Xylan Deconstruction by Thermophilic Thermoanaerobacterium bryantii Hemicellulases Is Stimulated by Two Oxidoreductases
Abstract
1. Introduction
2. Results
2.1. Isolation and Characterization of the T. bryantii mel9T Strain
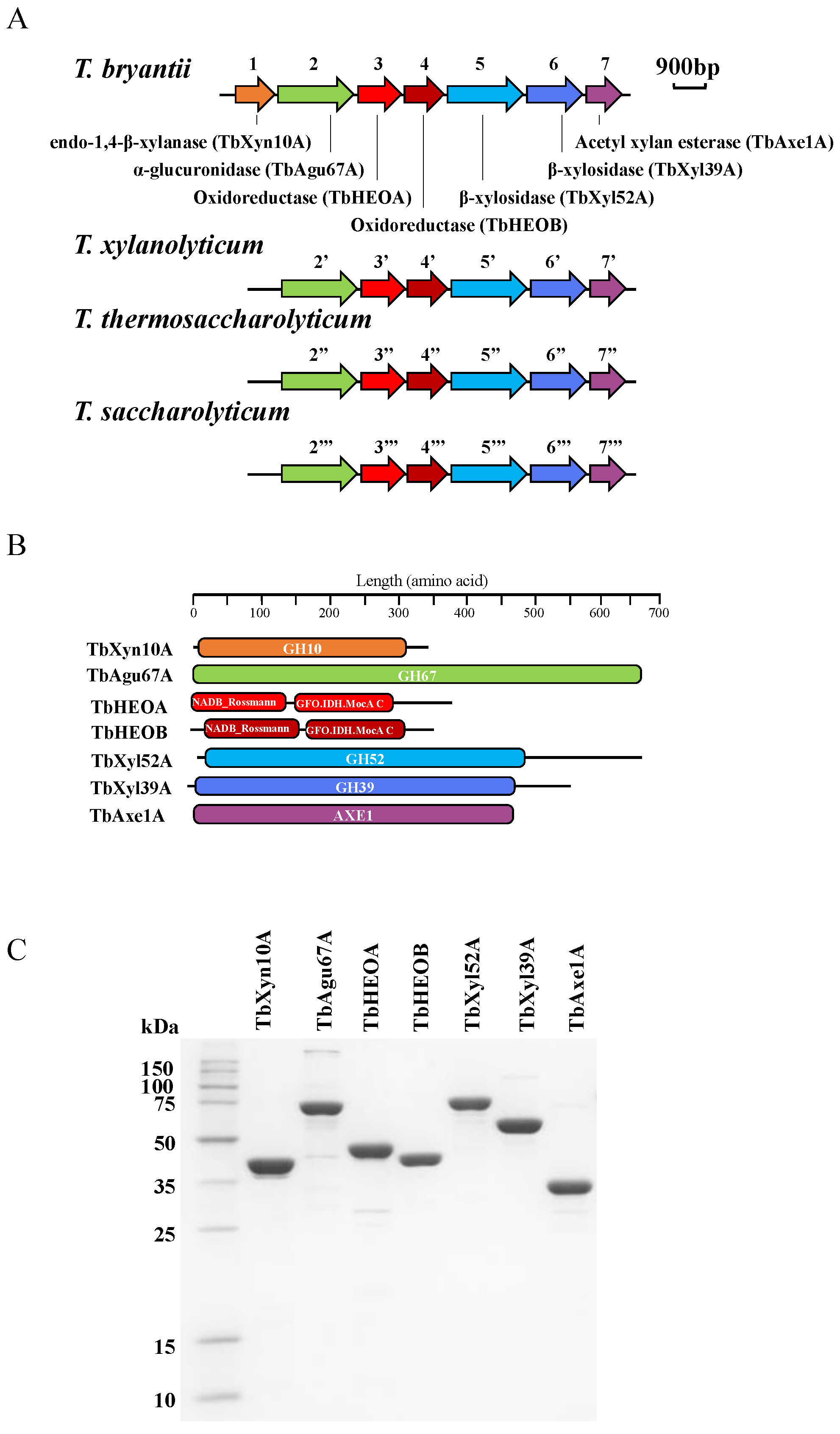
2.2. Sequencing the Genome of T. bryantii mel9T
2.3. Gene Cloning, Expression, and Protein Purification
2.4. Kinetic Parameters of TbXyn10A, TbXyl39A, TbXyl52A, and TbAxe1A
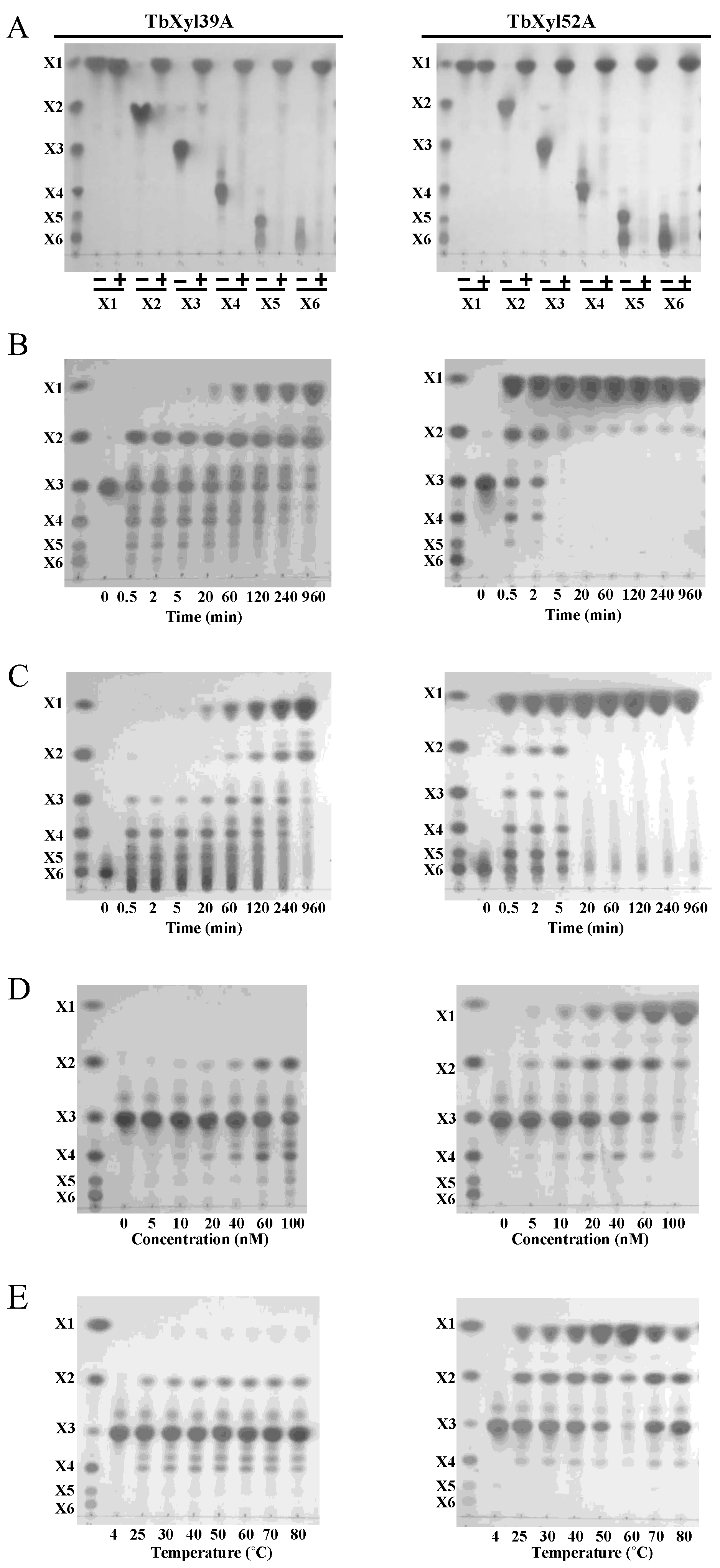
2.5. Hydrolytic Pattern of TbXyl39A and TbXyl52A on Xylo-Oligosaccharides
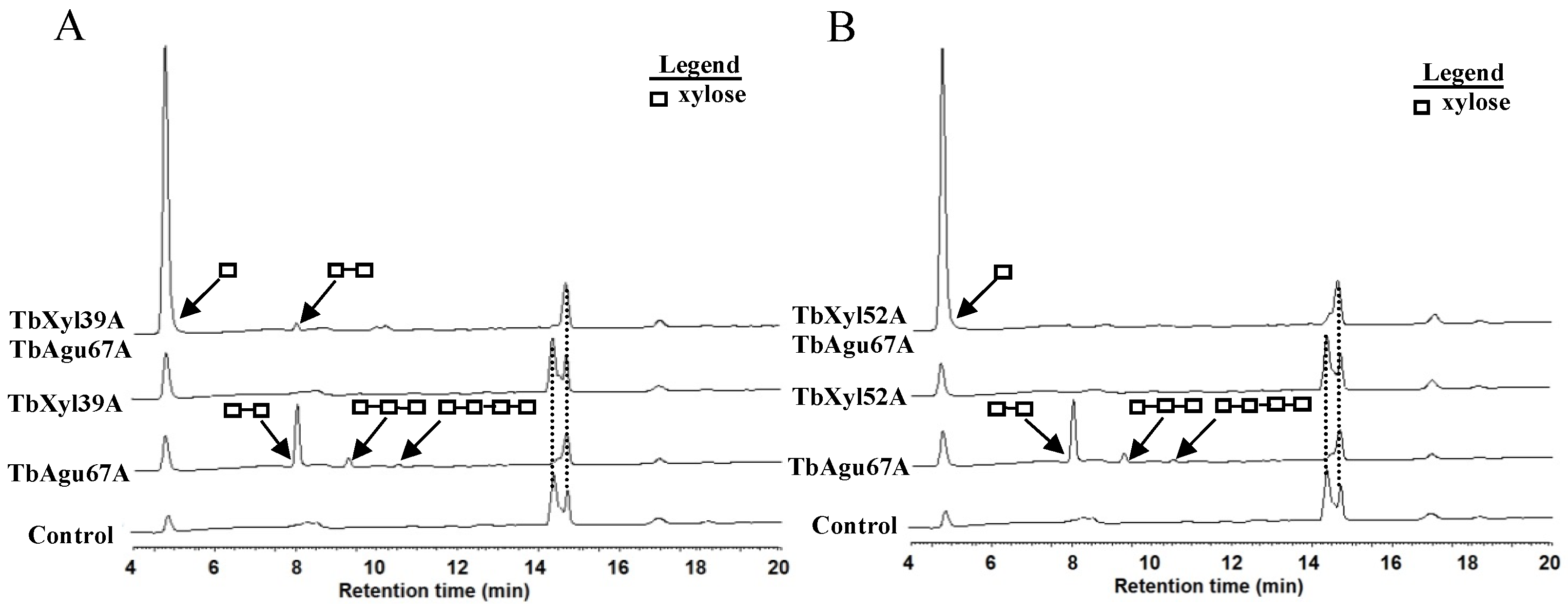
2.6. Synergistic Hydrolysis of Aldouronic Acid Mixture by TbAgu67A and the Two β-Xylosidases
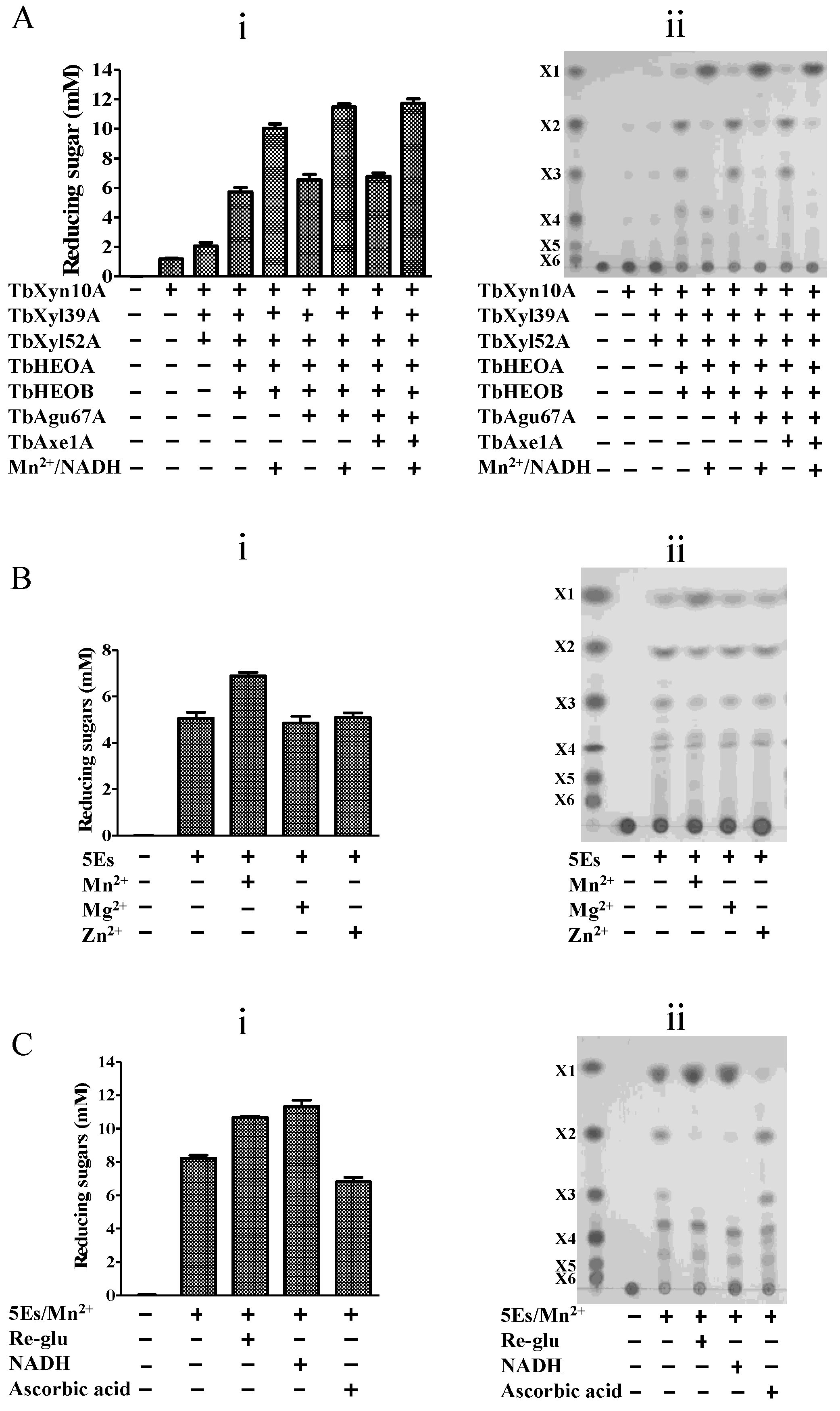
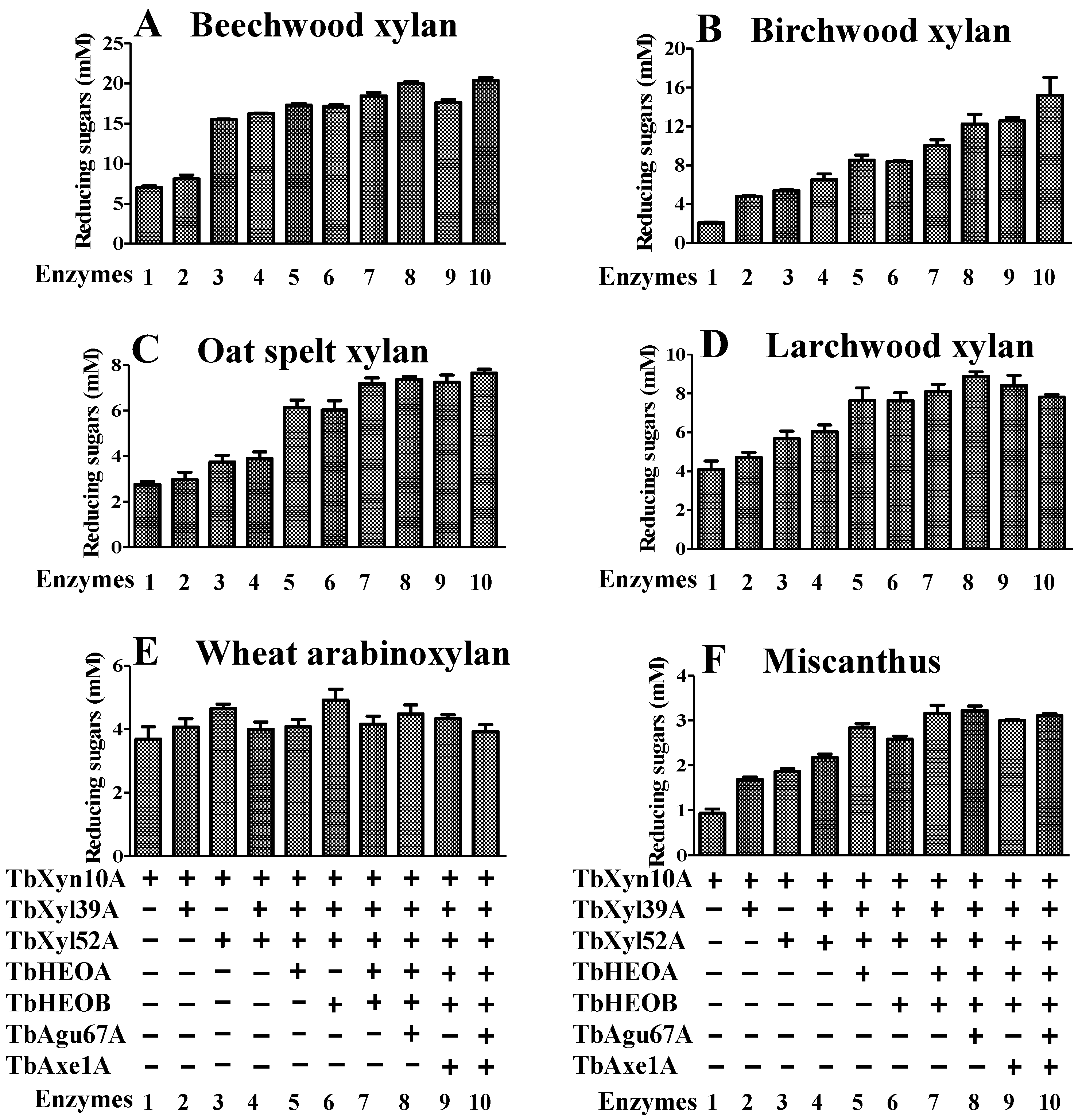
2.7. The Two Oxidoreductases Improve Hydrolysis of Xylans by the T. bryantii Hemicellulases
2.8. Hydrolysis of Xylan by the Seven Enzymes Encoded by the Hemicellulase Gene Cluster
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Characterization of T. bryantii mel9T
4.3. Genome Sequencing
4.4. Gene Cloning, Gene Expression, and Protein Purification
4.5. Determination of Optimal pH and Temperature
4.6. Kinetic Studies of TbXyn10A, TbXyl39A, TbXyl52A, and TbAxe1A
4.7. Reaction Patterns of TbXyl39A and TbXyl52A on Xylose and Xylo-Oligosaccharides
4.8. Hydrolysis of Aldouronic Acid Mixture by TbXyl39A or TbXyl52A Supplemented with TbAgu67A
4.9. Identification of Potential Redox-Active and Metal Ion Cofactors for Hemicellulase-Enhancing Oxidoreductases (HEOs)
4.10. Synergistic Effect of the Seven Enzymes in Hydrolyzing Different Xylan-Containing Plant Biomass
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dodd, D.; Cann, I.K. Enzymatic deconstruction of xylan for biofuel production. Glob. Change Biol. Bioenergy 2009, 1, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Cann, I.; Pereira, G.V.; Abdel-Hamid, A.M.; Kim, H.; Wefers, D.; Kayang, B.B.; Kanai, T.; Sato, T.; Bernardi, R.C.; Atomi, H.; et al. Thermophilic Degradation of Hemicellulose, a Critical Feedstock in the Production of Bioenergy and Other Value-Added Products. Appl. Environ. Microbiol. 2020, 86, 86. [Google Scholar] [CrossRef] [PubMed]
