Covalent Immobilization of Proteases on Polylactic Acid for Proteins Hydrolysis and Waste Biomass Protein Content Valorization
Abstract
1. Introduction
2. Results and Discussion
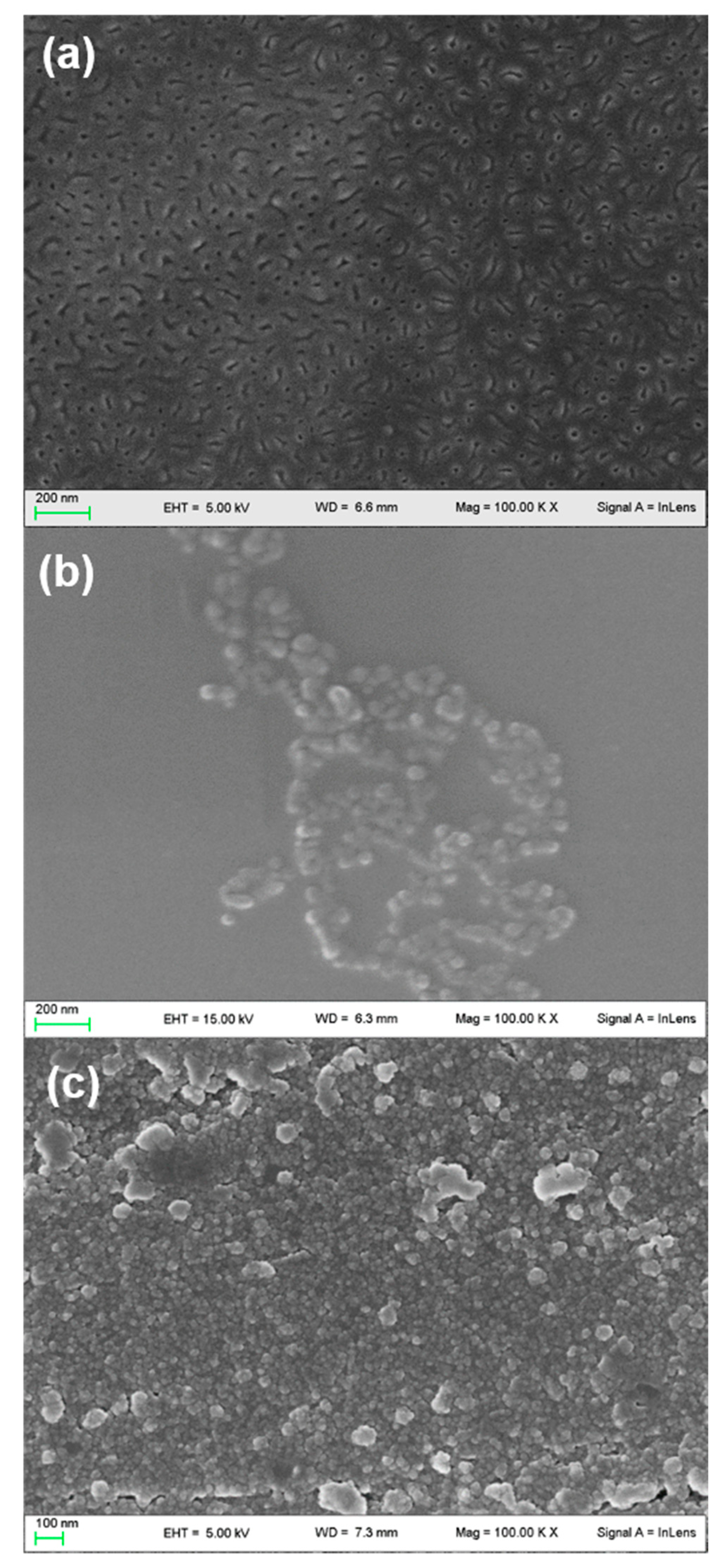
