Dye Decoloring Peroxidase Structure, Catalytic Properties and Applications: Current Advancement and Futurity
Abstract
:1. Introduction
2. Composition and Structural Characteristics of DyPs
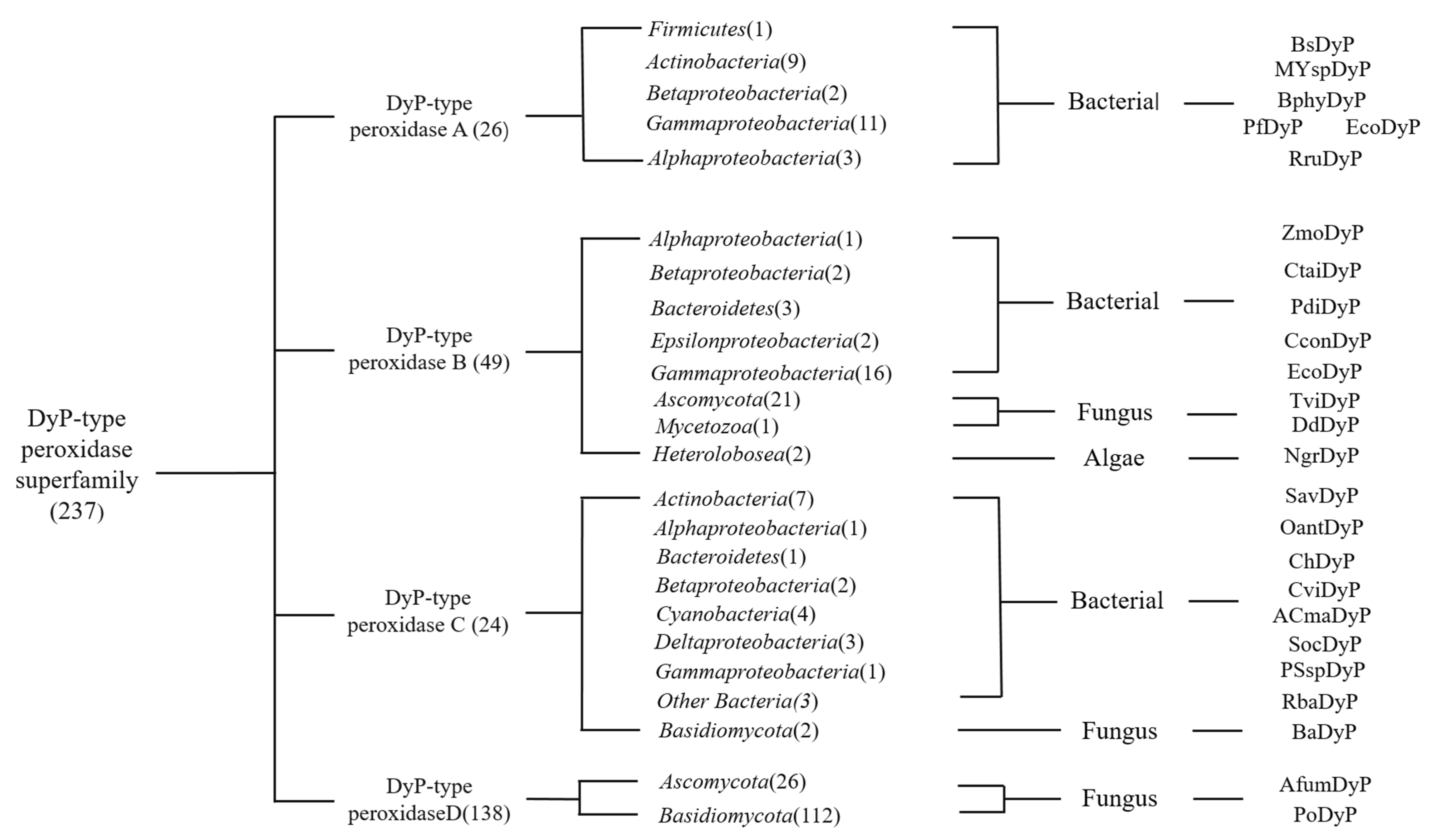
2.1. Sources and Classification of DyPs
2.2. Characteristics of DyPs
2.3. Structural Characteristics of DyP
2.4. The Physical and Chemical Properties of DyPs
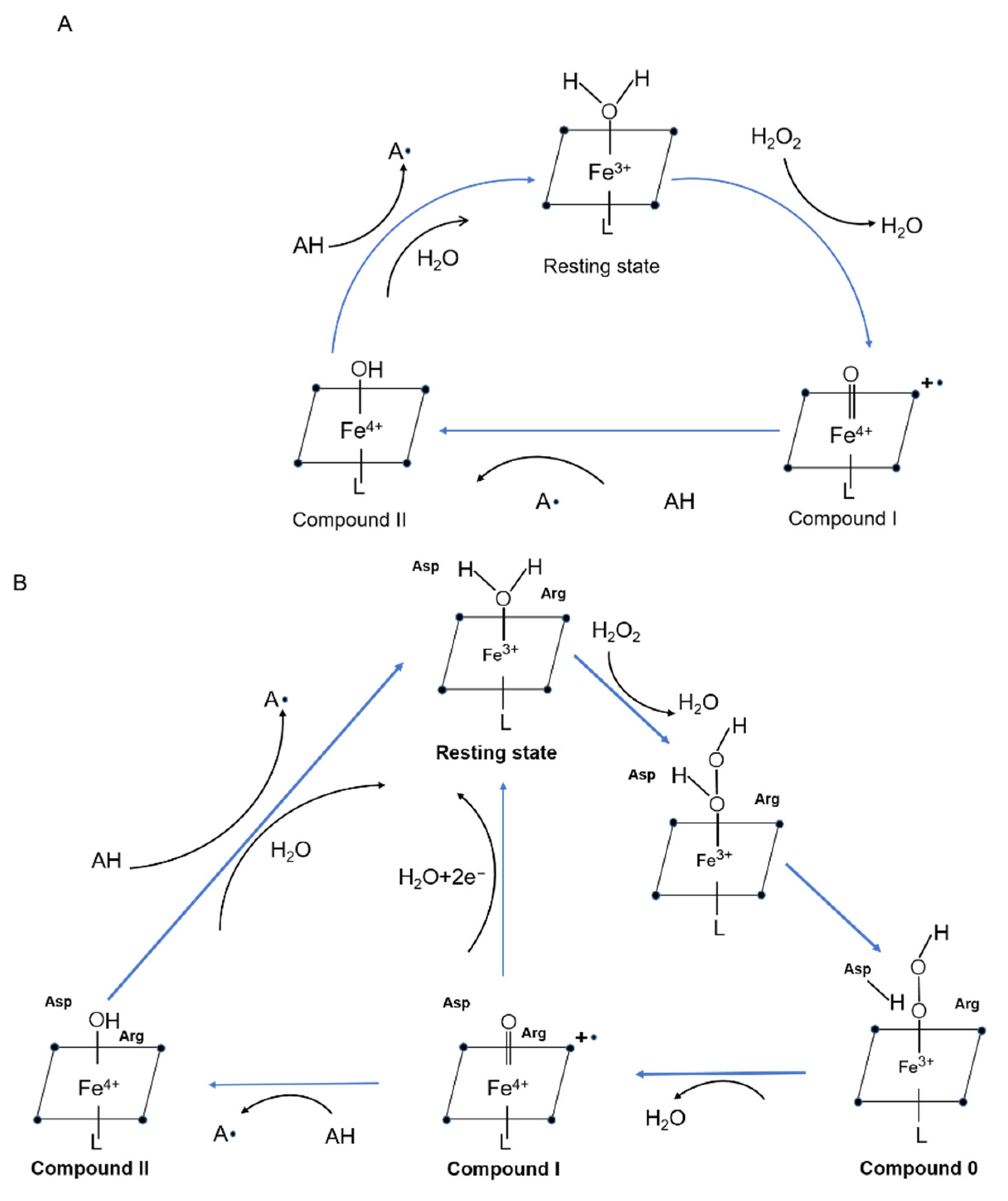
2.5. Catalytic Characteristics and Mechanism
2.6. Unrepresentative Characteristics, a Multifunctional Enzyme
3. Application of Dye Decoloring Enzyme
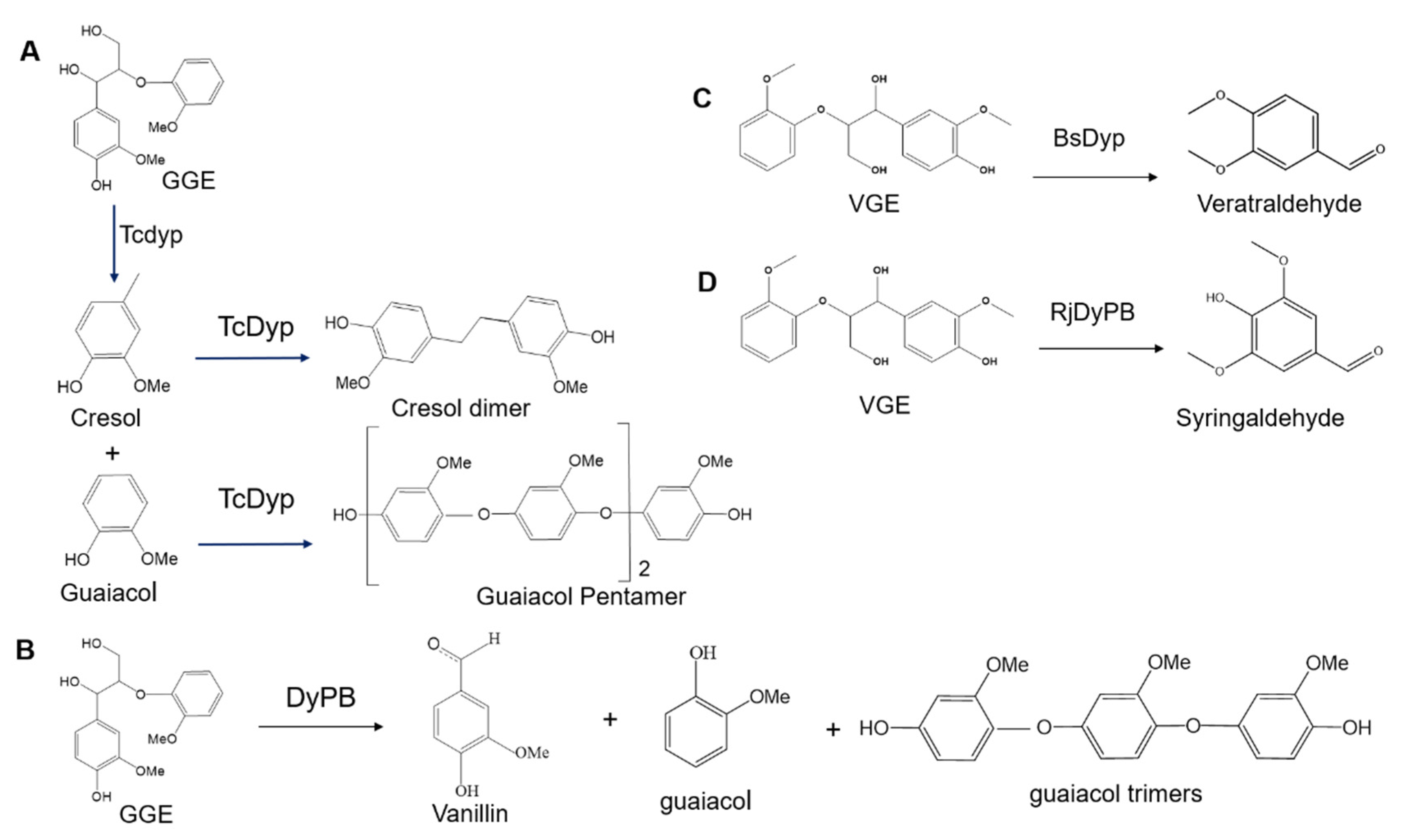
3.1. Lignin Degradation
3.2. Dye Decolorization
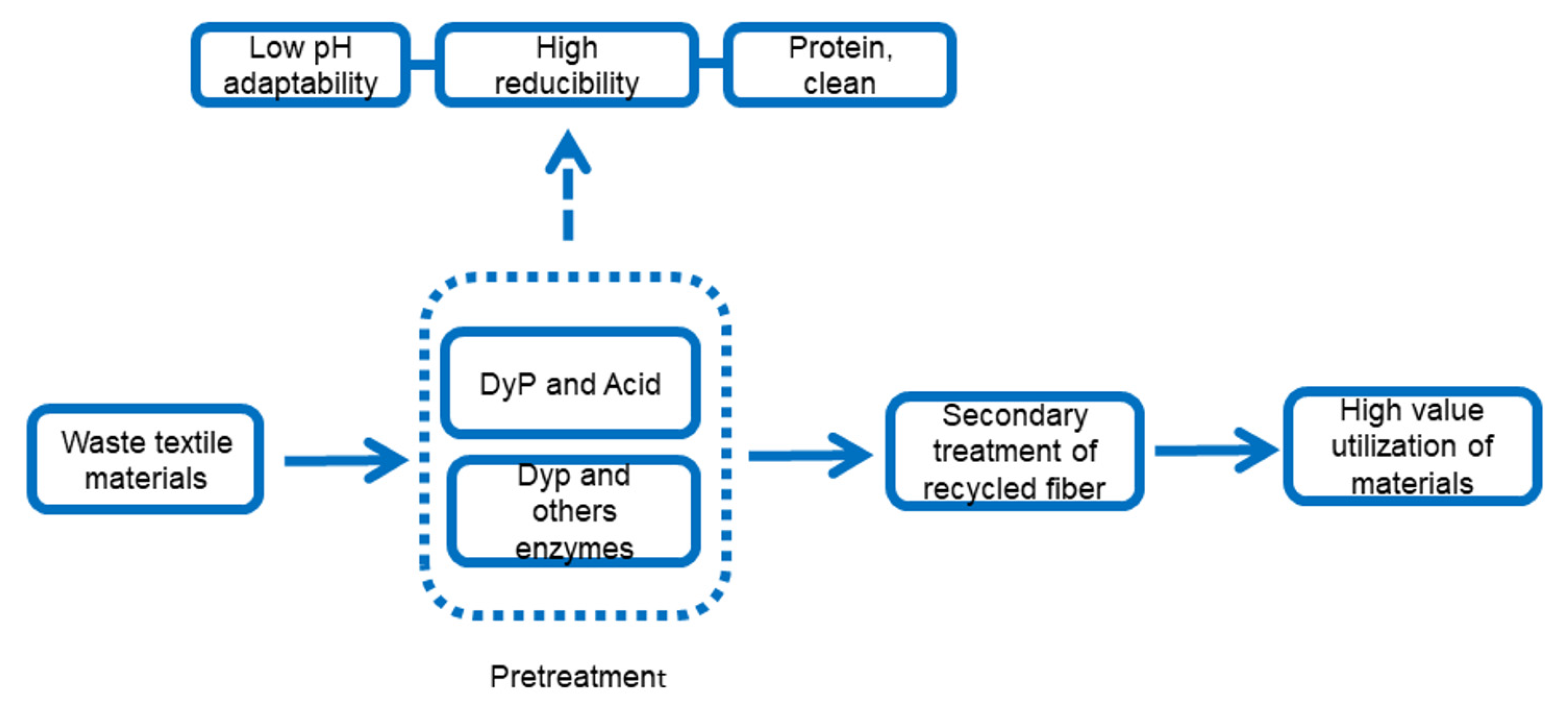
3.3. Other Potential Applications
4. New Technology Promotes DyP Industrialization (Application) Research
4.1. Improvement of DyPs Properties by Gene-Editing Technology
4.2. Choose Suitable Host Cells to Promote the Industrial Application of Enzymes
4.3. The Rise of Synthetic Protein Technology
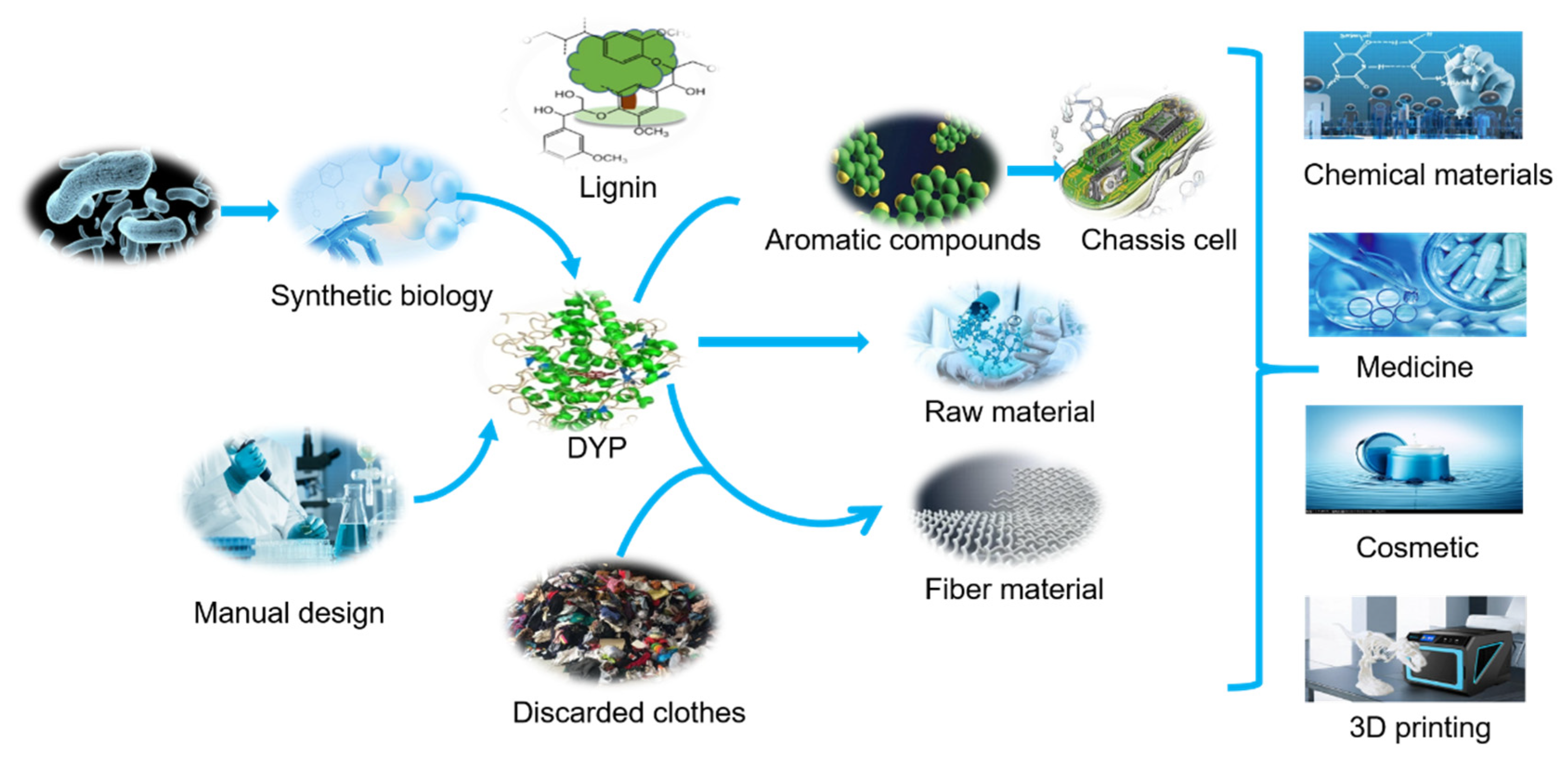
5. Prospective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Davies, M.J.; Hawkins, C.L.; Pattison, D.I.; Rees, M.D. Mammalian heme peroxidases: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 1199–1234. [Google Scholar] [CrossRef] [PubMed]
- Heinfling, A.; Martínez, M.A.J.; Martínez, A.T.; Bergbauer, M.; Szewzyk, U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol. Lett. 1998, 165, 43–50. [Google Scholar] [CrossRef]
- Sugano, Y.; Muramatsu, R.; Ichiyanagi, A.; Sato, T.; Shoda, M. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family: ASP171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 2007, 282, 36652–36658. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Sugano, Y. A structural and functional perspective of DyP-type peroxidase family. Arch. Biochem. Biophys. 2015, 574, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Shoda, M. Purification and characterization of a novel peroxidase from Geotrichum candidum dec 1 involved in decolorization of dyes. Appl. Environ. Microbiol. 1999, 65, 1029–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 871–897. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Pandey, V.P.; Naaz, H.; Dwivedi, U.N. Phylogenetic analysis, molecular modeling, substrate-inhibitor specificity, and active site comparison of bacterial, fungal, and plant heme peroxidases. Biotechnol. Appl. Biochem. 2012, 59, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Zámocký, M. Phylogenetic relationships in class I of the superfamily of bacterial, fungal, and plant peroxidases. Eur. J. Biochem. 2004, 271, 3297–3309. [Google Scholar] [CrossRef] [PubMed]