- Harris, P.V.; Welner, D.; McFarland, K.C.; Re, E.; Navarro Poulsen, J.C.; Brown, K.; Salbo, R.; Ding, H.; Vlasenko, E.; Merino, S.; et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: Structure and function of a large, enigmatic family. Biochemistry 2010, 49, 3305–3316. [Google Scholar] [CrossRef] [PubMed]
- Beeson, W.T.; Phillips, C.M.; Cate, J.H.D.; Marletta, M.A. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 2011, 134, 890–892. [Google Scholar] [CrossRef]
- Langston, J.A.; Shaghasi, T.; Abbate, E.; Xu, F.; Vlasenko, E.; Sweeney, M.D. Oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 2011, 77, 7007–7015. [Google Scholar] [CrossRef]
- Phillips, C.M.; Beeson, W.T.; Cate, J.H.; Marletta, M.A. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 2011, 6, 1399–1406. [Google Scholar] [CrossRef]
- Quinlan, R.J.; Sweeney, M.D.; Lo Leggio, L.; Otten, H.; Poulsen, J.-C.N.; Johansen, K.S.; Krogh, K.B.R.M.; Jørgensen, C.I.; Tovborg, M.; Anthonsen, A.; et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. USA 2011, 108, 15079–15084. [Google Scholar] [CrossRef]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Schuster-Bockler, B.; Griffiths-Jones, S.; Hollich, V.; Lassmann, T.; Moxon, S.; Marshall, M.; Khanna, A.; Durbin, R.; et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006, 34, D247–D251. [Google Scholar] [CrossRef]
- Kingston, R.L.; Scopes, R.K.; Baker, E.N. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: An osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure 1996, 4, 1413–1428. [Google Scholar] [CrossRef]
- Whitby, F.G.; Phillips, J.D.; Hill, C.P.; McCoubrey, W.; Maines, M.D. Crystal structure of a biliverdin IXα reductase enzyme–cofactor complex. J. Mol. Biol. 2002, 319, 1199–1210. [Google Scholar] [CrossRef]
- Thoden, J.B.; Sellick, C.A.; Reece, R.J.; Holden, H.M. Understanding a Transcriptional Paradigm at the Molecular Level. J. Biol. Chem. 2007, 282, 1534–1538. [Google Scholar] [CrossRef]
- Moon, Y.H.; Iakiviak, M.; Bauer, S.; Mackie, R.I.; Cann, I.K. Biochemical analyses of multiple endoxylanases from the rumen bacterium Ruminococcus albus 8 and their synergistic activities with accessory hemicellulose-degrading enzymes. Appl. Environ. Microbiol. 2011, 77, 5157–5169. [Google Scholar] [CrossRef]