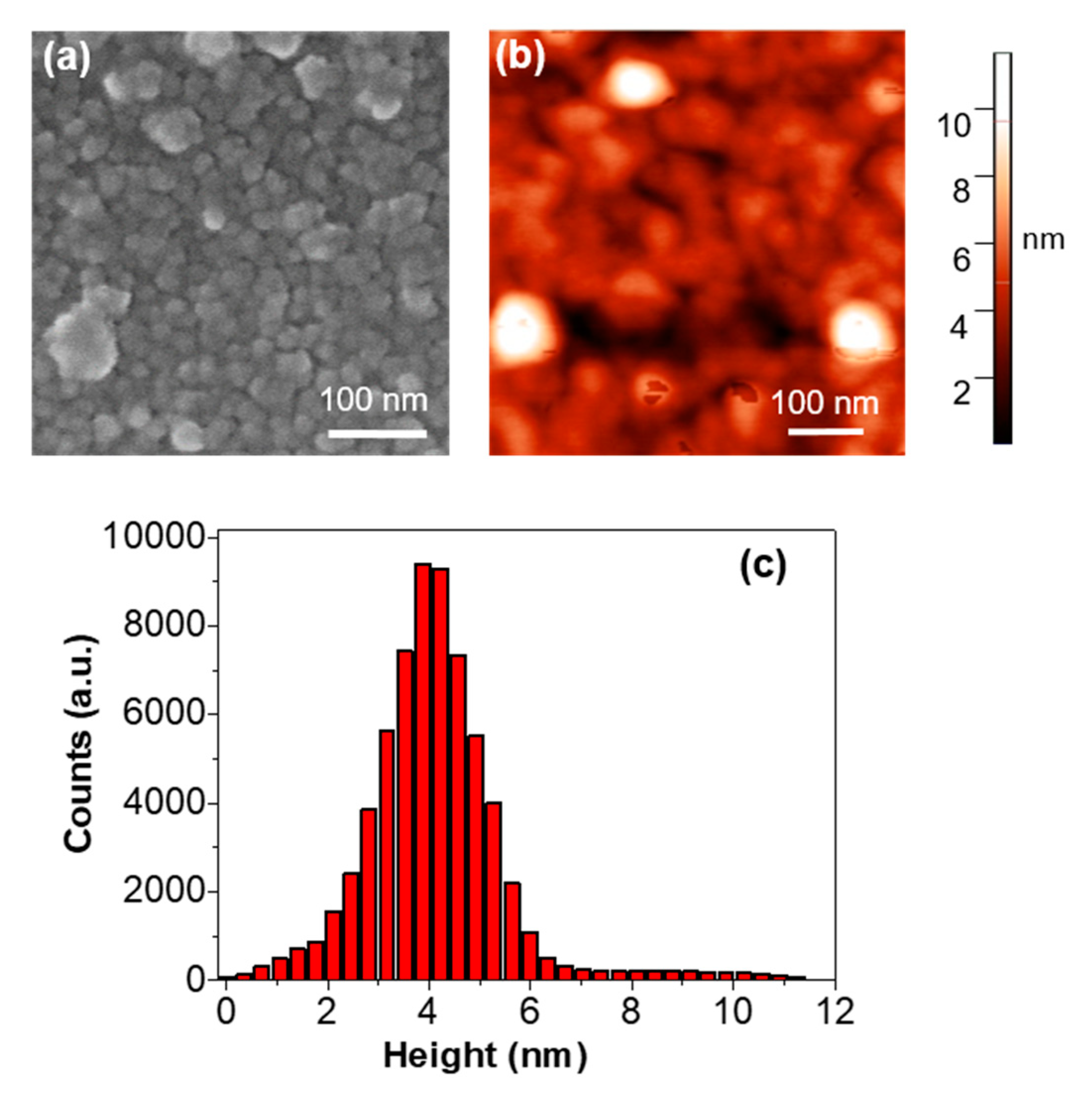
2.1. SEM and AFM Analyses
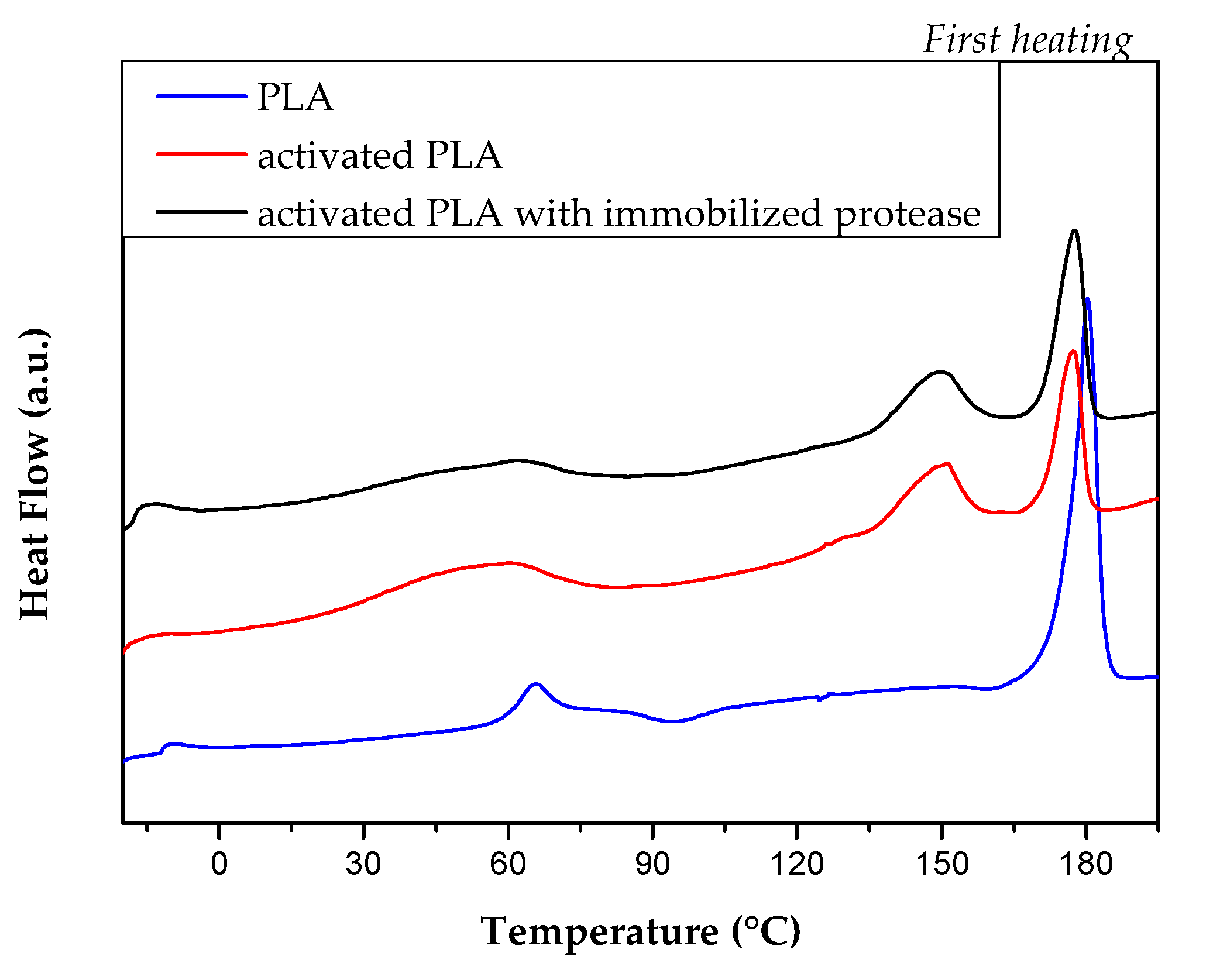