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Tsuge, H.; Konno, H.; Hisabori, T.; Sugano, Y. The catalytic mechanism of dye-decolorizing peroxidase DyP may require the swinging movement of an aspartic acid residue. FEBS J. 2011, 278, 2387–2394. [Google Scholar] [CrossRef]
- Brown, M.E.; Barros, T.; Chang, M.C. Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem. Biol. 2012, 7, 2074–2081. [Google Scholar] [CrossRef] [PubMed]
- Ogola, H.J.; Kamiike, T.; Hashimoto, N.; Ashida, H.; Ishikawa, T.; Shibata, H.; Sawa, Y. Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 2009, 75, 7509–7518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvachúa, D.; Prieto, A.; Martínez, Á.T.; Martínez, M.J. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl. Environ. Microbiol. 2013, 79, 4316–4324. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.; Schwarz, T.; Nguyen, Q.K.; Lorenz, P.; Lochnit, G.; Zorn, H. Identification, heterologous expression and characterization of a dye-decolorizing peroxidase of Pleurotus sapidus. AMB Express 2017, 7, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liers, C.; Pecyna, M.J.; Kellner, H.; Worrich, A.; Zorn, H.; Steffen, K.T.; Hofrichter, M.; Ullrich, R. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl. Microbiol. Biotechnol. 2013, 97, 5839–5849. [Google Scholar] [CrossRef]
- Krahe, N.K.; Berger, R.G.; Ersoy, F. A DyP-Type Peroxidase of Pleurotus sapidus with Alkene Cleaving Activity. Molecules 2020, 25, 1536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras, H.; Joens, M.S.; McMath, L.M.; Le, V.P.; Tullius, M.V.; Kimmey, J.M.; Bionghi, N.; Horwitz, M.A.; Fitzpatrick, J.A.; Goulding, C.W. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J. Biol. Chem. 2014, 289, 18279–18289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Létoffé, S.; Heuck, G.; Delepelaire, P.; Lange, N.; Wandersman, C. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. USA 2009, 106, 11719–11724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Du, Q.; Wang, Z.; Zhu, D.; Huang, Y.; Li, N.; Wei, T.; Xu, S.; Gu, L. Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes. J. Biol. Chem. 2011, 286, 14922–14931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colpa, D.I.; Fraaije, M.W.; van Bloois, E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014, 41, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zámocký, M.; Hofbauer, S.; Schaffner, I.; Gasselhuber, B.; Nicolussi, A.; Soudi, M.; Pirker, K.F.; Furtmüller, P.G.; Obinger, C. Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 2015, 574, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; Krishna, S.S.; Kapoor, M.; Kozbial, P.; McMullan, D.; Axelrod, H.L.; Miller, M.D.; Abdubek, P.; Ambing, E.; Astakhova, T.; et al. Crystal structures of two novel dye-decolorizing peroxidases reveal a beta-barrel fold with a conserved heme-binding motif. Proteins 2007, 69, 223–233. [Google Scholar] [CrossRef]
