Abstract
Recently, we described the preparation of the recombinant oleate hydratase from Lactobacillus rhamnosus ATCC 53103. We observed that the purified C-terminal His-tagged enzyme was completely inactive and the catalytic activity was partially restored only in presence of a large amount of flavin adenine dinucleotide (FAD). In the present work, we assess that this hydratase in the presence of the reduced form of flavin adenine dinucleotide (FADH2) is at least one hundred times as active as in the presence of the same concentration of FAD. By means of two different biochemical processes, we demonstrated unambiguously that oleate hydratase from Lactobacillus rhamnosus ATCC 53103 is a FADH2-dependent enzyme. As a first relevant application of this discovery, we devised a preparative procedure for the stereoselective synthesis of (R)-10-hydroxystearic acid. Accordingly, the hydration of oleic acid (up to 50 g/L) is performed on a multigram scale using the recombinant hydratase and FADH2 generated in situ as cofactor. The produced (R)-10-hydroxystearic acid (ee > 97%) precipitates from the reaction solvent (water/glycerol/ethanol) and is conveniently recovered by simple filtration (>90% yield).
1. Introduction
The hydration of unsaturated fatty acids (UFAs) is a very relevant reaction, potentially exploitable for the synthesis of different compounds of industrial interest. Indeed, the hydroxy-fatty acids (HFAs) produced by this process are widely used for a number of applications such as starting materials for biodegradable polymers, lubricants, emulsifier, drugs, cosmetic ingredients, and flavors [1,2,3,4,5]. A very large number of HFAs have been identified in nature but only a few of them are available in an industrially significant amount. Consequently, the preparation of many HFAs is usually achieved by UFA hydration, which is straightforwardly available as glycerides from vegetable oils or fish fats. Unfortunately, even though a number of chemical means can efficiently perform this kind of reaction, the latter processes lack stereochemical control and afford complex mixtures of isomeric HFAs. In fact, the chemical reactions available to perform the hydration step allow neither the selective functionalization of a specific position of the fatty acid chain nor the enantioselective formation of the secondary alcohol functional group.
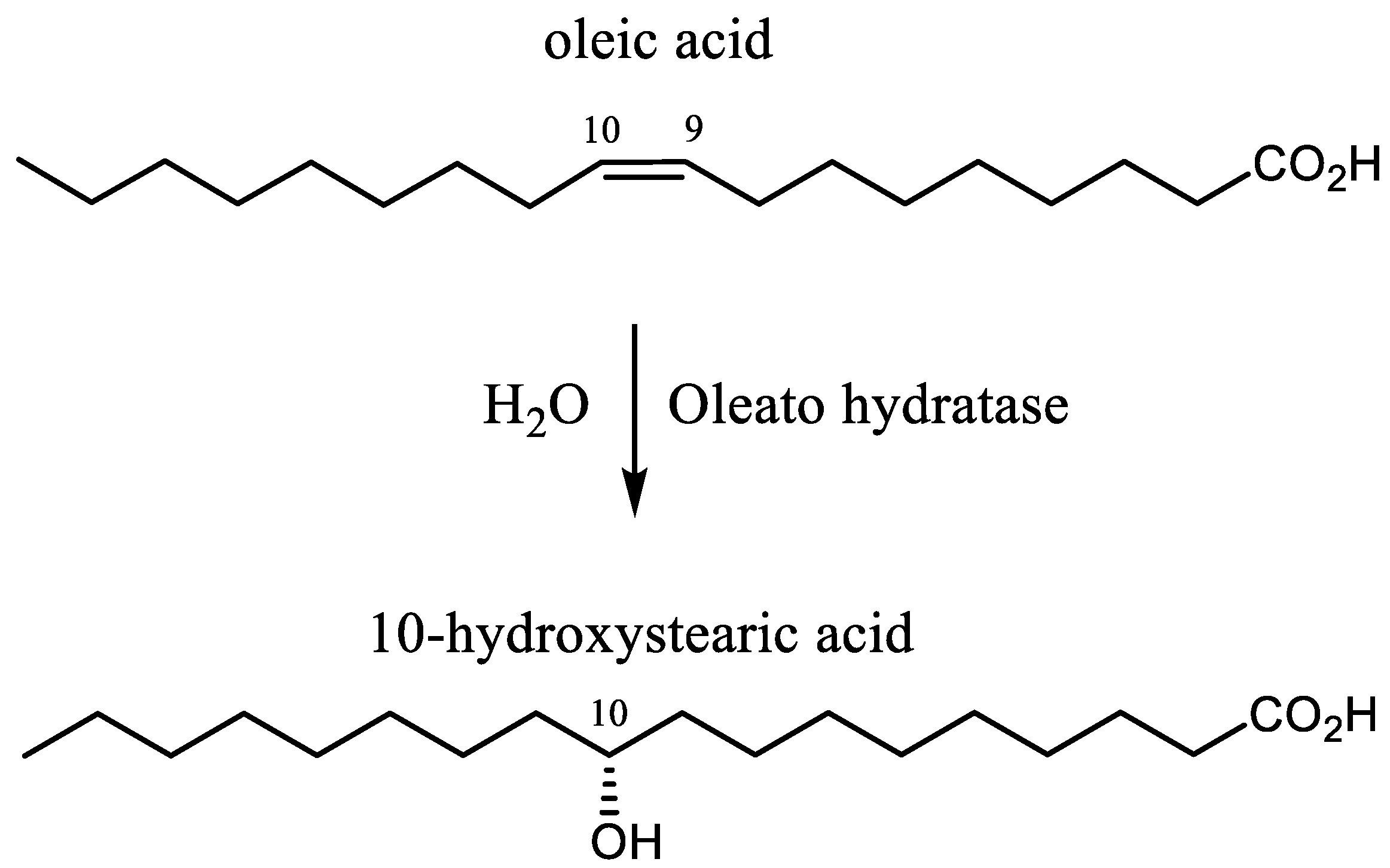
In contrast, the biocatalyzed reactions proceed with complete regioselectivity and enantioselectivity, even using polyunsaturated fatty acids (PUFAs) as substrates. These biochemical transformations were discovered in the early 1960s, during a study on the hydration of oleic acid (Figure 1) using a Pseudomonas strain [6,7]. Afterwards, a number of other microorganisms proved to be able to perform this transformation [8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23] but the enzymes responsible for the hydration step (oleate hydratases) have been characterized only recently [24].
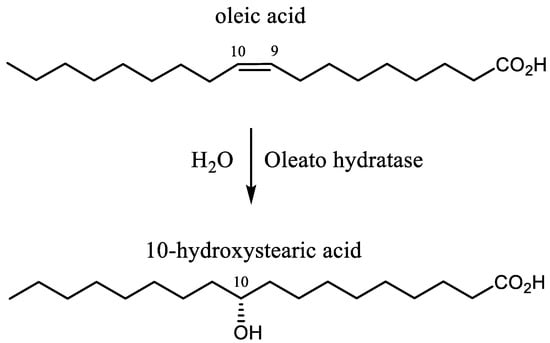
Figure 1.
The hydration of oleic acid catalyzed by oleate hydratase.
Oleate hydratases (OLH, EC 4.2.1.53) convert oleic acid (OA) into (R)-10-hydroxystearic acid (10-HSA) [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39], a high value commercial molecule due to its potential application as surfactant, lubricant, additive in cosmetics industries, and as a starting material in polymer and flavor chemistry. In addition, this enzyme can catalyze the hydration of different UFAs [36,40,41,42,43,44,45,46,47] with the same regio- and stereoselectivity, affording exclusively the corresponding 10-hydroxy derivatives. The most common UFAs, with a chain length of 18 carbon atoms, are oleic, linoleic, and linolenic acid, and they are the main components (as triglycerides) of the vegetable oils used for human consumption.
Being involved in a research project aimed at the valorization of the vegetable oils waste, we studied the biocatalytic hydration of the latter fatty acids using both microbial transformations [21,22,23] and isolated hydratase [26]. In this context, we demonstrated the versatility of the probiotic bacteria Lactobacillus rhamnosus ATCC 53103 [22], which is able to hydrate oleic, linoleic, and linolenic acid affording (R)-10-hydroxystearic acid, (S)-(12Z)-10-hydroxy-octadecenoic acid, and (S)-(12Z,15Z)-10-hydroxy-octadecadienoic acid, respectively, in very high enantiomeric purity (ee > 95%), without the formation of other regioisomers or polyhydroxylated fatty acids.
Since the biocatalytic activity of this Lactobacillus strain is due to the expression of an oleate hydratase previously identified from the bacteria genome [41], we cloned and overexpressed the latter enzyme using Escherichia coli BL21(DE3) as a heterologous host [26]. Our study demonstrated that the obtained recombinant C-terminal His-tagged hydratase retains the catalytic properties of the Lactobacillus strain and we devised a reliable whole-cell-based procedure for the hydration of oleic acid [26]. Unfortunately, we also observed that the purified OLH is completely inactive and the catalytic activity can be restored only marginally through the addition of flavin adenine dinucleotide (FAD) to the reaction mixture.