- Sun, H.J.; Yoshida, S.; Park, N.H.; Kusakabe, I. Preparation of (1→4)-beta-D-xylooligosaccharides from an acid hydrolysate of cotton-seed xylan: Suitability of cotton-seed xylan as a starting material for the preparation of (1→4)-beta-D-xylooligosaccharides. Carbohydr. Res. 2002, 337, 657–661. [Google Scholar] [CrossRef]
- Teleman, A.; Tenkanen, M.; Jacobs, A.; Dahlman, O. Characterization of O-acetyl-(4-O-methylglucurono)xylan isolated from birch and beech. Carbohydr. Res. 2002, 337, 373–377. [Google Scholar] [CrossRef]
- Timell, T.E. Recent progress in the chemistry of wood hemicelluloses. Wood Sci. Technol. 1967, 1, 45–70. [Google Scholar] [CrossRef]
- Kormelink, F.J.M.; Voragen, A.G.J. Degradation of different [(glucurono)arabino]xylans by a combination of purified xylan-degrading enzymes. Appl. Microbiol. Biotechnol. 1993, 38, 688–695. [Google Scholar] [CrossRef]
- Vaaje-Kolstad, G.; Westereng, B.; Horn, S.J.; Liu, Z.; Zhai, H.; Sorlie, M.; Eijsink, V.G. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 2010, 330, 219–222. [Google Scholar] [CrossRef]
- Forsberg, Z.; Vaaje-Kolstad, G.; Westereng, B.; Bunaes, A.C.; Stenstrom, Y.; MacKenzie, A.; Sorlie, M.; Horn, S.J.; Eijsink, V.G. Cleavage of cellulose by a CBM33 protein. Protein Sci. 2011, 20, 1479–1483. [Google Scholar] [CrossRef]
- Khan, A.W.; Lamb, K.A.; Overend, R.P. Comparison of natural hemicellulose and chemically acetylated xylan as substrates for the determination of acetyl-xylan esterase activity in Aspergilli. Enzyme Microb. Technol. 1990, 12, 127–131. [Google Scholar] [CrossRef]
- Gruppen, H.; Hamer, R.J.; Voragen, A.G.J. Water-unextractable cell wall material from wheat flour. 2. fractionation of alkali-extracted polymers and comparison with water-extractable arabinoxylans. J. Cereal Sci. 1992, 16, 53–67. [Google Scholar] [CrossRef]
- Somerville, C. Biofuels. Curr. Biol. 2007, 17, R115–R119. [Google Scholar] [CrossRef]
- Lygin, A.V.; Upton, J.; Dohleman, F.G.; Juvik, J.; Zabotina, O.A.; Widholm, J.M.; Lozovaya, V.V. Composition of cell wall phenolics and polysaccharides of the potential bioenergy crop—Miscanthus. GCB Bioenergy 2011, 3, 333–345. [Google Scholar] [CrossRef]
- Solomon, V.; Teplitsky, A.; Shulami, S.; Zolotnitsky, G.; Shoham, Y.; Shoham, G. Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus. Acta Crystallogr. D 2007, 63, 845–859. [Google Scholar] [CrossRef]
