identification of a microsatellite on chromosome 7B showing a strong linkage with yellow pigment in durum wheat (Triticum turgidum L. var. durum). Hereditas 2001, 135, 255–261. [Google Scholar] [CrossRef]
- Pozniak, C.J.; Knox, R.E.; Clarke, F.R.; Clarke, J.M. Identification of QTL and association of a phytoene synthase gene with endosperm colour in durum wheat. Theor. Appl. Genet. 2007, 114, 525–537. [Google Scholar] [CrossRef]
- Patil, R.M.; Oak, M.D.; Tamhankar, S.A.; Sourdille, P.; Rao, V.S. Mapping and validation of a major QTL for yellow pigment content on 7AL in durum wheat (Triticum turgidum L. ssp. durum). Mol. Breed. 2008, 21, 485–496. [Google Scholar] [CrossRef]
- Zhang, W.; Dubcovsky, J. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor. Appl. Genet. 2008, 116, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, Y.; Xiao, Y.; He, Z.; Zhang, Y.; Yan, J.; Zhang, Y.; Xia, X.; Ma, C. QTL mapping for flour and noodle colour components and yellow pigment content in common wheat. Euphytica 2009, 165, 435. [Google Scholar] [CrossRef]
- Blanco, A.; Colasuonno, P.; Gadaleta, A.; Mangini, G.; Schiavulli, A.; Simeone, R.; Digesù, A.M.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L. Quantitative trait loci for yellow pigment concentration and individual carotenoid compounds in durum wheat. J. Cereal Sci. 2011, 54, 255–264. [Google Scholar] [CrossRef]
- Crawford, A.C.; Stefanova, K.; Lambe, W.; McLean, R.; Wilson, R.; Barclay, I.; Francki, M.G. Functional relationships of phytoene synthase 1 alleles on chromosome 7A controlling flour colour variation in selected Australian wheat genotypes. Theor. Appl. Genet. 2011, 123, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Roncallo, P.F.; Cervigni, G.L.; Jensen, C.; Miranda, R.; Carrera, A.D.; Helguera, M.; Echenique, V. QTL analysis of main and epistatic effects for flour color traits in durum wheat. Euphytica 2012, 185, 77–92. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, H.Y.; Wang, Y.Y.; Pu, Y.Y.; Kong, F.M.; Li, S.S. QTL mapping for the color, carotenoids and polyphenol oxidase activity of flour in recombinant inbred lines of wheat. Aust. J. Crop Sci. 2013, 7, 328–337. [Google Scholar]
- Colasuonno, P.; Gadaleta, A.; Giancaspro, A.; Nigro, D.; Giove, S.; Incerti, O.; Mangini, G.; Signorile, A.; Simeone, R.; Blanco, A. Development of a high-density SNP-based linkage map and detection of yellow pigment content QTLs in durum wheat. Mol. Breed. 2014, 34, 1563–1578. [Google Scholar] [CrossRef]
- Kuchel, H.; Langridge, P.; Mosionek, L.; Williams, K.; Jefferies, S.P. The genetic control of milling yield, dough rheology and baking quality of wheat. Theor. Appl. Genet. 2006, 112, 1487. [Google Scholar] [CrossRef]
- Zhai, S.; He, Z.; Wen, W.; Jin, H.; Liu, J.; Zhang, Y.; Liu, Z.; Xia, X. Genome-wide linkage mapping of flour color-related traits and polyphenol oxidase activity in common wheat. Theor. Appl. Genet. 2016, 129, 377–394. [Google Scholar] [CrossRef]
- He, X.Y.; Zhang, Y.L.; He, Z.H.; Wu, Y.P.; Xiao, Y.G.; Ma, C.X.; Xia, X.C. Characterization of phytoene synthase 1 gene (Psy1) located on common wheat chromosome 7A and development of a functional marker. Theor. Appl. Genet. 2008, 116, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Ficco, D.B.; Mastrangelo, A.M.; Trono, D.; Borrelli, G.M.; De Vita, P.; Fares, C.; Beleggia, R.; Platani, C.; Papa, R. The colours of durum wheat: A review. Crop Pasture Sci. 2014, 65, 1–15. [Google Scholar] [CrossRef]