Due to the pivotal relevance of this enzymatic transformation, the possibility of identifying the key-points that affect the OLH catalytic activity could lead to a new and efficient industrial process for HFAs production.
According to recent studies [27,31,33,36], OLH is a FAD-dependent enzyme, in which FAD is noncovalently bound to the protein. However, FAD is not involved in the reaction mechanism whilst it seems to stabilize the active conformation of the enzyme. The first crystal structure of a recombinant oleate hydratase originating from Elizabethkingia meningoseptica in the presence of flavin adenine dinucleotide (FAD) provided the first reaction mechanism for this enzyme class that explains the requirement for the flavin cofactor [48]. Later, the determination of the crystal structure of the oleate hydratase from Stenotrophomonas sp. KCTC 12332 confirmed that this enzyme also requires FAD as a cofactor. Moreover, the study indicates a remarkable conformational change of the loop region surrounding the FAD molecule upon binding of FAD [49].
Concerning the hydratase activity of strains belonging to the Lactobacillus genus, we observed that the transformation of UFAs into HFAs can be properly accomplished using anaerobic conditions and adding the suitable UFA during the exponential phase of growth [22,23]. According to Lactobacillus metabolism, these conditions lead to the production of the reduced forms of nicotinamide adenine dinucleotide (NADH) or nicotinamide adenine dinucleotide phosphate (NADPH), which, in absence of oxygen, lower the oxidation-reduction potential of the medium. This clue suggested to us the possibility that a reduced form of FAD [50], namely FAD●−, FADH●, FADH−, and FADH2, could be the actual activating agent of the enzyme.
In this context, some relevant works on the characterization of the linoleic acid hydratase from Lactobacillus plantarum AKU 1009a underlined that the investigated enzyme required FAD as cofactor and its activity was enhanced by NADH addition [44,45]. These pioneering studies, as well as some recent reports on other hydratase [33], pointed to possible FADH2 involvement in the catalytic step but unfortunately, they reported no proof to support this hypothesis.
2. Results and Discussion
Herein, we unambiguously demonstrated that the C-terminal His-tagged oleate hydratase from Lactobacillus rhamnosus ATCC 53103 is a FADH2-dependent enzyme.
To this end, we first ran a number of experiments using different formulations of the above-mentioned recombinant OLH (Table 1). As previously reported [26], a PBS buffer containing glycerol and ethanol effectively stabilize the enzyme. In these experimental conditions, the protein is highly stable and can be easily handled by the operator without observing any precipitation or denaturation, even after different freeze/thaw cycles. Accordingly, we tested the hydration reaction of oleic acid (3 g/L) using this medium.

Table 1.
Hydration of oleic acid using different forms of recombinant OLH from L. rhamnosus as catalyst.
Each experiment was performed in a sealed vial, to which FAD (0.06 mM) was previously added and which was flushed with nitrogen.
The use of recombinant E. coli whole cell gave promising results, as we obtained almost complete conversion of OA to 10-HSA (entry 1).
In contrast, the use of the clear lysate of the same E. coli cell batch gave disappointing results either using glycerol (entry 2) or sorbitol (entry 3) as enzyme stabilizers. The experiment performed using glycerol afforded a minor amount of the hydroxy acid (2% yield) using recombinant E. coli lysate whereas the hydration reaction did not proceed with the use of sorbitol, using the same form of catalyst.
Even worse results were obtained using purified His-tagged OLH. The enzyme contained in the above-described clear lysate was purified through elution on a Ni Sepharose resin (Ni-NTA). Then, it was dialyzed against a PBS/glycerol/ethanol buffer in order to remove the inorganic salt and the imidazole present in the elution buffer. We observed that neither purified OLH containing imidazole nor dialyzed OLH catalyzed the hydration reaction (entry 4 and 5). A slight hydratase activity was detected increasing the FAD content from 0.06 mM to 0.2 mM (entry 6), as indicated by the formation of a minor amount of 10-HSA (<1% yield). Overall, the experiments indicated that recombinant OLH is very active only inside cells and its activity is dramatically decreased by purification. As previously demonstrated [26], the presence of FAD is necessary for the catalytic activity but it does not seem to be the only factor affecting enzyme activity. We suppose that NAD(P)H, present both inside cells and to a minor extent in the lysate, can act as reducing agent able to transform FAD into FADH2, which is the actual activating agent of the hydratase.
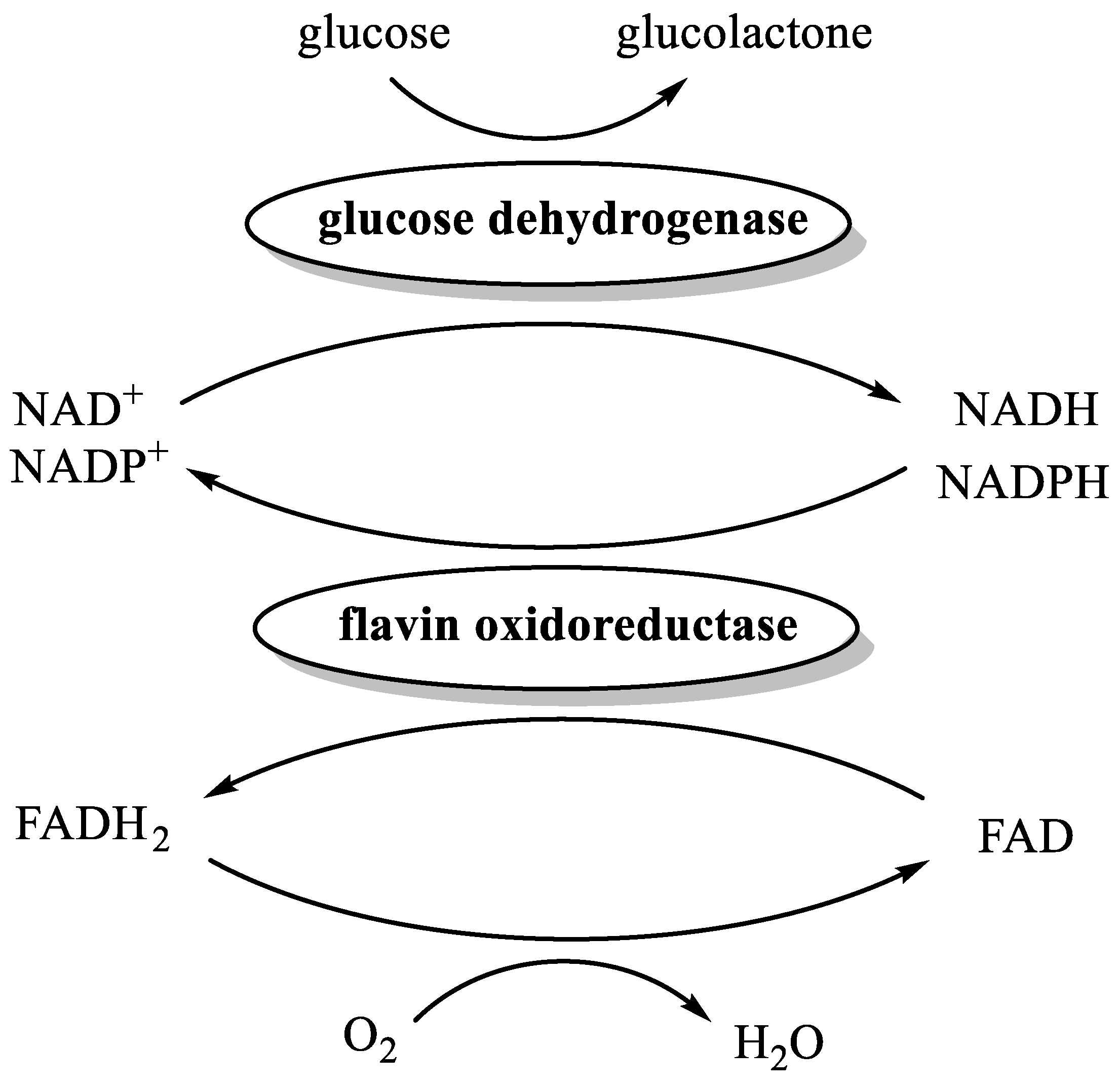
The reduction of FAD to FADH2 by NAD(P)H is catalyzed by flavin oxidoreductases. A member of this class of enzymes, named Fre [51], is present in Escherichia coli, thus corroborating the possible formation of traces of FADH2 in the experiments performed using whole cells and cells lysate as catalysts. In order to demonstrate our hypothesis, we devised a new set of biochemical experiments based on the combined activity of three enzymes (Figure 2 and Table 2).