- Gallardo, Ó.; Pastor, F.I.J.; Polaina, J.; Diaz, P.; Łysek, R.; Vogel, P.; Isorna, P.; González, B.; Sanz-Aparicio, J. Structural insights into the specificity of Xyn10B from Paenibacillus barcinonensis and its improved stability by forced protein evolution. J. Biol. Chem. 2010, 285, 2721–2733. [Google Scholar] [CrossRef]
- Kim do, Y.; Han, M.K.; Oh, H.W.; Bae, K.S.; Jeong, T.S.; Kim, S.U.; Shin, D.H.; Kim, I.H.; Rhee, Y.H.; Son, K.H.; et al. Novel intracellular GH10 xylanase from Cohnella laeviribosi HY-21: Biocatalytic properties and alterations of substrate specificities by site-directed mutagenesis of Trp residues. Bioresour. Technol. 2010, 101, 8814–8821. [Google Scholar] [CrossRef]
- Shi, P.; Tian, J.; Yuan, T.; Liu, X.; Huang, H.; Bai, Y.; Yang, P.; Chen, X.; Wu, N.; Yao, B. Paenibacillus sp. strain E18 bifunctional xylanase-glucanase with a single catalytic domain. Appl. Environ. Microbiol. 2010, 76, 3620–3624. [Google Scholar] [CrossRef]
- Wang, J.; Bai, Y.; Yang, P.; Shi, P.; Luo, H.; Meng, K.; Huang, H.; Yin, J.; Yao, B. A new xylanase from thermoalkaline Anoxybacillus sp. E2 with high activity and stability over a broad pH range. World J. Microb. Biot. 2010, 26, 917–924. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, Y.; Tang, X.; Li, J.; Xu, B.; Wu, Q.; Gao, Y.; Pan, L.; Huang, Z. Molecular and biochemical characterization of a novel intracellular low-temperature-active xylanase. J. Microbiol. Biotechnol. 2012, 22, 501–509. [Google Scholar] [CrossRef]
- Luthi, E.; Jasmat, N.B.; Bergquist, P.L. Xylanase from the extremely thermophilic bacterium “Caldocellum saccharolyticum”: Overexpression of the gene in Escherichia coli and characterization of the gene product. Appl. Environ. Microbiol. 1990, 56, 2677–2683. [Google Scholar] [CrossRef] [PubMed]
- Lüthi, E.; Love, D.R.; McAnulty, J.; Wallace, C.; Caughey, P.A.; Saul, D.; Bergquist, P.L. Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile “Caldocellum saccharolyticum”. Appl. Environ. Microbiol. 1990, 56, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, O.; Diaz, P.; Pastor, F.I. Characterization of a Paenibacillus cell-associated xylanase with high activity on aryl-xylosides: A new subclass of family 10 xylanases. Appl. Microbiol. Biotechnol. 2003, 61, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Tabernero, C.; Sanchez-Torres, J.; Perez, P.; Santamaria, R.I. Cloning and DNA sequencing of xyaA, a gene encoding an endo-beta-1,4-xylanase from an alkalophilic Bacillus strain (N137). Appl. Environ. Microbiol. 1995, 61, 2420–2424. [Google Scholar] [CrossRef]