- Singh, R.; Eltis, L.D. The multihued palette of dye-decolorizing peroxidases. Arch. Biochem. Biophys. 2015, 574, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Klare, J.P.; Reinke, P.Y.; Englmaier, F.; Fohrer, J.; Fedorov, R.; Taft, M.H.; Chizhov, I.; Curth, U.; Plettenburg, O. Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum. Int. J. Mol. Sci. 2021, 22, 6265. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Shrestha, R.; Jia, K.; Gao, P.F.; Geisbrecht, B.V.; Bossmann, S.H.; Shi, J.; Li, P. Characterization of Dye-decolorizing Peroxidase (DyP) from Thermomonospora curvata Reveals Unique Catalytic Properties of A-type DyPs. J. Biol. Chem. 2015, 290, 23447–23463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, A.; Mendes, S.; Brissos, V.; Martins, L.O. New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: Towards biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2053–2065. [Google Scholar] [CrossRef] [PubMed]
- Rahmanpour, R.; Rea, D.; Jamshidi, S.; Fülöp, V.; Bugg, T.D. Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Arch. Biochem. Biophys. 2016, 594, 54–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, A.M.; Martins, V.C.; Prazeres, D.M.; Vojinović, V.; Cabral, J.M.; Fonseca, L.P. Horseradish peroxidase: A valuable tool in biotechnology. Biotechnol. Annu. Rev. 2003, 9, 199–247. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Roberts, J.N.; Hardiman, E.M.; Singh, R.; Eltis, L.D.; Bugg, T.D. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 2011, 50, 5096–5107. [Google Scholar] [CrossRef]
- Shrestha, R.; Chen, X.; Ramyar, K.X.; Hayati, Z.; Carlson, E.A.; Bossmann, S.H.; Song, L.; Geisbrecht, B.V.; Li, P. Identification of Surface-Exposed Protein Radicals and A Substrate Oxidation Site in A-Class Dye-Decolorizing Peroxidase from Thermomonospora curvata. ACS Catal. 2016, 6, 8036–8047. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Grigg, J.C.; Armstrong, Z.; Murphy, M.E.P.; Eltis, L.D. Distal heme pocket residues of B-type dye-decolorizing peroxidase: Arginine but not aspartate is essential for peroxidase activity. J. Biol. Chem. 2012, 287, 10623–10630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbosa, C.; Silveira, C.M.; Silva, D.; Brissos, V.; Hildebrandt, P.; Martins, L.O.; Todorovic, S. Immobilized dye-decolorizing peroxidase (DyP) and directed evolution variants for hydrogen peroxide biosensing. Biosens. Bioelectron. 