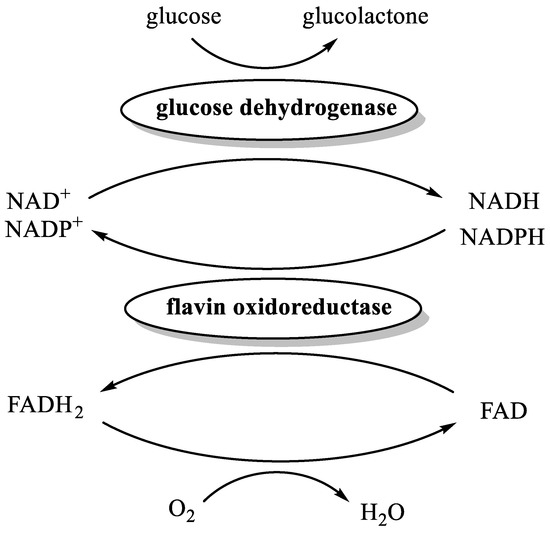
Figure 2.
The enzyme-catalyzed reduction of FAD to FADH2.

Table 2.
Oleate hydratase catalyzed hydration of oleic acid in presence of enzymatically generated NADH, NADPH, and FADH2.
Accordingly, each experiment was performed in a nitrogen-flushed vial, containing PBS/glycerol/ethanol buffer, FAD, glucose, purified His-tagged OLH, purified His-tagged glucose dehydrogenase (GDH), oleic acid, and flavin oxidoreductase. As a source of the latter enzyme, we employed either the lysate of Bacillus subtilis or the lysate of recombinant E. coli expressing GDH [52]. Indeed, both bacteria produce two different flavin oxidoreductases, namely YcnD [53] and Fre [51], respectively able to catalyze the reduction of FAD to FADH2 using both NADH and NADPH. The reasoning behind the choice of recombinant E. coli expressing GDH as a source of Fre lies in the fact that this host strain, namely BL21 (DE3) [52], is free of hydratase activity but is the same as that used for the production of recombinant OLH [26].
Overall, GDH catalyzes the oxidation of glucose to glucolactone with concomitant reduction of NAD(P)+ to NAD(P)H, which in turn reduces FAD to FADH2 by means of flavin oxidoreductases catalyst. In addition, the anaerobiosis is warranted by the presence of FADH2 itself that efficiently reduces oxygen to water.
The establishment of the catalytic cycle was confirmed by the discoloration of the reaction from bright yellow (FAD color) to colorless (FADH2). Accordingly, in a first experiment performed using OLH, GDH, FAD+, and NADP+ but without flavin oxidoreductase (Table 2, entry 1), we observed neither discoloration nor 10-HSA formation.
In contrast, in the same experimental conditions, the addition of the bacterial lysate (B. subtilis or E. coli) triggered the rapid discoloration of the reaction mixture followed by the precipitation of the hydroxy-acid. Regardless of the use of NAD+ instead of NADP+, the hydration reactions were almost quantitative (entries 2–4), as shown by the formation of 10-HSA in yields superior to 95%. A further blank experiment in which OLH was not added (entry 5) confirmed that the hydration reaction does not occur in absence of the hydratase.
Finally, the exclusion of FAD from the reaction dramatically reduced the OLH activity (entry 6 and 7). Most likely, the minor production of 10-HSA obtained for entry 6 was due to the trace amount of FAD contained in the microbial lysate.
Overall, these experiments demonstrate that the presence of NAD(P)H and FAD are not enough to ensure an effective hydration reaction, which proceeds straightforwardly only in the presence of FADH2. A further confirmation of our hypothesis was obtained by means of a series of experiments in which we tuned the oxidation-reduction potential of the reaction medium (Table 3). Accordingly, we added different reducing agents (entries 1–5, Table 3) to the reaction mixtures that do not contain FAD. No experiments showed hydratase activity with the exception of that containing titanium (III) citrate. This trial gave 10-HSA in a trace amount (entry 5, <1% yield). The latter reducing agent is a well-known nontoxic reagent applicable for preparing low oxidation media for anaerobic cultures [54]. Since this titanium salt is also used for the preparation of FADH2 from FAD [55], we suppose that traces of FAD present in purified OLH were reduced, triggering the hydration reaction. The following experiments (entry 6 and 7, Table 3) clearly demonstrated this assumption. The addition of FAD in the presence of OLH and titanium (III) salt produced a rapid and almost quantitative transformation of OA into 10-HSA. Further confirmation was given by the experiment performed in the presence of FAD and titanium salt but in the absence of OLH (entry 8). The latter reaction did not afford 10-HSA, clearly indicating that neither FADH2 nor titanium (III) citrate were responsible for the hydration step. Furthermore, the reaction performed with OLH and FAD but without reducing agent afforded only a trace of 10-HSA (entry 9, <1% yield). It is worth noting that we had to halve the oleic acid concentration (from 6 to 3 g/L) to obtain a measurable value of 10-HSA concentration.

Table 3.
Oleate hydratase catalyzed hydration of unsaturated fatty acids in presence of the indicated reducing agents and with or without addition of FAD.
The latter experiment demonstrated that, using oleic acid as substrate, the hydratase activity of the enzyme in the presence of FADH2 was at least one hundred times higher as in the presence of the same concentration of FAD.
Finally, we checked the hydratase activity of the enzyme using linoleic and linolenic acid as substrates (entries 10 and 11, Table 3). We observed the decrease of the activity in the order oleic acid, linoleic acid, and linolenic acid (98% oleic acid conversion, 58% of linoleic acid conversion, and 31% of linolenic acid conversion). These results are consistent with those previously reported for other recombinant hydratases from Lactobacillus sp. [26,44]. The hydroxy derivatives formed were 10-hydroxystearic acid, (12Z)-10-hydroxy-octadecenoic acid, and (12Z,15Z)-10-hydroxy-octadecadienoic acid, respectively. Only the 10-hydroxyderivatives were detected in the reaction mixtures. These results confirmed the very high regioselectivity of the His-tagged recombinant OLH from Lactobacillus rhamnosus ATCC 53103 as previous described in the experiments performed using wild type microorganism [22].
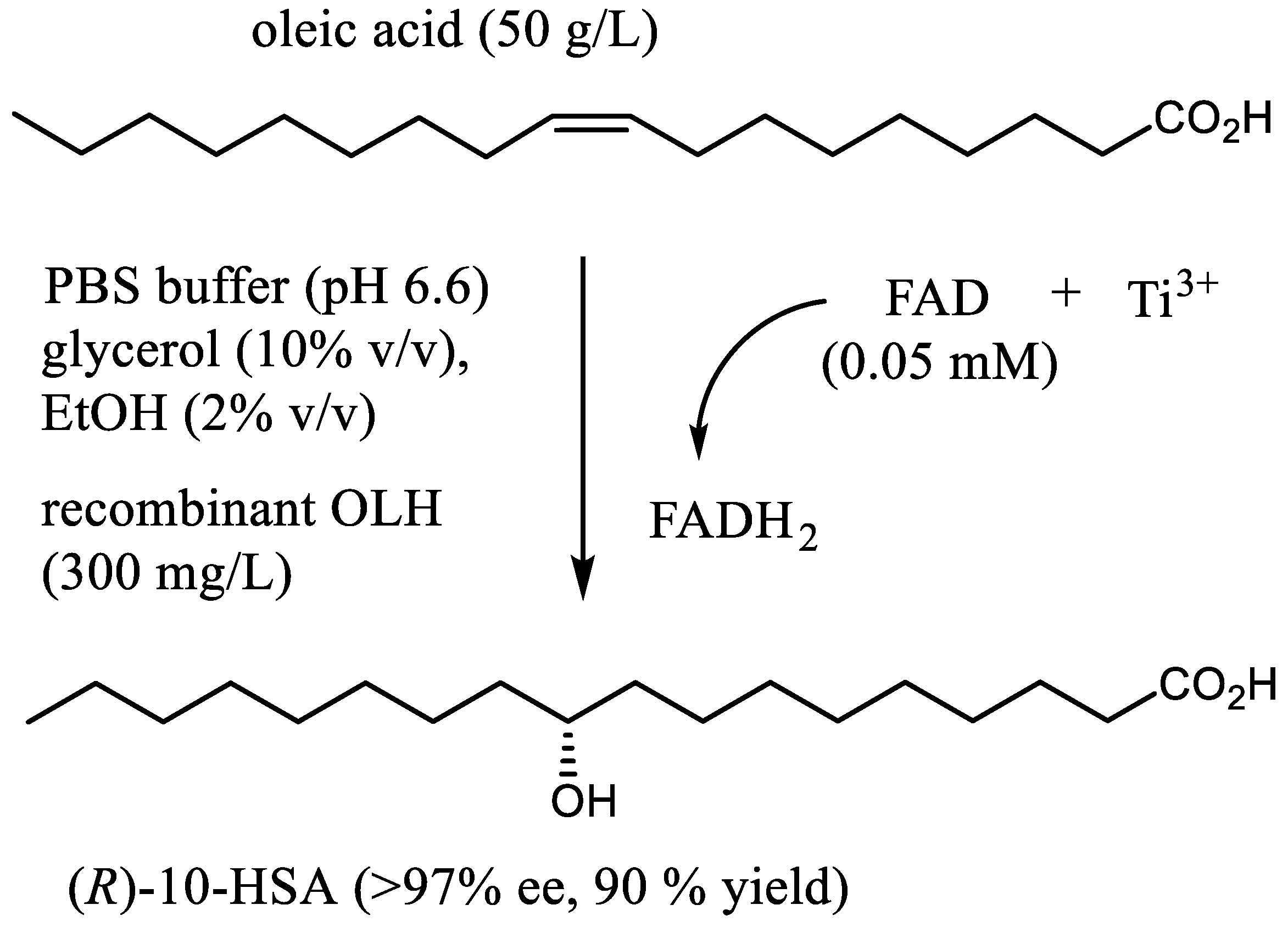
The presented results point to the potential of the recombinant OLH for the large-scale preparation of 10-HSA. To evaluate the feasibility of this goal, we devised a scalable and user-friendly procedure for the stereoselective synthesis of (R)-10-hydroxystearic acid. Accordingly, the hydration of oleic acid was performed on a multigram scale and with high substrate concentration (up to 50 g/L) using in situ generated FADH2 (0.05 mM) and recombinant hydratase (300 mg/L) as catalysts (Figure 3 and Figure 4).