- Dodd, D.; Kiyonari, S.; Mackie, R.I.; Cann, I.K. Functional diversity of four glycoside hydrolase family 3 enzymes from the rumen bacterium Prevotella bryantii B14. J. Bacteriol. 2010, 192, 2335–2345. [Google Scholar] [CrossRef][Green Version]
- Bravman, T.; Zolotnitsky, G.; Shulami, S.; Belakhov, V.; Solomon, D.; Baasov, T.; Shoham, G.; Shoham, Y. Stereochemistry of family 52 glycosyl hydrolases: A beta-xylosidase from Bacillus stearothermophilus T-6 is a retaining enzyme. FEBS Lett. 2001, 495, 39–43. [Google Scholar] [CrossRef]
- Yang, J.K.; Yoon, H.J.; Ahn, H.J.; Lee, B.I.; Pedelacq, J.D.; Liong, E.C.; Berendzen, J.; Laivenieks, M.; Vieille, C.; Zeikus, G.J.; et al. Crystal structure of b-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase. J. Mol. Biol. 2004, 335, 155–165. [Google Scholar] [CrossRef]
- Withers, S.G. Mechanisms of glycosyl transferases and hydrolases. Carbohyd. Polym. 2001, 44, 325–337. [Google Scholar] [CrossRef]
- Zachariou, M.; Scopes, R.K. Glucose-fructose oxidoreductase, a new enzyme isolated from Zymomonas mobilis that is responsible for sorbitol production. J. Bacteriol. 1986, 167, 863–869. [Google Scholar] [CrossRef]
- Fujita, Y.; Shindo, K.; Miwa, Y.; Yoshida, K. Bacillus subtilis inositol dehydrogenase-encoding gene (idh): Sequence and expression in Escherichia coli. Gene 1991, 108, 121–125. [Google Scholar]
- Rossbach, S.; Kulpa, D.A.; Rossbach, U.; de Bruijn, F.J. Molecular and genetic characterization of the rhizopine catabolism (mocABRC) genes of Rhizobium meliloti L5-30. Mol. Gen. Genet. 1994, 245, 11–24. [Google Scholar] [CrossRef]
- Wiebe, M.G.; Nygård, Y.; Oja, M.; Andberg, M.; Ruohonen, L.; Koivula, A.; Penttilä, M.; Toivari, M. A novel aldose-aldose oxidoreductase for co-production of D-xylonate and xylitol from D-xylose with Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2015, 99, 9439–9447. [Google Scholar] [CrossRef][Green Version]
- Hardman, M.J.; Scopes, R.K. The kinetics of glucose–fructose oxidoreductase from Zymomonas mobilis. Eur. J. Biochem. 1988, 173, 203–208. [Google Scholar] [CrossRef]
