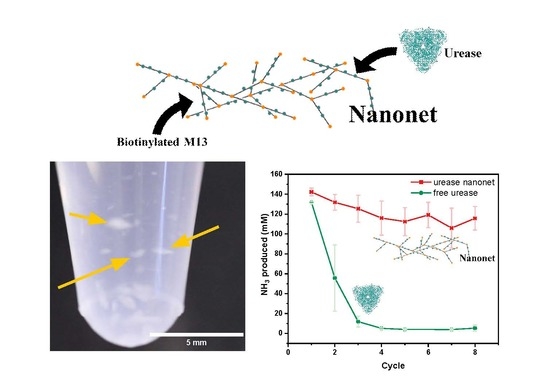
Genetically Modified M13 Bacteriophage Nanonets for Enzyme Catalysis and Recovery
Abstract
1. Introduction
2. Results and Discussion
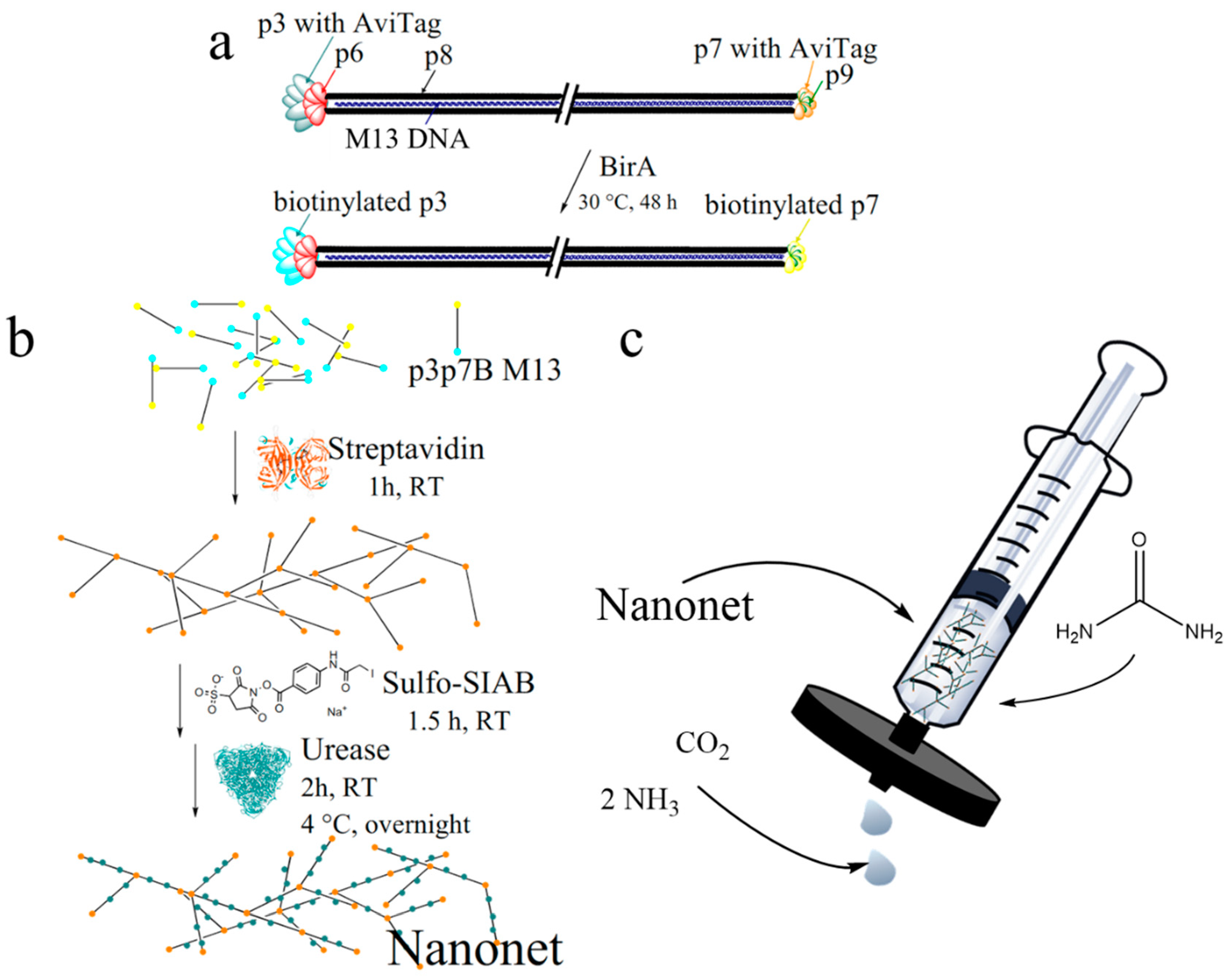
2.1. Biotinylation and Characterization of M13 Bacteriophages
2.2. Nanonet Synthesis and Characterization
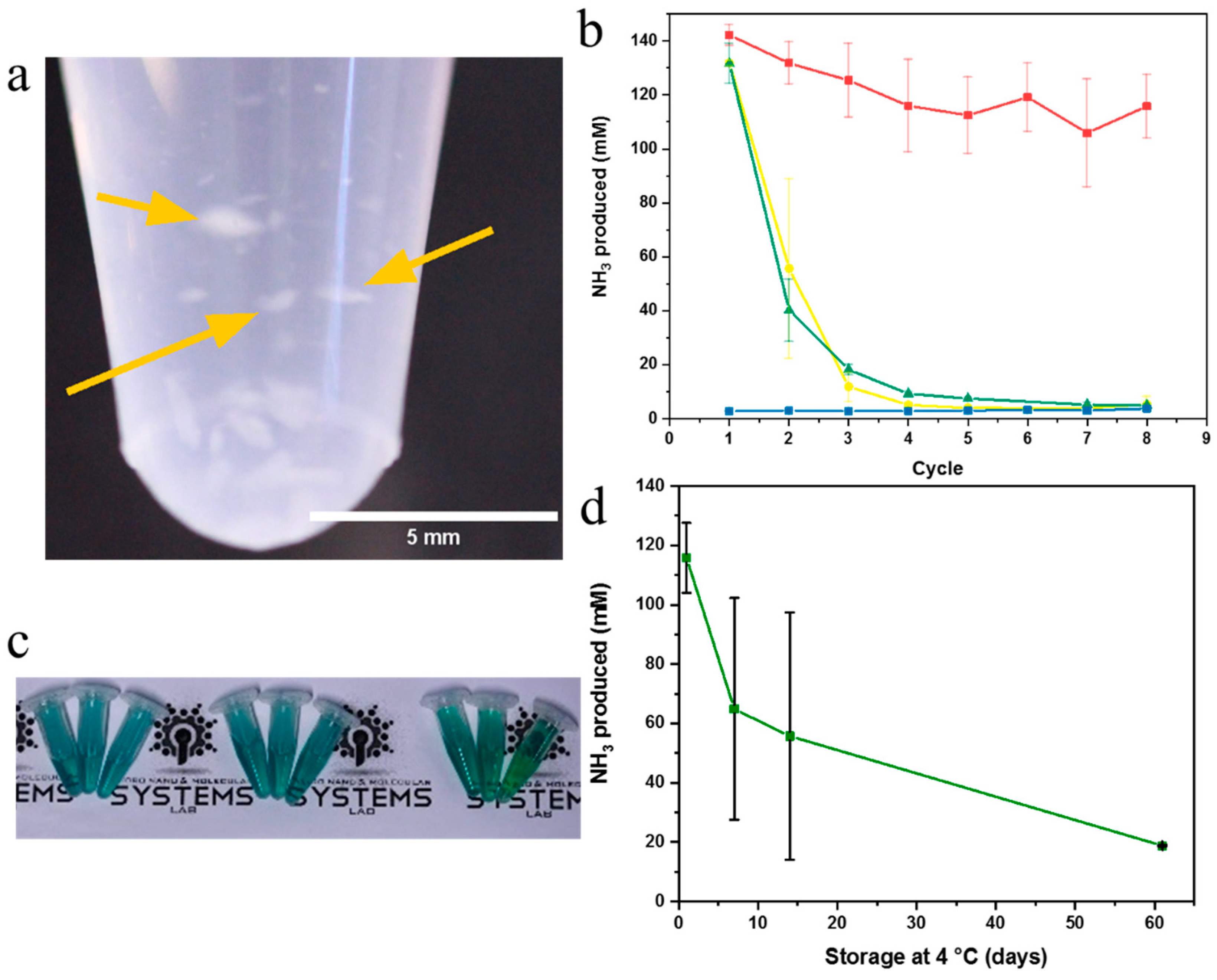
2.3. Enzyme Activity Assays of Nanonets
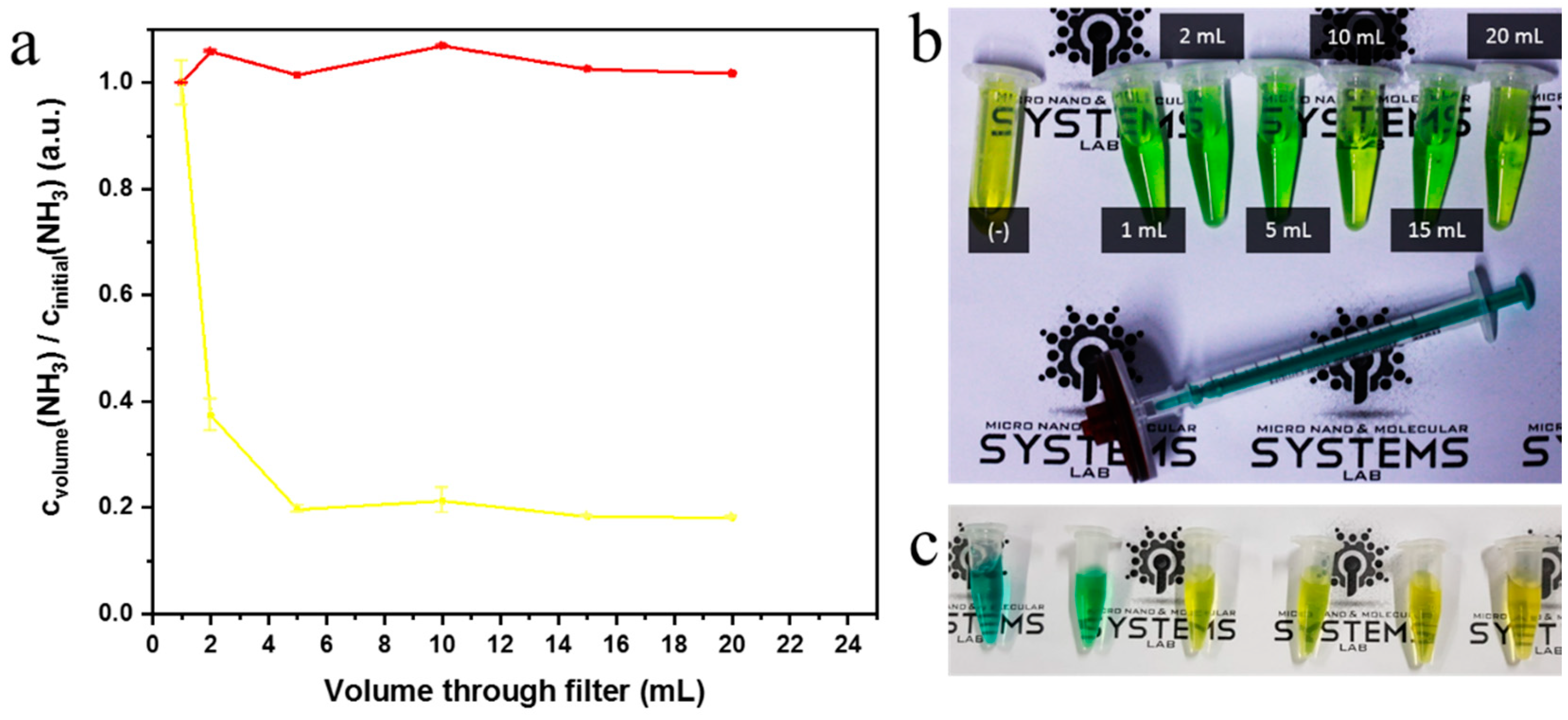
2.4. Syringe Filter Nanonet Reactor
3. Materials and Methods
3.1. Materials
3.2. M13 Bacteriophage Biotinylation
3.3. Characterization of Biotinylation Yield of M13 Bacteriophage
3.4. M13 Bacteriophage Nanonet
3.5. Sulfo-SIAB Cross-Linked Urease Nanonet (Enzyme-Nanonets)
3.6. Glutaraldehyde Cross-Linked Urease Nanonet
3.7. Fluorescence Imaging
3.8. Urease Activity Assays
3.9. Syringe Filter Reactor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lucena, G.N.; dos Santos, C.C.; Pinto, G.C.; da Rocha, C.O.; Brandt, J.V.; de Paula, A.V.; Jafelicci Júnior, M.; Marques, R.F.C. Magnetic cross-linked enzyme aggregates (MCLEAs) applied to biomass conversion. J. Solid State Chem. 2019, 270, 58–70. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Schoevaart, R.; Van Langen, L.M. Cross-linked enzyme aggregates (CLEAs): A novel and versatile method for enzyme immobilization (a review). Biocatal. Biotransform. 2005, 23, 141–147. [Google Scholar] [CrossRef]
- Touahar, I.E.; Haroune, L.; Ba, S.; Bellenger, J.-P.; Cabana, H. Characterization of combined cross-linked enzyme aggregates from laccase, versatile peroxidase and glucose oxidase, and their utilization for the elimination of pharmaceuticals. Sci. Total Environ. 2014, 481, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Lozano, S.; López-Gallego, F.; Mateos-Díaz Juan, C.; Favela-Torres, E. Cross-linked enzyme aggregates (CLEA) in enzyme improvement—A review. Biocatalysis 2016, 1, 166–177. [Google Scholar] [CrossRef]