- Wang, J.; He, X.; He, Z.; Wang, H.; Xia, X. Cloning and phylogenetic analysis of phytoene synthase 1 (Psy1) genes in common wheat and related species. Hereditas 2009, 146, 208–256. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Reimer, S.; Pozniak, C.J.; Clarke, F.R.; Clarke, J.M.; Knox, R.E.; Singh, A.K. Allelic variation at Psy1-A1 and association with yellow pigment in durum wheat grain. Theor. Appl. Genet. 2009, 118, 1539–1548. [Google Scholar] [CrossRef]
- Zhai, S.; Xia, X.; He, Z. Carotenoids in staple cereals: Metabolism, regulation, and genetic manipulation. Front. Plant Sci. 2016, 7, 1197. [Google Scholar] [CrossRef]
- Zhang, W.; Lukaszewski, A.J.; Kolmer, J.; Soria, M.A.; Goyal, S.; Dubcovsky, J. Molecular characterization of durum and common wheat recombinant lines carrying leaf rust resistance (Lr19) and yellow pigment (Y) genes from Lophopyrum ponticum. Theor. Appl. Genet. 2005, 111, 573–582. [Google Scholar] [CrossRef]
- Mérida-García, R.; Liu, G.; He, S.; Gonzalez-Dugo, V.; Dorado, G.; Gálvez, S.; Solís, I.; Zarco-Tejada, P.J.; Reif, J.C.; Hernandez, P. Genetic dissection of agronomic and quality traits based on association mapping and genomic selection approaches in durum wheat grown in Southern Spain. PLoS ONE 2019, 14, 0211718. [Google Scholar] [CrossRef]
- Lolas, G.M.; Palmidis, N.; Markakis, P. The Phytic Acid-Total Phosphorus Relationship in Barley, Oats, Soybeans, and Wheat. Cereal Chem. 1976, 53, 867–871. [Google Scholar]
- Raboy, V.; Noaman, M.W.; Taylor, G.A.; Pickett, S.G. Grain phytic acid and protein are highly correlated in winter wheat. Crop Sci. 1991, 31, 631–635. [Google Scholar] [CrossRef]
- Reddy, N.R.; Pierson, M.D. Reduction in antinutritional and toxic components in plant foods by fermentation. Food Res. Int. 1994, 27, 281–290. [Google Scholar] [CrossRef]
- Cook, J.D.; Reddy, M.B.; Burri, J.; Juillerat, M.A.; Hurrell, R.F. The influence of different cereal grains on iron absorption from infant cereal foods. Am. J. Clin. Nutr. 1997, 65, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Barbro, N.; Brittmarie, S.; Cederblad, Å.K.E. Reduction of the phytate content of bran by leavening in bread and its effect on zinc absorption in man. Br. J. Nutr. 1985, 53, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Ram, S.; Verma, A.; Sharma, S. Large variability exits in phytase levels among Indian wheat varieties and synthetic hexaploids. J. Cereal Sci. 2010, 52, 486–490. [Google Scholar] [CrossRef]
- Shitre, A.S.; Gadekar, D.A.; Vikas, V.R.; Bakshi, S.; Kumar, V.; Vishwakarma, G.; Das, B.K. Genotypic variation for phytic acid, inorganic phosphate and mineral contents in advanced breeding lines of wheat (Triticum aestivum L.). Electron. J. Plant Breed. 2015, 6, 395–402. [Google Scholar]
- Bhati, K.K.; Aggarwal, S.; Sharma, S.; Mantri, S.; Singh, S.P.; Bhalla, S.; Kaur, J.; Tiwari, S.; Roy, J.K.; Tuli, R.; et al. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Sci. 2014, 224, 74–85. [Google Scholar] [CrossRef]
- Bhati, K.K.; Alok, A.; Kumar, A.; Kaur, J.; Tiwari, S.; Pandey, A.K. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 2016, 67, 4379–4389. [Google Scholar] [CrossRef]
- Naidoo, R.; Watson, G.M.F.; Derera, J.; Tongoona, P.; Laing, M.D. Marker-assisted selection for low phytic acid (lpa1-1) with single nucleotide polymorphism marker and amplified fragment length polymorphisms for background selection in a maize backcross breeding programme. Mol. Breed. 2012, 30, 1207–1217. [Google Scholar] [CrossRef]
- Kris-Etherton, P.M.; Lefevre, M.; Beecher, G.R.; Gross, M.D.; Keen, C.L.; Etherton, T.D. Bioactive compounds in nutrition and health-research methodologies for establishing biological function: The antioxidant and anti-inflammatory effects of flavonoids on atherosclerosis. Annu. Rev. Nutr. 2004, 24, 511–538. [Google Scholar] [CrossRef]