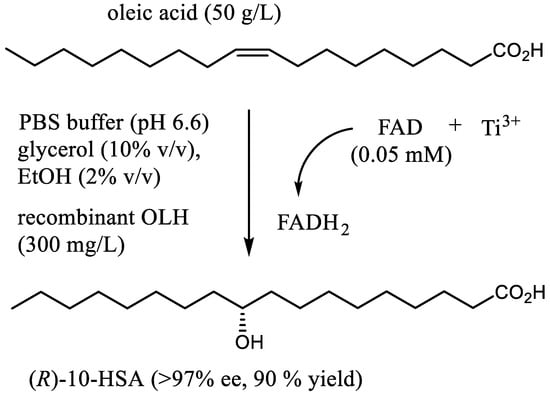
Figure 3.
The preparative synthesis of (R)-10-hydroxystearic acid through hydration of oleic acid by means of recombinant oleate hydratase and FADH2 as cofactor.
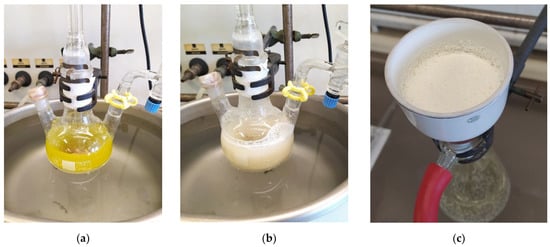
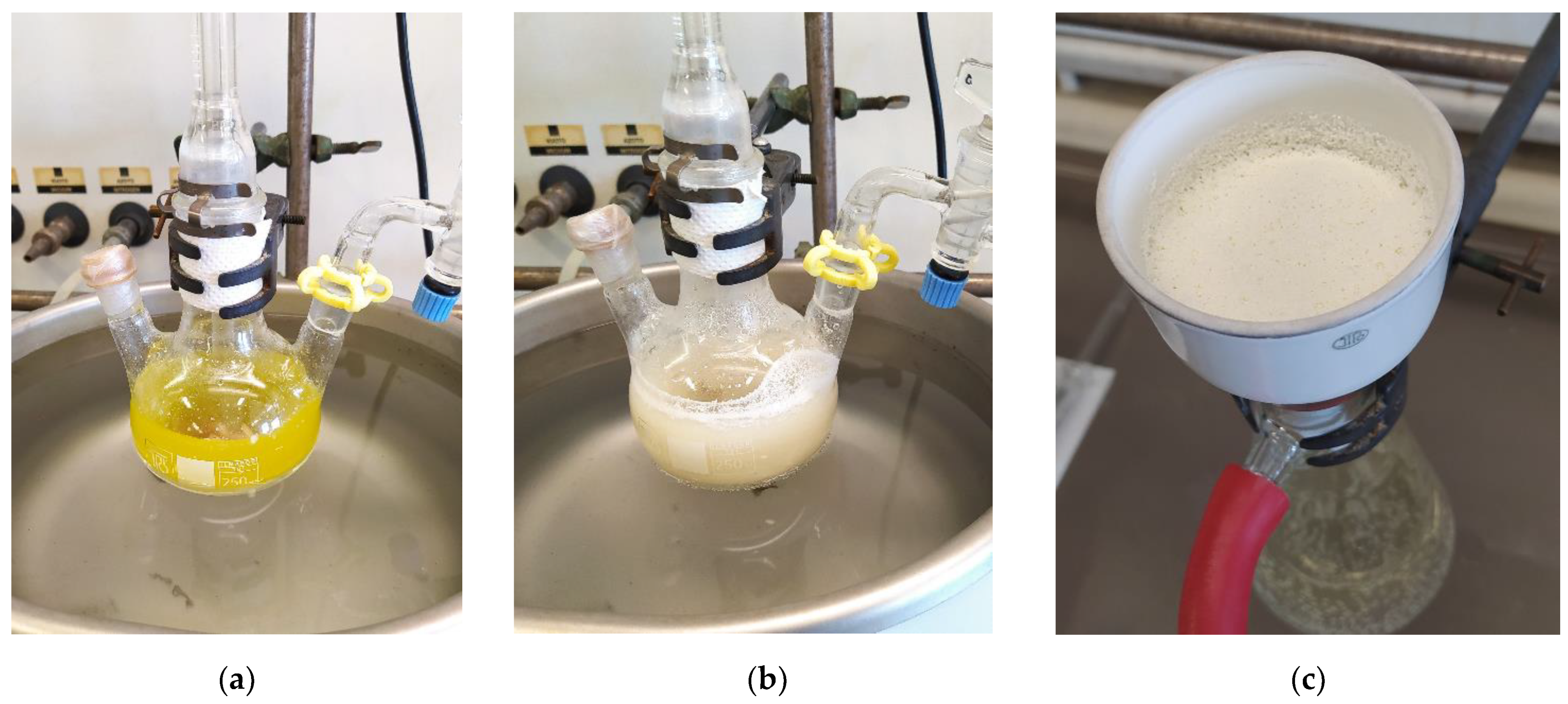
Figure 4.
Laboratory scale, multigram synthesis of (R)-10-hydroxystearic acid through hydration of oleic acid (50 g/L) by means of recombinant oleate hydratase and FADH2 as a cofactor: (a) reaction mixture before titanium (III) citrate addition; (b) reaction mixture six hours after titanium (III) citrate addition; (c) (R)-10-hydroxystearic acid obtained by filtration of the reaction mixture, after 48 h of reaction time.
From a practical standpoint, a nitrogen-flushed reactor was loaded with oleic acid, FAD, PBS/glycerol/ethanol buffer, the lysate of recombinant E. coli expressing OLH and the minimum amount of titanium citrate solution necessary to reduce FAD and to warrant an anaerobic environment (Figure 4).
The complete reduction of the flavin was indicated by the vanishing of the yellow color (Figure 4a,b). Due to the high oleic acid load, the reaction was run in a biphasic medium, with help of vigorous stirring. The precipitation of 10-HSA, which has low solubility in the water/glycerol/ethanol mixture, started after a few hours. After 48 h, the hydroxy acid was isolated by simple filtration (Figure 4c). Since the collected crystals held traces of starting oleic acid, a single recrystallization provided pure (R)-10-hydroxystearic acid (90% yield, 99.7% chemical purity), showing very high enantiomeric purity (ee > 97%).
In summary, our work provides the first demonstration that His-tagged oleate hydratase from Lactobacillus rhamnosus ATCC 53103 is a FADH2-dependent enzyme. These findings pave the way for the exploitation of other already-isolated hydratases whose activity has been erroneously classified as too low to deserve further studies.
As a first relevant application of this discovery, we devised a preparative procedure for the stereoselective synthesis of (R)-10-hydroxystearic acid starting from oleic acid. The proposed process can be led on a multigram scale and using high OA concentration (up to 50 g/L) even as a technical grade reagent. Moreover, the easily available lysate of recombinant E. coli cells can be conveniently employed as a source of OLH, which is activated by a very low concentration of FADH2 generated in situ. (R)-10-hydroxystearic acid was obtained in high yield, in high enantiomeric purity, and was isolated by simple filtration from the reaction mixture. Overall, our procedure can be included among the best preparative syntheses of 10-HSA, based on the use of recombinant OLH, thus pointing to its prospective industrial development.
3. Materials and Methods
3.1. Materials and General Methods
All air- and moisture-sensitive reactions were carried out using dry solvents and under a static atmosphere of nitrogen. All solvents and reagents, including oleic acid (technical grade lot. MKBZ2615V, 91% by GC), linoleic acid (99%, lot. SLBT2627), (S)-mandelic acid, acetic anhydride, pyridine, 4-dimethylaminopyridine (DMAP), N,N′-dicyclohexylcarbodiimide (DCC), kanamycin sulphate, and flavin adenine dinucleotide disodium salt hydrate (FAD, purity: ≥95% HPLC) were of commercial quality and were purchased from Sigma-Aldrich (St. Louis, MO, USA).