- Lee, Y.-E.; Jain, M.K.; Lee, C.; Zeikus, J.G. Taxonomic distinction of saccharolytic thermophilic anaerobes: Description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int. J. Syst. Bacteriol. 1993, 43, 41–51. [Google Scholar] [CrossRef]
- Liu, S.Y.; Gherardini, F.C.; Matuschek, M.; Bahl, H.; Wiegel, J. Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J. Bacteriol. 1996, 178, 1539–1547. [Google Scholar] [CrossRef][Green Version]
- Kozianowski, G.; Canganella, F.; Rainey, F.A.; Hippe, H.; Antranikian, G. Purification and characterization of thermostable pectate-lyases from a newly isolated thermophilic bacterium, Thermoanaerobacter italicus sp. nov. Extremophiles 1997, 1, 171–182. [Google Scholar] [CrossRef]
- Larsen, L.; Nielsen, P.; Ahring, B.K. Thermoanaerobacter mathranii sp. nov., an ethanol-producing, extremely thermophilic anaerobic bacterium from a hot spring in Iceland. Arch. Microbiol. 1997, 168, 114–119. [Google Scholar] [CrossRef]
- Ganghofner, D.; Kellermann, J.; Staudenbauer, W.L.; Bronnenmeier, K. Purification and properties of an amylopullulanase, a glucoamylase, and an alpha-glucosidase in the amylolytic enzyme system of Thermoanaerobacterium thermosaccharolyticum. Biosci. Biotechnol. Biochem. 1998, 62, 302–308. [Google Scholar] [CrossRef]
- Warnick, T.A.; Methé, B.A.; Leschine, S.B. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int. J. Syst. Evol. Microbiol. 2002, 52, 1155–1160. [Google Scholar] [CrossRef]
- StJohn, F.J.; Rice, J.D.; Preston, J.F. Paenibacillus sp. Strain JDR-2 and XynA1: A Novel System for Methylglucuronoxylan Utilization. Appl. Environ. Microbiol. 2006, 72, 1496–1506. [Google Scholar] [CrossRef]
- Foster, B.; Pukall, R.; Abt, B.; Nolan, M.; Glavina Del Rio, T.; Chen, F.; Lucas, S.; Tice, H.; Pitluck, S.; Cheng, J.F.; et al. Complete genome sequence of Xylanimonas cellulosilytica type strain (XIL07). Stand. Genomic Sci. 2010, 2, 1–8. [Google Scholar] [CrossRef]
- Bi, C.; Zhang, X.; Ingram, L.O.; Preston, J.F. Genetic engineering of Enterobacter asburiae strain JDR-1 for efficient production of ethanol from hemicellulose hydrolysates. Appl. Environ. Microbiol. 2009, 75, 5743–5749. [Google Scholar] [CrossRef]