- Patel, K.; Jain, A.; Patel, D.K. Medicinal significance, pharmacological activities, and analytical aspects of anthocyanidins ‘delphinidin’: A concise report. J. Acute Dis. 2013, 2, 169–178. [Google Scholar] [CrossRef]
- Arts, I.C.; Hollman, P.C. Polyphenols and disease risk in epidemiologic studies. Am. J. Clin. Nutr. 2005, 81, 317S–325S. [Google Scholar] [CrossRef]
- Tsuda, T.; Horio, F.; Uchida, K.; Aoki, H.; Osawa, T. Dietary cyanidin 3-O-β-D-glucoside-rich purple corn color prevents obesity and ameliorates hyperglycemia in mice. J. Nutr. 2003, 133, 2125–2130. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaya, O.; Arbuzova, V.S.; Lohwasser, U.; Röder, M.S.; Börner, A. Microsatellite mapping of complementary genes for purple grain colour in bread wheat (Triticum aestivum) L. Euphytica 2006, 150, 355–364. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Röder, M.S.; Börner, A. Mapping genes controlling anthocyanin pigmentation on the glume and pericarp in tetraploid wheat (Triticum durum L.). Euphytica 2010, 171, 65–69. [Google Scholar] [CrossRef]
- Tereshchenko, O.; Gordeeva, E.; Arbuzova, V.; Börner, A.; Khlestkina, E. The D genome carries a gene determining purple grain colour in wheat. Cereal Res. Commun. 2012, 40, 334–341. [Google Scholar] [CrossRef]
- Liu, D.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Liu, B.; Zhang, H. Transcriptome analysis of purple pericarps in common wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0155428. [Google Scholar] [CrossRef]
- Shoeva, O.; Gordeeva, E.; Khlestkina, E. The regulation of anthocyanin synthesis in the wheat pericarp. Molecules 2014, 19, 20266–20279. [Google Scholar] [CrossRef]
- Zong, Y.; Xi, X.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Liu, B.; Wang, D.; Zhang, H. Allelic variation and transcriptional isoforms of wheat TaMYC1 gene regulating anthocyanin synthesis in pericarp. Front. Plant Sci. 2017, 8, 1645. [Google Scholar] [CrossRef]
- Li, N.; Zong, Y.; Liu, B.L.; Chen, W.J.; Zhang, B. TaMYB3, encoding a functional MYB transcriptor, isolated from the purple pericarp of Triticum aestivum. Cereal Res. Commun. 2017, 45, 369–380. [Google Scholar] [CrossRef]
- Jiang, W.; Liu, T.; Nan, W.; Jeewani, D.C.; Niu, Y.; Li, C.; Wang, Y.; Shi, X.; Wang, C.; Wang, J.; et al. Two transcription factors TaPpm1 and TaPpb1 co-regulate anthocyanin biosynthesis in purple pericarps of wheat. J. Exp. Bot. 2018, 69, 2555–2567. [Google Scholar] [CrossRef]
- Burešová, V.; Kopecký, D.; Bartoš, J.; Martinek, P.; Watanabe, N.; Vyhnánek, T.; Doležel, J. Variation in genome composition of blue-aleurone wheat. Theor. Appl. Genet. 2015, 128, 273–282. [Google Scholar] [CrossRef]
- Martinek, P.; Jirsa, O.; Vaculová, K.; Chrpová, J.; Watanabe, N.; Burešová, V.; Kopecký, D.; Štiasna, K.; Vyhnánek, T.; Trojan, V. Use of wheat gene resources with different grain colour in breeding. In Proceedings of the Tagungsband der 64, Raumberg-Gumpenstein, Austria, 25–26 November 2013. [Google Scholar]
- Neeraja, C.N.; Ravindra, B.V.; Ram, S.; Hossain, F.; Hariprasanna, K.; Rajpurohit, B.S.; Prabhakar, T.L.; Prasad, K.S.; Sandhu, J.S.; Datta, S.K. Biofortification in cereals—Progress and prospects. Curr. Sci. 2017, 113, 1050–1057. [Google Scholar] [CrossRef]
- PTI. M S Swaminathan: India Must Focus on Nutrition Security. Available online: https://www.thehindubusinessline.com/economy/agri-business/m-s-swaminathanindia-must-focus-onnutrition-security/article24739207.ece (accessed on 12 October 2018).
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).