NADP (sodium salt hydrate, 97% purity, lot. A0404841) was purchased from Acros Organics (Geel, Belgium).
NAD (lot. K51804342018) was purchased from Merck Millipore (Milan, Italy).
Riboflavin was purchased from Health Leads UK Ltd., (Horeb, UK).
Linolenic acid (85% purity, lot. 81003) was purchased from Nissan—Nippon Oil and Fats Co. (Tokyo, Japan), LTD.
(S)-O-acetyl mandelic acid was prepared starting from (S)-mandelic acid and using acetic anhydride, pyridine, and cat. dimethylaminopyridine (DMAP), as described previously [22].
A reference standard sample of (R)-10-hydroxystearic acid (ee > 95%) was prepared by Lactobacillus rhamnosus mediated hydration of oleic acid [22].
A reference standard sample of 10-(R)-hydroxystearic acid, showing 21% ee, was prepared by baker’s yeast-mediated hydration of oleic acid [21].
3.2. Microorganisms and Growth Media
The growth and expression media used in this work were LB and ZYM-5052 implemented with 20 mg/L of riboflavin (FAD precursor).
ZYM-5052 medium composition: ZYM-0, 50X 5052 solution (20 mL/L) and trace elements solution (1 mL).
The bulk composition of ZYM medium (ZYM-0) was casein peptone (10 g/L), yeast extract (5 g/L), Na2HPO4 (3.6 g/L), KH2PO4 (3.5 g/L), NH4Cl (2.7g/L), Na2SO4 (0.7 g/L), and MgSO4 (0.25 g/L). pH was adjusted to 7.0. ZYM-0 was then autoclaved for 15 min at 121 °C.
Trace elements solution: FeCl3 (50 mM), CaCl2 (20 mM), MnCl2 (10 mM), ZnSO4 (10 mM), CoCl2 (2 mM), CuCl2 (2 mM), NiCl2 (2 mM), Na2MoO4 (2 mM), Na2SeO3 (2 mM), and H3BO3 (2 mM).
50X 5052 solution composition: glycerol (250 g/L), glucose (25 g/L), and α-lactose (100 g/L). Once prepared, the solution was filtered on Sartorius™ Minisart™ Plus Syringe Filters 0.2 µm pore size and stored at 4 °C.
LB medium composition: NaCl (5 g/L), tryptone (10 g/L), and yeast extract (5 g/L). pH was adjusted to 7.0.
Tryptic soy broth composition: casein peptone (17 g/L), glucose (2.5 g/L), soya peptone (3 g/L), NaCl (5 g/L), and K2HPO4 (2.5 g/L).
Bacillus subtilis (DSM 1088) was grown in tryptic soy broth at 30 °C.
3.3. Protein Expression and Purification of Oleate Hydratase (OLH)
Recombinant E. coli BL21(DE3) cells, containing the pETite C-His Kan plasmid [26], harboring the gene of Lactobacillus rhamnosus ATCC 53103 coding for oleate hydratase (GenBank: WP_005714981), were inoculated in a 1 L Erlenmeyer flask containing LB medium (100 mL). The flask was supplemented with kanamycin (30 µg/mL) and was incubated at 37 °C overnight with shaking (220 rpm). Hence, the culture was centrifuged at 3220× g for 5 min (4 °C), the supernatant was removed, and the cells were resuspended in 10 mL of fresh LB medium. The obtained suspension was added to a fermenter vessel of the 5L bioreactor containing ZYM-0 medium (3 L) supplemented with kanamycin (30 µg/mL), 50X 5052 solution (60 mL), and trace elements solution (3 mL). The temperature, the stirring speed, and the pH were set to 20 °C, 250 rpm, and 6.8, respectively. The pH was controlled by dropwise addition of sterilized aqueous solutions (10% w/w in water) of either sulfuric acid or ammonia. The fermentation was stopped 72 h after the inoculum.
The centrifugation (8000 rpm, 15 min, 4 °C) of the whole fermentation broth allowed obtaining 36.3 g of wet cells. The pellet was resuspended in 20 mM potassium phosphate buffer, pH 7.0, containing 500 mM NaCl and 20 mM imidazole. At this point, cells were disrupted by ultrasonication (15 cycles, 20″ON/120″ OFF pulses in ice).
Clear lysate (222 mL) was obtained by centrifugation (11,000 rpm, 30 min) and the protein content titer was assessed by Bradford assay (17.3 mg/mL). The presence of soluble oleate hydratase was assessed by SDS-PAGE (12% T, 2.6% C), where T = concentration (g/100 mL) of acrylamide + bis-acrylamide; and C = bis-acrylamide/acrylamide + bis-acrylamide ratio (w/w). A sample of the aforementioned clear lysate (55.5 mL) was purified by incubation with Ni Sepharose 6 Fast Flow agarose resin (Ni-NTA) (GE Healthcare, Milan, Italy) under mild shaking for 90 min at 4 °C. The resin was then loaded onto a glass column (10 mm × 110 mm) and washed with 20 mL of wash buffer (20 mM imidazole, 500 mM NaCl, and 20 mM potassium phosphate buffer). Elution of His-tagged proteins was achieved by using a 3-step gradient (15 mL washing buffer containing 100, 200, and 300 mM imidazole). To the purified protein solution was added glycerol (10% v/v) and ethanol (2% v/v) and the resulting mixture was dialyzed for 24 h at 4 °C against 15 mM potassium phosphate buffer, pH 6.6 (1.5 L) containing glycerol (10% v/v) and ethanol (2% v/v). The dialyzed solution (145 mL) contained 320 mg of oleate hydratase, which appeared as a single band by SDS-PAGE analysis. The aforementioned solution was stored at −80 °C.
Protein concentration was determined by using the Bio-Rad Protein Assay according to the method of Bradford, and SDS-PAGE analyses (12% T, 2.6% C) were performed to assess protein purity. Molecular weight protein standards were from Bio-Rad (Karlsruhe, Germany), and gels were stained with Coomassie Brilliant Blue for protein detection.
3.4. Protein Expression and Purification of Glucose Dehydrogenase (GDH)
Recombinant E. coli BL21(DE3) cells, containing the pKTS plasmid harboring the gene of Bacillus megaterium DSM 509 coding for glucose dehydrogenase (UniProtKB accession number P40288) [52], were inoculated in a 1 L Erlenmeyer flask containing LB medium (100 mL). The flask was supplemented with ampicillin (100 µg/mL) and was incubated at 37 °C overnight with shaking (220 rpm). Hence, the culture was centrifuged at 3220× g for 5 min (4 °C), the supernatant was removed, and the cells were resuspended in 10 mL of fresh LB medium. The obtained suspension was added to a fermenter vessel of the 5L bioreactor containing ZYM-0 medium (2 L) supplemented with ampicillin (100 µg/mL), 50X 5052 solution (40 mL), and trace elements solution (2 mL). The temperature, the stirring speed, and the pH were set to 22 °C, 250 rpm, and 7.0, respectively. The pH was controlled by dropwise addition of sterilized aqueous solutions (10% w/w in water) of either sulphuric acid or ammonia. The fermentation was stopped 72 h after the inoculum.
The centrifugation (8000 rpm, 15 min, 4 °C) of the whole fermentation broth allowed obtaining 25.1 g of wet cells. The pellet was resuspended in 20 mM potassium phosphate buffer, pH 7.0, containing 500 mM NaCl and 20 mM imidazole. At this point, cells were disrupted by ultrasonication (10 cycles, 20″ON/120″ OFF pulses in ice).
Clear lysate (228 mL) was obtained by centrifugation (11,000 rpm, 30 min) and the protein content titer was assessed by Bradford assay (11.7 mg/mL). The presence of soluble glucose dehydrogenase was assessed by SDS-PAGE (12% T, 2.6% C). A sample of the aforementioned clear lysate (29 mL) was purified by incubation with Ni Sepharose 6 Fast Flow agarose resin (Ni-NTA) (GE Healthcare, Milan, Italy) under mild shaking for 90 min at 4 °C. The resin was then loaded onto a glass column (10 mm × 110 mm) and washed with 20 mL of wash buffer (20 mM imidazole, 500 mM NaCl, 20 mM potassium phosphate buffer). Elution of His-tagged proteins was achieved using a 3-step gradient (15 mL washing buffer containing 100, 200, and 300 mM imidazole). To the purified protein solution (130 mg of GDH) was added glycerol (10% v/v) and was stored at −80 °C.