2020, 153, 112055. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yuan, Z.; Wang, J.; Cui, Y.; Liu, S.; Ma, Y.; Gu, L.; Xu, S. Crystal structure and biochemical features of dye-decolorizing peroxidase YfeX from Escherichia coli O157 Asp(143) and Arg(232) play divergent roles toward different substrates. Biochem. Biophys. Res. Commun. 2017, 484, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Woodhall, M.R.; Alvarez, J.; Cartron, M.L.; Andrews, S.C. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 2007, 65, 857–875. [Google Scholar] [CrossRef]
- Kong, L.; Guo, D.; Zhou, S.; Yu, X.; Hou, G.; Li, R.; Zhao, B. Cloning and expression of a toxin gene from Pseudomonas fluorescens GcM5-1A. Arch. Microbiol. 2010, 192, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liu, W.; Huang, H.; Zheng, F.; Wang, X.; Wu, Y.; Li, K.; Xie, X.; Jin, Y. Application of a novel alkali-tolerant thermostable DyP-type peroxidase from Saccharomonospora viridis DSM 43017 in biobleaching of eucalyptus kraft pulp. PLoS ONE 2014, 9, e110319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, K.; Gong, G.; Woo, H.M.; Kim, Y.; Um, Y. A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and β-ether lignin dimer. Sci. Rep. 2015, 5, 8245. [Google Scholar] [CrossRef] [PubMed]
- Field, M.J.; Bains, R.K.; Warren, J.J. Using an artificial tryptophan “wire” in cytochrome c peroxidase for oxidation of organic substrates. Dalton Trans. 2017, 46, 11078–11083. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.K.; Hyeon, J.E.; Joo, Y.C.; Jeong, D.W.; You, S.K.; Han, S.O. Effective melanin degradation by a synergistic laccase-peroxidase enzyme complex for skin whitening and other practical applications. Int. J. Biol. Macromol. 2019, 129, 181–186. [Google Scholar] [CrossRef]
- Ralph, J.; Lapierre, C.; Boerjan, W. Lignin structure and its engineering. Curr. Opin. Biotechnol. 2019, 56, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Sulej, J.; Swiderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczynski, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Zhang, P.; Xie, C.; Zhang, W.; Sun, J.; Qian, W.J.; Yang, B. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol. Biofuels 2017, 10, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Zhang, S.; Chen, G. Mechanisms of Lignin-Degrading Enzymes. Protein Pept. Lett. 2020, 27, 574–581. [Google Scholar] [CrossRef]