- Xu, M.-Q.; Wang, S.-S.; Li, L.-N.; Gao, J.; Zhang, Y.-W. Combined cross-linked enzyme aggregates as biocatalysts. Catalysts 2018, 8, 460. [Google Scholar] [CrossRef]
- Zhang, Q.; Zha, X.; Zhou, N.; Tian, Y. Preparation of crosslinked enzyme aggregates (CLEAs) of acid urease with urethanase activity and their application. J. Basic Microbiol. 2016, 56, 422–431. [Google Scholar] [CrossRef]
- Sheldon, R.A. CLEAs, Combi-CLEAs and ‘Smart’Magnetic CLEAs: Biocatalysis in a Bio-Based Economy. Catalysts 2019, 9, 261. [Google Scholar] [CrossRef]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef]
- Cao, L.; Langen, L.; Sheldon, R.A. Immobilised enzymes: Carrier-bound or carrier-free? Curr. Opin. Biotechnol. 2003, 14, 387–394. [Google Scholar] [CrossRef]
- Sheldon, R.A. Enzyme Immobilization: The Quest for Optimum Performance. Adv. Synth. Catal. 2007, 349, 1289–1307. [Google Scholar] [CrossRef]
- Magne, V.; Amounas, M.; Innocent, C.; Dejean, E.; Seta, P. Enzyme textile for removal of urea with coupling process: Enzymatic reaction and electrodialysis. Desalination 2002, 144, 163–166. [Google Scholar] [CrossRef]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of Different Enzyme Immobilization Strategies to Improve Enzyme Performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Cuenca, S.; Mansilla, C.; Aguado, M.; Yuste-Calvo, C.; Sánchez, F.; Sánchez-Montero, J.M.; Ponz, F. Nanonets Derived from Turnip Mosaic Virus as Scaffolds for Increased Enzymatic Activity of Immobilized Candida antarctica Lipase B. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, D.; Carette, N.; Michon, T. Virus scaffolds as enzyme nano-carriers. Trends Biotechnol. 2012, 30, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Hooker, J.M.; Kovacs, E.W.; Francis, M.B. Interior Surface Modification of Bacteriophage MS2. J. Am. Chem. Soc. 2004, 126, 3718–3719. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.S.; Kirshenbaum, K. Nano-Tailoring: Stitching Alterations on Viral Coats. Chem. Biol. 2004, 11, 418–420. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Comellas-Aragonès, M.; Engelkamp, H.; Claessen, V.I.; Sommerdijk, N.A.J.M.; Rowan, A.E.; Christianen, P.C.M.; Maan, J.C.; Verduin, B.J.M.; Cornelissen, J.J.L.M.; Nolte, R.J.M. A virus-based single-enzyme nanoreactor. Nat. Nanotechnol. 2007, 2, 635. [Google Scholar] [CrossRef] [PubMed]
- Röder, J.; Fischer, R.; Commandeur, U. Engineering Potato Virus X Particles for a Covalent Protein Based Attachment of Enzymes. Small 2017, 13, 1702151. [Google Scholar] [CrossRef]
- Eiben, S.; Koch, C.; Altintoprak, K.; Southan, A.; Tovar, G.; Laschat, S.; Weiss, I.M.; Wege, C. Plant virus-based materials for biomedical applications: Trends and prospects. Adv. Drug Deliv. Rev. 2018. [Google Scholar] [CrossRef]
- Carette, N.; Engelkamp, H.; Akpa, E.; Pierre, S.J.; Cameron, N.R.; Christianen, P.C.M.; Maan, J.C.; Thies, J.C.; Weberskirch, R.; Rowan, A.E.; et al. A virus-based biocatalyst. Nat. Nanotechnol. 2007, 2, 226. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Chen, Y.; Li, S.; Nguyen, H.G.; Niu, Z.; You, S.; Mello, C.M.; Lu, X.; Wang, Q. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug. Chem. 2010, 21, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Mohan, K.; Weiss, G.A. Chemically Modifying Viruses for Diverse Applications. ACS Chem. Biol. 2016, 11, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, M.; Strych, U.; Kim, J.; Goux, H.; Dhamane, S.; Poongavanam, M.-V.; Hagström, A.E.V.; Kourentzi, K.; Conrad, J.C.; Willson, R.C. Aptamer-Phage Reporters for Ultrasensitive Lateral Flow Assays. Anal. Chem. 2015, 87, 11660–11665. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, M.; Dhamane, S.; Hagstrom, A.E.V.; Garvey, G.; Chen, W.-H.; Kourentzi, K.; Strych, U.; Willson, R.C. Functionalized viral nanoparticles as ultrasensitive reporters in lateral-flow assays. Analyst 2013, 138, 5584–5587. [Google Scholar] [CrossRef]
- Litvinov, J.; Hagström, A.E.V.; Lopez, Y.; Adhikari, M.; Kourentzi, K.; Strych, U.; Monzon, F.A.; Foster, W.; Cagle, P.T.; Willson, R.C. Ultrasensitive immuno-detection using viral nanoparticles with modular assembly using genetically-directed biotinylation. Biotechnol. Lett. 2014, 36, 1863–1868. [Google Scholar] [CrossRef]
- Smelyanski, L.; Gershoni, J.M. Site directed biotinylation of filamentous phage structural proteins. Virol. J. 2011, 8, 495. [Google Scholar] [CrossRef]
- Rothenstein, D.; Claasen, B.; Omiecienski, B.; Lammel, P.; Bill, J. Isolation of ZnO-Binding 12-mer Peptides and Determination of Their Binding Epitopes by NMR Spectroscopy. J. Am. Chem. Soc. 2012, 134, 12547–12556. [Google Scholar] [CrossRef]
- Rothenstein, D.; Shopova-Gospodinova, D.; Bakradze, G.; Jeurgens, L.P.H.; Bill, J. Generation of luminescence in biomineralized zirconia by zirconia-binding peptides. CrystEngComm 2015, 17, 1783–1790. [Google Scholar] [CrossRef]