3.5. Oleic Acid Hydration Experiments
3.5.1. Evaluation of the OLH Activity with Different Enzyme Preparations and in Presence of FAD (Table 1)
The suitable OLH preparation was added to a reaction mixture (6 mL) made up of PB buffer (pH 6.6, 20 mM) containing oleic acid (20 μL), FAD (50 μL of a 10 mM solution in water), ethanol (120 μL), and the suitable enzyme stabilizer (glycerol or sorbitol; 10% v/v). The mixture was placed in a hungate tube that was nitrogen-flushed and sealed. The reaction was shaken (100 rpm) at 26 °C for 24 h, then was acidified with concentrated HCl aq. (500 μL) and was extracted with ethyl acetate (6 mL). The organic phase was separated and was analyzed (paragraph 6).
3.5.2. Evaluation of the OLH Activity in Presence of Enzymatically Generated NADH, NADPH and FADH2 (Table 2)
A reaction mixture (6 mL) made up of PB buffer (pH 6.6, 20 mM) containing oleic acid (20 μL), glucose (180 mg), glycerol (10% v/v) and ethanol (2% v/v) was placed in a hungate tube, which was nitrogen-flushed and sealed. The suitable enzyme (OLH, GDH, or the E. coli/B. subtilis lysate, Table 2) and FAD were injected into the sealed tube using a syringe. Each trial was shaken at 100 rpm for 24 h. After this time, the reaction was acidified with concentrated HCl aq. (500 μL), then was extracted with ethyl acetate (6 mL). The organic phase was separated and was analyzed (paragraph 6).
3.5.3. Evaluation of the OLH Activity in Presence of Reducing Agents and with or without Addition of FAD (Table 3)
A reaction mixture (10 mL) made up of PB buffer (pH 6.6, 20 mM) unsaturated fatty acid (Table 3), glycerol (10% v/v) and ethanol (2% v/v), OLH (2 mg), and FAD was placed in a hungate tube, which was nitrogen-flushed and sealed. Cysteine, DTT (1,4-dithiothreitol), sodium thioglycolate, and formaldehyde sulfoxylate were added before treatment with nitrogen gas whereas the solution of titanium citrate was injected into the sealed tube using a syringe. The reaction was shaken (100 rpm) at 26 °C for 24 h, then was acidified with concentrated HCl aq. (500 μL) and was extracted with ethyl acetate (6 mL). The organic phase was separated and was analyzed (paragraph 6).
3.6. Analytical Methods and Characterization of the Products Deriving from the Biotransformation Experiments
Instruments and Analytic Condition
Nuclear Magnetic Resonance spectroscopy (NMR): 1H- and 13C-NMR Spectra and DEPT experiments: CDCl3 solutions at RT using a Bruker-AC-400 spectrometer (Billerica, MA, USA) at 400, 100, and 100 MHz, respectively; 13C spectra were proton decoupled; chemical shifts in ppm relative to internal SiMe4 (=0 ppm).
TLC: Merck silica gel 60 F254 plates (Merck Millipore, Milan, Italy).
Column chromatography: silica gel.
GC-MS analyses: A HP-6890 gas chromatograph equipped with a 5973 mass detector and using a HP-5MS column (30 m × 0.25 mm, 0.25 μm film thickness; Hewlett Packard, Palo Alto, CA, USA) was used with the following temperature program: 120 °C (3 min)—12 °C/min—195 °C (10 min)—12 °C/min—300 °C (10 min); carrier gas: He; constant flow 1 mL/min; split ratio: 1/30; tR given in min.
The biotransformations of oleic acid, linoleic acid, and linolenic acid to give 10-hydroxystearic acid, (12Z)-10-hydroxy-octadecenoic acid, and (12Z,15Z)-10-hydroxy-octadecadienoic acid, respectively, were monitored by means of GC-MS analysis. To this end, the biotransformation mixture was acidified at pH 4 and filtered on celite. The aqueous phase was then extracted three times with ethyl acetate and the combined organic layer was washed with brine and dried on Na2SO4. The solvent was then removed under reduced pressure and the residue was treated at 0 °C with an excess of an ethereal solution of freshly-prepared diazomethane. As soon as the evolution of nitrogen ceased, the solvent was eliminated and the residue was treated at RT with a 1:1 mixture of pyridine/acetic anhydride (4 mL for about 100 mg of residue) and DMAP (10 mg). After five hours, the excess of reagents was removed in vacuo and the residue was analyzed by GC-MS in order to determine the fatty acid/hydrated fatty acid ratio.
Oleic acid methyl ester: tR 18.95
GC-MS (EI): m/z (%) = 296 [M+] (7), 264 (49), 235 (6), 222 (30), 180 (19), 166 (10), 152 (12), 137 (17), 123 (26), 110 (32), 97 (62), 83 (68), 69 (79), 55 (100).
Linoleic acid methyl ester: tR 18.52
GC-MS (EI): m/z (%) = 294 [M+] (18), 263 (15), 234 (1), 220 (4), 178 (6), 164 (10), 150 (16), 135 (15), 123 (18), 109 (36), 95 (70), 81 (93), 67 (100), 55 (56).
Linolenic acid methyl ester: tR 18.79
GC-MS (EI): m/z (%) = 292 [M+] (7), 261 (4), 249 (2), 236 (5), 191 (3), 173 (5), 149 (13), 135 (15), 121 (20), 108 (34), 95 (56), 79 (100), 67 (66), 55 (43).
Methyl 10-acetoxystearate: tR 24.47
GC-MS (EI): m/z (%) = 313 [M+-MeCO] (6), 296 [M+-AcOH] (3), 281 (17), 264 (31), 243 (11), 222 (9), 201 (100), 169 (64), 157 (16), 125 (21), 97 (18), 83 (19), 69 (21), 55 (27).
Methyl (12Z)-10-acetoxy-octadecenoate: tR 24.28
GC-MS (EI): m/z (%) 311 [M+-MeCO] (<1), 294 [M+-AcOH] (39), 279 (1), 263 (24), 220 (7), 201 (46), 169 (100), 150 (13), 136 (9), 123 (15), 109 (21), 95 (37), 81 (53), 67 (46), 55 (32).
Methyl (12Z,15Z)-10-acetoxy-octadecadienoate: tR 24.33
GC-MS (EI): m/z (%) 292 [M+-AcOH] (76), 277 (1), 261 (20), 201 (33), 169 (100), 149 (19), 135 (28), 121 (41), 108 (42), 93 (57), 79 (87), 55 (39).
The enantiomeric composition of the isolated 10-hydroxystearic acid samples was determined by 1H-NMR analysis [22]. Hence, each one of the hydroxy acid samples (100 mg, 0.33 mmol) was treated with an excess of an ethereal solution of freshly prepared diazomethane. As soon as the evolution of nitrogen ceased, the solvent was eliminated and the resulting methyl ester was dissolved in dry CH2Cl2 (5 mL) treated with (S)-O-acetylmandelic acid (130 mg, 0.67 mmol), DCC (140 mg, 0.68 mmol), and DMAP (10 mg), stirring at RT for 6 h. The reaction was then quenched by the addition of water and diethyl ether (60 mL). The formed dicyclohexylurea was removed by filtration on celite and the organic phase was washed with aq. NaHCO3, brine and dried on Na2SO4. The solvent was then removed under reduced pressure and the residue was roughly purified by chromatography, collecting every fraction containing the fatty acid mandelates. The 1H-NMR analysis of the obtained (S)-O-acetylmandelates allowed the determination of the absolute configuration of the starting hydroxy acid as well as the measurement of their enantiomeric purity. The methyl ester signals due to (R) and (S)-10-hydroxystearic acid derivatives gives two well-resolved singlets at 3.66 and 3.67 ppm, respectively, whose relative peak areas indicate the corresponding isomeric ratio.
3.7. Preparative Synthesis of (R)-10-Hydroxystearic Acid
A solution of titanium III citrate (3 mL of a 80 mM solution in water) was added under nitrogen to a mechanically stirred mixture (100 mL) made up of PB buffer (pH 6.6, 20 mM), technical grade oleic acid (5.3 g, 92% chemical purity, 17.3 mMol, GC-MS n°1, Figure S1), FAD (500 μL of a 10 mM solution in water), ethanol (2 mL), glycerol (10 mL), and the lysate of recombinant E. coli (3 mL, containing 10 mg/mL of OLH). The yellow color disappeared immediately after the titanium salt addition. The temperature was kept at 26 °C by means of a thermostatic bath whilst the anaerobiosis was warranted to keep the reaction under a static atmosphere of nitrogen. The precipitation of hydroxystearic acid started a few hours after titanium addition and stirring was prolonged for further 48 h. Hence, the reaction was quenched by addition of HCl aq. (5 mL of a 3% w/v solution) followed by cooling with an ice bath. The hydroxy-fatty acid was filtered and the filtrated was washed with ice-cooled water (3 × 25 mL). The collected crystals were dried in vacuo to afford crude (R)-10-hydroxystearic acid (5.6 g, GC-MS n° 2, Figure S2). A single recrystallization from hexane/ethyl acetate (2:1, 80 mL) afforded pure (R)-10-hydroxystearic acid (4.68 g, 90% yield, GC-MS n° 3, Figure S3) showing 99.7% chemical purity and ee > 97% (determined by NMR analysis, Figure S4).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/catal11091051/s1. Figure S1: GC-MS analysis of the starting oleic acid, Figure S2: GC-MS analysis of the crude (R)-10-hydroxystearic acid obtained after filtration, Figure S3: GC-MS analysis of recrystallized (R)-10-hydroxystearic acid, Figure S4: Determination of the enantiomeric purity of the obtained 10-hydroxystearic acid by means of 1H-NMR analysis of its (S)-O-acetylmandelates ester.