- Khudyakov, J.I.; D’haeseleer, P.; Borglin, S.E.; DeAngelis, K.M.; Woo, H.; Lindquist, E.A.; Hazen, T.C.; Simmons, B.A.; Thelen, M.P. Global transcriptome response to ionic liquid by a tropical rain forest soil bacterium, Enterobacter lignolyticus. Proc. Natl. Acad. Sci. USA 2012, 109, E2173–E2182. [Google Scholar] [CrossRef]
- Piriya, P.S.; Vasan, P.T.; Padma, V.S.; Vidhyadevi, U.; Archana, K.; Vennison, S.J. Cellulosic ethanol production by recombinant cellulolytic bacteria harbouring pdc and adh II genes of Zymomonas mobilis. Biotechnol. Res. Int. 2012, 2012, 817549. [Google Scholar] [CrossRef][Green Version]
- Cann, I.K.; Stroot, P.G.; Mackie, K.R.; White, B.A.; Mackie, R.I. Characterization of two novel saccharolytic, anaerobic thermophiles, Thermoanaerobacterium polysaccharolyticum sp. nov. and Thermoanaerobacterium zeae sp. nov., and emendation of the genus Thermoanaerobacterium. Int. J. Syst. Evol. Microbiol. 2001, 51, 293–302. [Google Scholar] [CrossRef]
- Mesbah, M.; Premachandran, U.; Whitman, W.B. Precise Measurement of the G + C Content of Deoxyribonucleic Acid by High-Performance Liquid Chromatography. Int. J. Syst. Bacteriol. 1989, 39, 159–167. [Google Scholar] [CrossRef]
- Arab, H.; Volker, H.; Thomm, M. Thermococcus aegaeicus sp. nov. and Staphylothermus hellenicus sp. nov., two novel hyperthermophilic archaea isolated from geothermally heated vents off Palaeochori Bay, Milos, Greece. Int. J. Syst. Evol. Microbiol. 2000, 50, 2101–2108. [Google Scholar] [CrossRef]
- Cann, I.K.; Kocherginskaya, S.; King, M.R.; White, B.A.; Mackie, R.I. Molecular cloning, sequencing, and expression of a novel multidomain mannanase gene from Thermoanaerobacterium polysaccharolyticum. J. Bacteriol. 1999, 181, 1643–1651. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lever, M. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 1972, 47, 273–279. [Google Scholar] [CrossRef]
- Dodd, D.; Kocherginskaya, S.A.; Spies, M.A.; Beery, K.E.; Abbas, C.A.; Mackie, R.I.; Cann, I.K.O. Biochemical analysis of a β-d-Xylosidase and a bifunctional xylanase-ferulic acid esterase from a xylanolytic gene cluster in Prevotella ruminicola 23. J. Bacteriol. 2009, 191, 3328–3338. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Y.; Rainey, F.A.; Morgan, H.W.; Mayer, F.; Wiegel, J. Thermoanaerobacterium aotearoense sp. nov., a Slightly Acidophilic, Anaerobic Thermophile Isolated from Various Hot Springs in New Zealand, and Emendation of the Genus Thermoanaerobacterium. Int. J. Syst. Bacteriol. 1996, 46, 388–396. [Google Scholar] [CrossRef][Green Version]