- Rajhans, G.; Sen, S.K.; Barik, A.; Raut, S. Elucidation of fungal dye-decolourizing peroxidase (DyP) and ligninolytic enzyme activities in decolourization and mineralization of azo dyes. J. Appl. Microbiol. 2020, 129, 1633–1643. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamimura, N.; Sakamoto, S.; Mitsuda, N.; Masai, E.; Kajita, S. Advances in microbial lignin degradation and its applications. Curr. Opin. Biotechnol. 2019, 56, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Rahman Pour, R.; Ehibhatiomhan, A.; Huang, Y.; Ashley, B.; Rashid, G.M.; Mendel-Williams, S.; Bugg, T.D.H. Protein engineering of Pseudomonas fluorescens peroxidase Dyp1B for oxidation of phenolic and polymeric lignin substrates. Enzym. Microb. Technol. 2019, 123, 21–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Bugg, T.D.; Rahmanpour, R. Enzymatic conversion of lignin into renewable chemicals. Curr. Opin. Chem. Biol. 2015, 29, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Routoula, E.; Patwardhan, S.V. Degradation of Anthraquinone Dyes from Effluents: A Review Focusing on Enzymatic Dye Degradation with Industrial Potential. Environ. Sci. Technol. 2020, 54, 647–664. [Google Scholar] [CrossRef]
- Piaskowski, K.; Świderska-Dąbrowska, R.; Zarzycki, P.K. Dye Removal from Water and Wastewater Using Various Physical, Chemical, and Biological Processes. J. AOAC Int. 2018, 101, 1371–1384. [Google Scholar] [CrossRef]
- Kaushik, P.; Malik, A. Fungal dye decolourization: Recent advances and future potential. Environ. Int. 2009, 35, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Mendes, S.; Brissos, V.; Gabriel, A.; Catarino, T.; Turner, D.L.; Todorovic, S.; Martins, L.O. An integrated view of redox and catalytic properties of B-type PpDyP from Pseudomonas putida MET94 and its distal variants. Arch. Biochem. Biophys. 2015, 574, 99–107. [Google Scholar] [CrossRef]
- Kadam, A.A.; Kamatkar, J.D.; Khandare, R.V.; Jadhav, J.P.; Govindwar, S.P. Solid-state fermentation: Tool for bioremediation of adsorbed textile dyestuff on distillery industry waste-yeast biomass using isolated Bacillus cereus strain EBT1. Environ. Sci. Pollut. Res. Int. 2013, 20, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Mendes, S.; Farinha, A.; Ramos, C.G.; Leitão, J.H.; Viegas, C.A.; Martins, L.O. Synergistic action of azoreductase and laccase leads to maximal decolourization and detoxification of model dye-containing wastewaters. Bioresour. Technol. 2011, 102, 9852–9859. [Google Scholar] [CrossRef] [PubMed]