- Kilper, S.; Facey, S.J.; Burghard, Z.; Hauer, B.; Rothenstein, D.; Bill, J. Macroscopic Properties of Biomimetic Ceramics Are Governed by the Molecular Recognition at the Bioorganic–Inorganic Interface. Adv. Funct. Mater. 2018, 28, 1705842. [Google Scholar] [CrossRef]
- Kilper, S.; Jahnke, T.; Aulich, M.; Burghard, Z.; Rothenstein, D.; Bill, J. Genetically Induced In Situ-Poling for Piezo-Active Biohybrid Nanowires. Adv. Mater. 2019, 31, 1805597. [Google Scholar] [CrossRef] [PubMed]
- Bruin, R.d.; Spelt, K.; Mol, J.; Koes, R.; Quattrocchio, F. Selection of high-affinity phage antibodies from phage display libraries. Nat. Biotechnol. 1999, 17, 397–399. [Google Scholar] [CrossRef] [PubMed]
- Merzlyak, A.; Indrakanti, S.; Lee, S.-W. Genetically Engineered Nanofiber-Like Viruses For Tissue Regenerating Materials. Nano Lett. 2009, 9, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Branston, S.D.; Stanley, E.C.; Ward, J.M.; Keshavarz-Moore, E. Determination of the survival of bacteriophage M13 from chemical and physical challenges to assist in its sustainable bioprocessing. Biotechnol. Bioprocess Eng. 2013, 18, 560–566. [Google Scholar] [CrossRef]
- Alarcón-Correa, M.; Günther, J.-P.; Troll, J.; Kadiri, V.M.; Bill, J.; Fischer, P.; Rothenstein, D. Self-Assembled Phage-Based Colloids for High Localized Enzymatic Activity. ACS Nano 2019, 13, 5810–5815. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Wabbel, K.; Eber, F.J.; Krolla-Sidenstein, P.; Azucena, C.; Gliemann, H.; Eiben, S.; Geiger, F.; Wege, C. Modified TMV Particles as Beneficial Scaffolds to Present Sensor Enzymes. Front. Plant Sci. 2015, 6, 1137. [Google Scholar] [CrossRef] [PubMed]