- Jones, D.T.; Woods, D.R. Solvent Production. In Biotechnology Handbooks. Clostridia; Minton, N.P., Clarke, D.J., Eds.; Plenum: New York, NY, USA, 1989; Volume 3, pp. 105–144. [Google Scholar]


| Enzyme | Substrate b | kcat (s−1) | Km (mg/mL) | kcat/Km (mL mg−1 s−1) |
|---|---|---|---|---|
| TbXyn10A | WAX | 853 ± 84 | 4.6 ± 1.2 | 184 ± 52 |
| OSX | 131 ± 9.4 | 7.4 ± 1.3 | 17.7 ± 3.4 | |
| BWX | 36.0 ± 0.9 | 1.8 ± 0.2 | 20.2 ± 2.1 | |
| BeeWX | 87.8 ± 3.5 | 1.6 ± 0.3 | 55.2 ± 9.0 | |
| LWX | 165 ± 9.2 | 7.1 ± 1.2 | 23.2 ± 4.0 | |
| TbXyl39A | pNP-β-D-xylopyanoside | 20.4 ± 0.7 | 0.5 ± 0.1 | 44.7 ± 7.2 |
| TbXyl52A | pNP-β-D-xylopyanoside | 97.0 ± 2.5 | 0.2 ± 0.0 | 443 ± 56 |
| TbAxe1A | pNP-acetate | 40.8 ± 2.2 | 0.2 ± 0.1 | 171 ± 46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Z.; Su, X.; Asangba, A.E.; Abdel-Hamid, A.M.; Chakraborty, S.; Dodd, D.; Stroot, P.G.; Mackie, R.I.; Cann, I. Xylan Deconstruction by Thermophilic Thermoanaerobacterium bryantii Hemicellulases Is Stimulated by Two Oxidoreductases. Catalysts 2022, 12, 182. https://doi.org/10.3390/catal12020182
Yi Z, Su X, Asangba AE, Abdel-Hamid AM, Chakraborty S, Dodd D, Stroot PG, Mackie RI, Cann I. Xylan Deconstruction by Thermophilic Thermoanaerobacterium bryantii Hemicellulases Is Stimulated by Two Oxidoreductases. Catalysts. 2022; 12(2):182. https://doi.org/10.3390/catal12020182
Chicago/Turabian StyleYi, Zhuolin, Xiaoyun Su, Abigail E. Asangba, Ahmed M. Abdel-Hamid, Siddhartha Chakraborty, Dylan Dodd, Peter G. Stroot, Roderick I. Mackie, and Isaac Cann. 2022. "Xylan Deconstruction by Thermophilic Thermoanaerobacterium bryantii Hemicellulases Is Stimulated by Two Oxidoreductases" Catalysts 12, no. 2: 182. https://doi.org/10.3390/catal12020182
APA StyleYi, Z., Su, X., Asangba, A. E., Abdel-Hamid, A. M., Chakraborty, S., Dodd, D., Stroot, P. G., Mackie, R. I., & Cann, I. (2022). Xylan Deconstruction by Thermophilic Thermoanaerobacterium bryantii Hemicellulases Is Stimulated by Two Oxidoreductases. Catalysts, 12(2), 182. https://doi.org/10.3390/catal12020182