- Linde, D.; Ruiz-Dueñas, F.J.; Fernández-Fueyo, E.; Guallar, V.; Hammel, K.E.; Pogni, R.; Martínez, A.T. Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance. Arch. Biochem. Biophys. 2015, 574, 66–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugano, Y.; Yoshida, T. DyP-Type Peroxidases: Recent Advances and Perspectives. Int. J. Mol. Sci. 2021, 22, 5556. [Google Scholar] [CrossRef] [PubMed]
- Jones, P. Roles of water in heme peroxidase and catalase mechanisms. J. Biol. Chem. 2001, 276, 13791–13796. [Google Scholar] [CrossRef] [Green Version]
- Sugano, Y.; Matsushima, Y.; Tsuchiya, K.; Aoki, H.; Hirai, M.; Shoda, M. Degradation pathway of an anthraquinone dye catalyzed by a unique peroxidase DyP from Thanatephorus cucumeris Dec 1. Biodegradation 2009, 20, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Asaadi, S.; Hummel, M.; Hellsten, S.; Härkäsalmi, T.; Ma, Y.; Michud, A.; Sixta, H. Renewable High-Performance Fibers from the Chemical Recycling of Cotton Waste Utilizing an Ionic Liquid. Chem. Sus. Chem. 2016, 9, 3250–3258. [Google Scholar] [CrossRef] [PubMed]
- Navone, L.; Moffitt, K.; Hansen, K.A.; Blinco, J.; Payne, A.; Speight, R. Closing the textile loop: Enzymatic fibre separation and recycling of wool/polyester fabric blends. Waste Manag. 2020, 102, 149–160. [Google Scholar] [CrossRef]
- Scheibner, M.; Hülsdau, B.; Zelena, K.; Nimtz, M.; de Boer, L.; Berger, R.G.; Zorn, H. Novel peroxidases of Marasmius scorodonius degrade beta-carotene. Appl. Microbiol. Biotechnol. 2008, 77, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.H.; Pourcher, A.M.; Klingelschmitt, F.; Le Roux, S.; Peu, P. Class P dye-decolorizing peroxidase gene: Degenerated primers design and phylogenetic analysis. J. Microbiol. Methods 2016, 130, 148–153. [Google Scholar] [CrossRef]
- Alessa, A.H.A.; Tee, K.L.; Gonzalez-Perez, D.; Omar Ali, H.E.M.; Evans, C.A.; Trevaskis, A.; Xu, J.H.; Wong, T.S. Accelerated directed evolution of dye-decolorizing peroxidase using a bacterial extracellular protein secretion system (BENNY). Bioresour. Bioprocess. 2019, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.; Horbal, L.; Starck, S.; Kohlstedt, M.; Luzhetskyy, A.; Wittmann, C. Enabling the valorization of guaiacol-based lignin: Integrated chemical and biochemical production of cis, cis-muconic acid using metabolically engineered Amycolatopsis sp ATCC 39116. Metab. Eng. 2018, 45, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.; Roy, I. Converting Enzymes into Tools of Industrial Importance. Recent Pat. Biotechnol. 2018, 12, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Li, D.; Alic, M.; Gold, M.H. Heat Shock Induction of Manganese Peroxidase Gene Transcription in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1993, 59, 4295–4299. [Google Scholar] [CrossRef] [Green Version]