- Verdouw, H.; Van Echteld, C.J.A.; Dekkers, E.M.J. Ammonia determination based on indophenol formation with sodium salicylate. Water Res. 1978, 12, 399–402. [Google Scholar] [CrossRef]
- Smith, G.P. Absorption Spectrum and Quantitation of Filamentous Phage. Available online: http://www.biosci.missouri.edu/smithgp/PhageDisplayWebsite/PhageDisplayWebsiteindex.html (accessed on 26 August 2019).
- Scholle, M.D.; Kriplani, U.; Pabon, A.; Sishtla, K.; Glucksman, M.J.; Kay, B.K. Mapping Protease Substrates by Using a Biotinylated Phage Substrate Library. ChemBioChem 2006, 7, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadiri, V.M.; Alarcón-Correa, M.; Ruppert, J.; Günther, J.-P.; Bill, J.; Rothenstein, D.; Fischer, P. Genetically Modified M13 Bacteriophage Nanonets for Enzyme Catalysis and Recovery. Catalysts 2019, 9, 723. https://doi.org/10.3390/catal9090723
Kadiri VM, Alarcón-Correa M, Ruppert J, Günther J-P, Bill J, Rothenstein D, Fischer P. Genetically Modified M13 Bacteriophage Nanonets for Enzyme Catalysis and Recovery. Catalysts. 2019; 9(9):723. https://doi.org/10.3390/catal9090723
Chicago/Turabian StyleKadiri, Vincent Mauricio, Mariana Alarcón-Correa, Jacqueline Ruppert, Jan-Philipp Günther, Joachim Bill, Dirk Rothenstein, and Peer Fischer. 2019. "Genetically Modified M13 Bacteriophage Nanonets for Enzyme Catalysis and Recovery" Catalysts 9, no. 9: 723. https://doi.org/10.3390/catal9090723
APA StyleKadiri, V. M., Alarcón-Correa, M., Ruppert, J., Günther, J.-P., Bill, J., Rothenstein, D., & Fischer, P. (2019). Genetically Modified M13 Bacteriophage Nanonets for Enzyme Catalysis and Recovery. Catalysts, 9(9), 723. https://doi.org/10.3390/catal9090723