Author Contributions
S.S. and S.M. conceived this study, designed and performed the experiments, and analyzed the data; D.D.S. and M.V. were responsible for the methodology and cloning the oleate hydratase in E. coli; S.S. wrote the paper and provided resources and funding; S.S., S.M. and D.D.S. did the review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Cariplo Foundation and Regione Lombardia within the project n° 2017-1015 SOAVE (Seed and vegetable Oils Active Valorization through Enzymes) and within the project POR-FESR 2014-2020 n° 228775 VIPCAT (Value Added Innovative Protocols for Catalytic Transformations), respectively.
Data Availability Statement
Data are contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kim, K.-R.; Oh, D.-K. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 2013, 31, 1473–1485. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, X. Production of long-chain hydroxy fatty acids by microbial conversion. Appl. Microbiol. Biotechnol. 2013, 97, 3323–3331. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Ness, J.E.; Xie, W.; Zhang, X.; Minshull, J.; Gross, R.A. Biosynthesis of Monomers for Plastics from Renewable Oils. J. Am. Chem. Soc. 2010, 132, 15451–15455. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Fuganti, C.; Brenna, M.E. Biocatalytic preparation of natural flavours and fragrances. Trends Biotechnol. 2005, 23, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.-S.; An, J.-U.; Oh, D.-K. γ-Dodecelactone Production from Safflower Oil via 10-Hydroxy-12(Z)-octadecenoic Acid Intermediate by Whole Cells of Candida boidinii and Stenotrophomonas nitritireducens. J. Agric. Food Chem. 2014, 62, 6736–6745. [Google Scholar] [CrossRef]
- Wallen, L.; Benedict, R.; Jackson, R. The microbiological production of 10-Hydroxystearic acid from oleic acid. Arch. Biochem. Biophys. 1962, 99, 249–253. [Google Scholar] [CrossRef]
- Davis, E.N.; Wallen, L.L.; Goodwin, J.C.; Rohwedder, W.K.; Rhodes, R.A. Microbial hydration of cis-9-alkenoic acids. Lipids 1969, 4, 356–362. [Google Scholar] [CrossRef]
- Seo, C.W.; Yamada, Y.; Takada, N.; Okada, H. Hydration of Squalene and Oleic Acid by Corynebacterium sp. S-401. Agric. Biol. Chem. 1981, 45, 2025–2030. [Google Scholar] [CrossRef]
- Sakata, K.; Takizawa, N.; Sankawa, U.; Ebizuka, Y.; Noguchi, H. Studies on changes of corn components in silage fermentation. Part VIII. Identification of 10-hydroxy-12Z-octadecenoic and 10-hydroxy-12Z,15Z-octadecadienoic acids in corn silage. Agric. Biol. Chem. 1986, 50, 1077–1079. [Google Scholar] [CrossRef][Green Version]
- Koritala, S.; Hou, C.; Hesseltine, C.; Bagby, M. Microbial conversion of oleic acid to 10-hydroxystearic acid. Appl. Microbiol. Biotechnol. 1989, 32, 299–304. [Google Scholar] [CrossRef]
- El-Sharkawy, S.H.; Yang, W.; Dostal, L.; Rosazza, J.P. Microbial oxidation of oleic acid. Appl. Environ. Microbiol. 1992, 58, 2116–2122. [Google Scholar] [CrossRef]
- Koritala, S.; Bagby, M.O. Microbial conversion of linoleic and linolenic acids to unsaturated hydroxy fatty acids. J. Am. Oil Chem. Soc. 1992, 69, 575–578. [Google Scholar] [CrossRef]
- Hou, C.T. Conversion of linoleic acid to 10-hydroxy-12(Z)-octadecenoic acid by Flavobacterium sp. (NRRL B-14859). J. Am. Oil Chem. Soc. 1994, 71, 975–978. [Google Scholar] [CrossRef]
- Kaneshiro, T.; Huang, J.-K.; Weisleder, D.; Bagby, M.O. 10(R)-Hydroxystearic acid production by a novel microbe, NRRL B-14797, isolated from compost. J. Ind. Microbiol. Biotechnol. 1994, 13, 351–355. [Google Scholar] [CrossRef]
- Hudson, J.; MacKenzie, C.A.M.; Joblin, K.N. Conversion of oleic acid to 10-hydroxystearic acid by two species of ruminal bacteria. Appl. Microbiol. Biotechnol. 1995, 44, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Morvan, B.; Joblin, K.N. Hydration of Oleic Acid by Enterococcus gallinarum, Pediococcus acidilactici and Lactobacillus sp. Isolated from the Rumen. Anaerobe 1999, 5, 605–611. [Google Scholar] [CrossRef]
- Kim, M.H.; Park, M.S.; Chung, C.H.; Kim, C.T.; Kim, Y.S.; Kyung, K.H. Conversion of unsaturated food fatty acids into hy-droxy fatty acids by lactic acid bacteria. J. Microbiol. Biotechnol. 2003, 13, 360–365. [Google Scholar]
- Kishimoto, N.; Yamamoto, I.; Toraishi, K.; Yoshioka, S.; Saito, K.; Masuda, H.; Fujita, T. Two distinct pathways for the formation of hydroxy FA from linoleic acid by lactic acid bacteria. Lipids 2003, 38, 1269–1274. [Google Scholar] [CrossRef]
- Kim, B.-N.; Yeom, S.-J.; Oh, D.-K. Conversion of oleic acid to 10-hydroxystearic acid by whole cells of Stenotrophomonas nitritireducens. Biotechnol. Lett. 2011, 33, 993–997. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kishino, S.; Tanabe, K.; Hirata, A.; Park, S.-B.; Shimizu, S.; Ogawa, J. Hydroxy fatty acid production by Pediococcus sp. Eur. J. Lipid Sci. Technol. 2013, 115, 386–393. [Google Scholar] [CrossRef]
- Serra, S.; De Simeis, D. New insights on the baker’s yeast-mediated hydration of oleic acid: The bacterial contaminants of yeast are responsible for the stereoselective formation of (R)-10-hydroxystearic acid. J. Appl. Microbiol. 2018, 124, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; De Simeis, D. Use of Lactobacillus rhamnosus (ATCC 53103) as Whole-Cell Biocatalyst for the Regio- and Stereoselective Hydration of Oleic, Linoleic, and Linolenic Acid. Catalysts 2018, 8, 109. [Google Scholar] [CrossRef]
- Serra, S.; De Simeis, D.; Castagna, A.; Valentino, M. The Fatty-Acid Hydratase Activity of the Most Common Probiotic Microorganisms. Catalysts 2020, 10, 154. [Google Scholar] [CrossRef]
- Bevers, L.E.; Pinkse, M.W.H.; Verhaert, P.D.E.M.; Hagen, W.R. Oleate Hydratase Catalyzes the Hydration of a Nonactivated Carbon-Carbon Bond. J. Bacteriol. 2009, 191, 5010–5012. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.-F.; Zheng, Y.-C.; Chen, Q.; Xu, J.-H.; Pan, J. Engineering of an oleate hydratase for efficient C10-Functionalization of oleic acid. Biochem. Biophys. Res. Commun. 2020, 537, 64–70. [Google Scholar] [CrossRef]
- Castagna, A.; De Simeis, D.; Ferrandi, E.E.; Marzorati, S.; Monti, D.; Serra, S.; Valentino, M. Recombinant Oleate Hydratase from Lactobacillus rhamnosus ATCC 53103: Enzyme Expression and Design of a Reliable Experimental Procedure for the Stereoselective Hydration of Oleic Acid. Catalysts 2020, 10, 1122. [Google Scholar] [CrossRef]
- Löwe, J.; Gröger, H. Fatty Acid Hydratases: Versatile Catalysts to Access Hydroxy Fatty Acids in Efficient Syntheses of Industrial Interest. Catalysts 2020, 10, 287. [Google Scholar] [CrossRef]
- Busch, H.; Tonin, F.; Alvarenga, N.; Broek, M.V.D.; Lu, S.; Daran, J.-M.; Hanefeld, U.; Hagedoorn, P.-L. Exploring the abundance of oleate hydratases in the genus Rhodococcus—discovery of novel enzymes with complementary substrate scope. Appl. Microbiol. Biotechnol. 2020, 104, 5801–5812. [Google Scholar] [CrossRef]