- Dartois, V.; Baulard, A.; Schanck, K.; Colson, C. Cloning, nucleotide sequence and expression in Escherichia coli of a lipase gene from Bacillus subtilis 168. Biochim. Biophys. Acta 1992, 1131, 253–260. [Google Scholar] [CrossRef]
- Reddy, G.V.; Gelpke, M.D.; Gold, M.H. Degradation of 2,4,6-trichlorophenol by Phanerochaete chrysosporium: Involvement of reductive dechlorination. J. Bacteriol. 1998, 180, 5159–5164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukihara, T.; Honda, Y.; Sakai, R.; Watanabe, T.; Watanabe, T. Exclusive overproduction of recombinant versatile peroxidase MnP2 by genetically modified white rot fungus, Pleurotus ostreatus. J. Biotechnol. 2006, 126, 431–439. [Google Scholar] [CrossRef]
- Zelena, K.; Eisele, N.; Berger, R.G. Escherichia coli as a production host for novel enzymes from basidiomycota. Biotechnol. Adv. 2014, 32, 1382–1395. [Google Scholar] [CrossRef]
- Wang, L.; Chang, A.K.; Yuan, W.; Bai, F. Recombinant expression, purification and characterization of a novel DyP-type peroxidase in Escherichia coli. Chin. J. Biotechnol. 2013, 29, 772–784. [Google Scholar]
- Pontrelli, S.; Chiu, T.Y.; Lan, E.I.; Chen, F.Y.; Chang, P.; Liao, J.C. Escherichia coli as a host for metabolic engineering. Metab. Eng. 2018, 50, 16–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linde, D.; Pogni, R.; Cañellas, M.; Lucas, F.; Guallar, V.; Baratto, M.C.; Sinicropi, A.; Sáez-Jiménez, V.; Coscolín, C.; Romero, A.; et al. Catalytic surface radical in dye-decolorizing peroxidase: A computational, spectroscopic and site-directed mutagenesis study. Biochem. J. 2015, 466, 253–262. [Google Scholar] [CrossRef]
- Sugano, Y.; Ishii, Y.; Shoda, M. Role of H164 in a unique dye-decolorizing heme peroxidase DyP. Biochem. Biophys. Res. Commun. 2004, 322, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Behrens, C.J.; Zelena, K.; Berger, R.G. Comparative Cold Shock Expression and Characterization of Fungal Dye-Decolorizing Peroxidases. Appl. Biochem. Biotechnol. 2016, 179, 1404–1417. [Google Scholar] [CrossRef]
- Lin, Y.W. Rational design of heme enzymes for biodegradation of pollutants toward a green future. Biotechnol. Appl. Biochem. 2020, 67, 484–494. [Google Scholar] [CrossRef]
- Lin, Y.W.; Nie, C.M.; Liao, L.F. Rational design of a nitrite reductase based on myoglobin: A molecular modeling and dynamics simulation study. J. Mol. Modeling 2012, 18, 4409–4415. [Google Scholar] [CrossRef]