- Zhang, J.; Bilal, M.; Liu, S.; Zhang, J.; Lu, H.; Luo, H.; Luo, C.; Shi, H.; Iqbal, H.M.N.; Zhao, Y. Sustainable Biotransformation of Oleic Acid to 10-Hydroxystearic Acid by a Recombinant Oleate Hydratase from Lactococcus garvieae. Processes 2019, 7, 326. [Google Scholar] [CrossRef]
- Engleder, M.; Strohmeier, G.A.; Weber, H.; Steinkellner, G.; Leitner, E.; Müller, M.; Mink, D.; Schürmann, M.; Gruber, K.; Pichler, H. Evolving the Promiscuity of Elizabethkingia meningoseptica Oleate Hydratase for the Regio- and Stereoselective Hydration of Oleic Acid Derivatives. Angew. Chem. Int. Ed. 2019, 58, 7480–7484. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.; Driller, R.; Waldow, A.; Qoura, F.; Loll, B.; Brück, T. Rhodococcus erythropolis Oleate Hydratase: A New Member in the Oleate Hydratase Family Tree-Biochemical and Structural Studies. ChemCatChem 2017, 10, 407–414. [Google Scholar] [CrossRef]
- Engleder, M.; Pichler, H. On the current role of hydratases in biocatalysis. Appl. Microbiol. Biotechnol. 2018, 102, 5841–5858. [Google Scholar] [CrossRef]
- Kang, W.-R.; Seo, M.-J.; Shin, K.-C.; Park, J.-B.; Oh, D.-K. Comparison of Biochemical Properties of the Original and Newly Identified Oleate Hydratases from Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 2017, 83, e03351-16. [Google Scholar] [CrossRef]
- Demming, R.M.; Otte, K.B.; Nestl, B.; Hauer, B. Optimized Reaction Conditions Enable the Hydration of Non-natural Substrates by the Oleate Hydratase from Elizabethkingia meningoseptica. ChemCatChem 2017, 9, 758–766. [Google Scholar] [CrossRef]
- Schmid, J.; Steiner, L.; Fademrecht, S.; Pleiss, J.; Otte, K.B.; Hauer, B. Biocatalytic study of novel oleate hydratases. J. Mol. Catal. B Enzym. 2016, 133, S243–S249. [Google Scholar] [CrossRef]
- Hiseni, A.; Arends, I.W.C.E.; Otten, L.G. New Cofactor-Independent Hydration Biocatalysts: Structural, Biochemical, and Biocatalytic Characteristics of Carotenoid and Oleate Hydratases. ChemCatChem 2014, 7, 29–37. [Google Scholar] [CrossRef]
- Jeon, E.-Y.; Lee, J.-H.; Yang, K.-M.; Joo, Y.-C.; Oh, D.-K.; Park, J.-B. Bioprocess engineering to produce 10-hydroxystearic acid from oleic acid by recombinant Escherichia coli expressing the oleate hydratase gene of Stenotrophomonas maltophilia. Process. Biochem. 2012, 47, 941–947. [Google Scholar] [CrossRef]
- Joo, Y.-C.; Jeong, K.-W.; Yeom, S.-J.; Kim, Y.-S.; Kim, Y.; Oh, D.-K. Biochemical characterization and FAD-binding analysis of oleate hydratase from Macrococcus caseolyticus. Biochimie 2012, 94, 907–915. [Google Scholar] [CrossRef]
- Kim, B.-N.; Joo, Y.-C.; Kim, Y.-S.; Kim, K.-R.; Oh, D.-K. Production of 10-hydroxystearic acid from oleic acid and olive oil hydrolyzate by an oleate hydratase from Lysinibacillus fusiformis. Appl. Microbiol. Biotechnol. 2011, 95, 929–937. [Google Scholar] [CrossRef]
- Oh, H.-J.; Shin, K.-C.; Oh, D.-K. Production of 10-hydroxy-12,15(Z,Z)-octadecadienoic acid from α-linolenic acid by permeabilized cells of recombinant Escherichia coli expressing the oleate hydratase gene of Stenotrophomonas maltophilia. Biotechnol. Lett. 2013, 35, 1487–1493. [Google Scholar] [CrossRef]
- Yang, B.; Chen, H.; Song, Y.; Chen, Y.Q.; Zhang, H.; Chen, W. Myosin-cross-reactive antigens from four different lactic acid bacteria are fatty acid hydratases. Biotechnol. Lett. 2012, 35, 75–81. [Google Scholar] [CrossRef]
- Hirata, A.; Kishino, S.; Park, S.-B.; Takeuchi, M.; Kitamura, N.; Ogawa, J. A novel unsaturated fatty acid hydratase toward C16 to C22 fatty acids from Lactobacillus acidophilus. J. Lipid Res. 2015, 56, 1340–1350. [Google Scholar] [CrossRef]
- Oh, H.-J.; Kim, S.-U.; Song, J.-W.; Lee, J.-H.; Kang, W.-R.; Jo, Y.-S.; Kim, K.-R.; Bornscheuer, U.T.; Oh, D.-K.; Park, J.-B. Biotransformation of Linoleic Acid into Hydroxy Fatty Acids and Carboxylic Acids Using a Linoleate Double Bond Hydratase as Key Enzyme. Adv. Synth. Catal. 2015, 357, 408–416. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kishino, S.; Hirata, A.; Park, S.-B.; Kitamura, N.; Ogawa, J. Characterization of the linoleic acid Δ9 hydratase catalyzing the first step of polyunsaturated fatty acid saturation metabolism in Lactobacillus plantarum AKU 1009a. J. Biosci. Bioeng. 2015, 119, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kishino, S.; Park, S.-B.; Kitamura, N.; Ogawa, J. Characterization of hydroxy fatty acid dehydrogenase involved in polyunsaturated fatty acid saturation metabolism in Lactobacillus plantarum AKU 1009a. J. Mol. Catal. B Enzym. 2015, 117, 7–12. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Liang, N.Y.; Curtis, J.M.; Gänzle, M.G. Characterization of Linoleate 10-Hydratase of Lactobacillus plantarum and Novel Antifungal Metabolites. Front. Microbiol. 2016, 7, 1561. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.-R.; Seo, M.-J.; Shin, K.-C.; Park, J.-B.; Oh, D.-K. Gene cloning of an efficiency oleate hydratase from Stenotrophomonas nitritireducens for polyunsaturated fatty acids and its application in the conversion of plant oils to 10-hydroxy fatty acids. Biotechnol. Bioeng. 2016, 114, 74–82. [Google Scholar] [CrossRef]
- Engleder, M.; Pavkov-Keller, T.; Emmerstorfer-Augustin, A.; Hromic, A.; Schrempf, S.; Steinkellner, G.; Wriessnegger, T.; Leitner, E.; Strohmeier, G.A.; Kaluzna, I.; et al. Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia meningoseptica. ChemBioChem 2015, 16, 1730–1734. [Google Scholar] [CrossRef]
- Park, A.K.; Lee, G.H.; Kim, D.W.; Jang, E.H.; Kwon, H.T.; Chi, Y.M. Crystal structure of oleate hydratase from Stenotrophomonas sp. KCTC 12332 reveals conformational plasticity surrounding the FAD binding site. Biochem. Biophys. Res. Commun. 2018, 499, 772–776. [Google Scholar] [CrossRef]
- Schwinn, K.; Ferré, N.; Huix-Rotllant, M. UV-visible absorption spectrum of FAD and its reduced forms embedded in a cryptochrome protein. Phys. Chem. Chem. Phys. 2020, 22, 12447–12455. [Google Scholar] [CrossRef]
- Nivière, V.; Fieschi, F.; Décout, J.-L.; Fontecave, M. The NAD(P)H:Flavin Oxidoreductase from Escherichia coli. J. Biol. Chem. 1999, 274, 18252–18260. [Google Scholar] [CrossRef]
- Bechtold, M.; Brenna, M.E.; Femmer, C.; Gatti, F.G.; Panke, S.; Parmeggiani, F.; Sacchetti, A. Biotechnological Development of a Practical Synthesis of Ethyl (S)-2-Ethoxy-3-(p-methoxyphenyl)propanoate (EEHP): Over 100-Fold Productivity Increase from Yeast Whole Cells to Recombinant Isolated Enzymes. Org. Process. Res. Dev. 2011, 16, 269–276. [Google Scholar] [CrossRef]
- Morokutti, A.; Lyskowski, A.; Sollner, S.; Pointner, E.; Fitzpatrick, T.B.; Kratky, C.; Gruber, K.; Macheroux, P. Structure and Function of YcnD from Bacillus subtilis, a Flavin-Containing Oxidoreductase†,‡. Biochemistry 2005, 44, 13724–13733. [Google Scholar] [CrossRef]
- Zehnder, A.J.; Wuhrmann, K. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 1976, 194, 1165–1166. [Google Scholar] [CrossRef]
- Unversucht, S.; Hollmann, F.; Schmid, A.; Van Pée, K.-H. FADH2-Dependence of Tryptophan 7-Halogenase. Adv. Synth. Catal. 2005, 347, 1163–1167. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).