- Yan, D.J.; Yuan, H.; Li, W.; Xiang, Y.; He, B.; Nie, C.M.; Wen, G.B.; Lin, Y.W.; Tan, X. How a novel tyrosine-heme crosslink fine-tunes the structure and functions of heme proteins: A direct comparitive study of L29H/F43Y myoglobin. Dalton Trans. 2015, 44, 18815–18822. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Yuan, H.; Liao, F.; He, B.; Gao, S.Q.; Wen, G.B.; Tan, X.; Lin, Y.W. Rational design of artificial dye-decolorizing peroxidases using myoglobin by engineering Tyr/Trp in the heme center. Dalton Trans. 2017, 46, 11230–11238. [Google Scholar] [CrossRef] [PubMed]


| DyP Type | Summary of Findings on DyP | Biotechnological Potential | Ref |
|---|---|---|---|
| PfuDyP | Has a toxic effect on cells | Higher prospects in the medical field | [35] |
| SviDyP | E. coli heterologous expression | Improved alkali resistance of DyP to lignin environment | [36] |
| BsDyP | Degradation of lignin derivatives | Production of Veratrum aldehyde and other substances | [37] |
| TcDyP | Degradation of lignin derivatives | Strong activity on a variety of lignin substrates | [25] |
| TfuDyP | Degradation of lignin derivatives | Oxidation of phenolic substances couples the substrate to produce dimers | [27] |
| rPsaDyP | Heterologous expression strains for degradation | ABTS oxidation and pigment bleaching | [14] |
| CcP | Artificial introduction of amino acid residues | Man-made structure speeds up the reaction rate | [38] |
| BsDyP | Synergistic with laccase Cue0 | Efficiently degrade melanin | [39] |
| PpDyP | Directed evolution technology transforms mutant DyP | High-sensitivity biosensor | [32] |
| PsaPOX | DyP can degrade a variety of olefins | The pyrolysis product will produce anisaldehyde | [16] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Sun, J.; Qaria, M.A.; Gao, L.; Zhu, D. Dye Decoloring Peroxidase Structure, Catalytic Properties and Applications: Current Advancement and Futurity. Catalysts 2021, 11, 955. https://doi.org/10.3390/catal11080955
Xu L, Sun J, Qaria MA, Gao L, Zhu D. Dye Decoloring Peroxidase Structure, Catalytic Properties and Applications: Current Advancement and Futurity. Catalysts. 2021; 11(8):955. https://doi.org/10.3390/catal11080955
Chicago/Turabian StyleXu, Lingxia, Jianzhong Sun, Majjid A. Qaria, Lu Gao, and Daochen Zhu. 2021. "Dye Decoloring Peroxidase Structure, Catalytic Properties and Applications: Current Advancement and Futurity" Catalysts 11, no. 8: 955. https://doi.org/10.3390/catal11080955
APA StyleXu, L., Sun, J., Qaria, M. A., Gao, L., & Zhu, D. (2021). Dye Decoloring Peroxidase Structure, Catalytic Properties and Applications: Current Advancement and Futurity. Catalysts, 11(8), 955. https://doi.org/10.3390/catal11080955