2.2. DSC Analysis
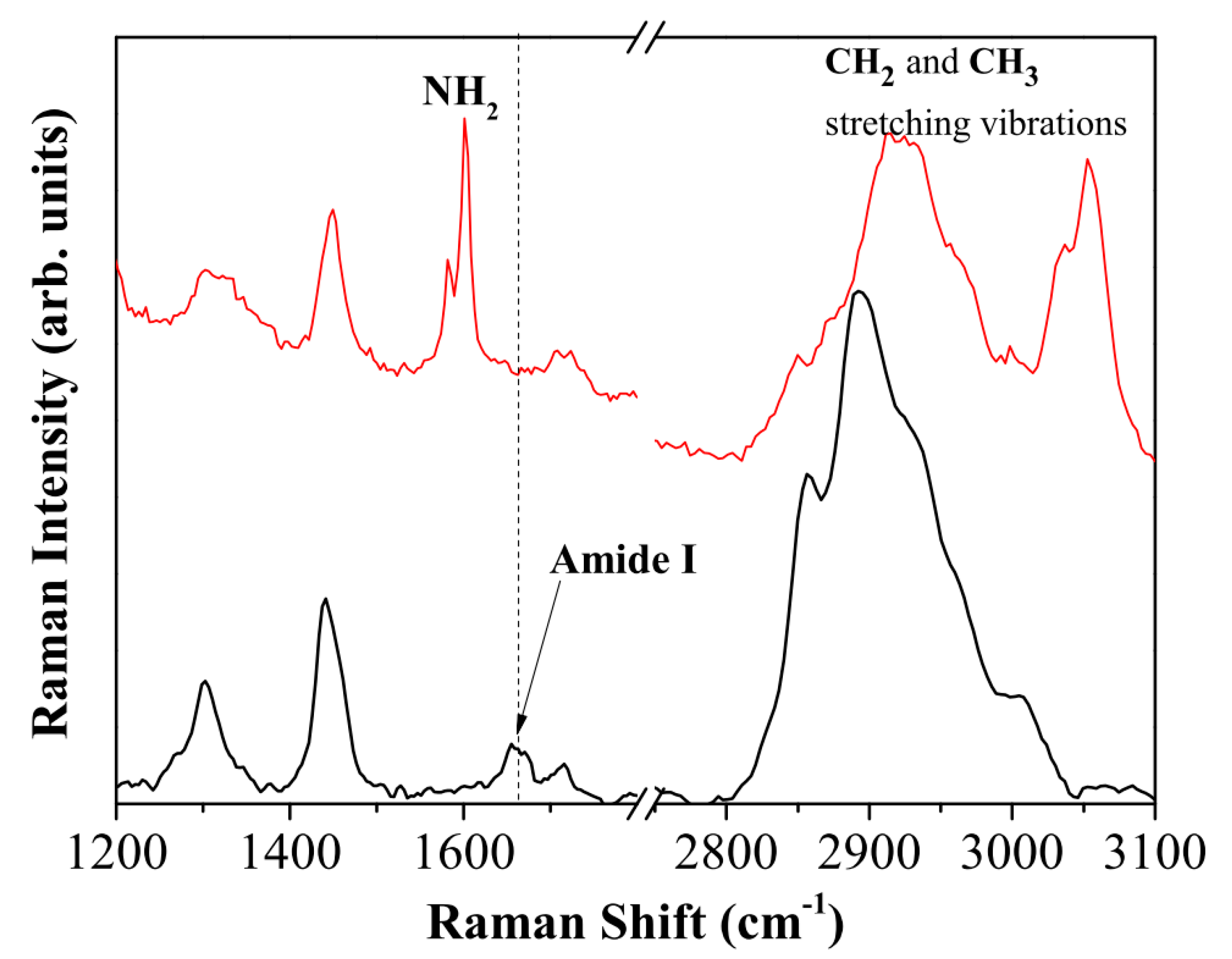
2.3. Raman Spectroscopy
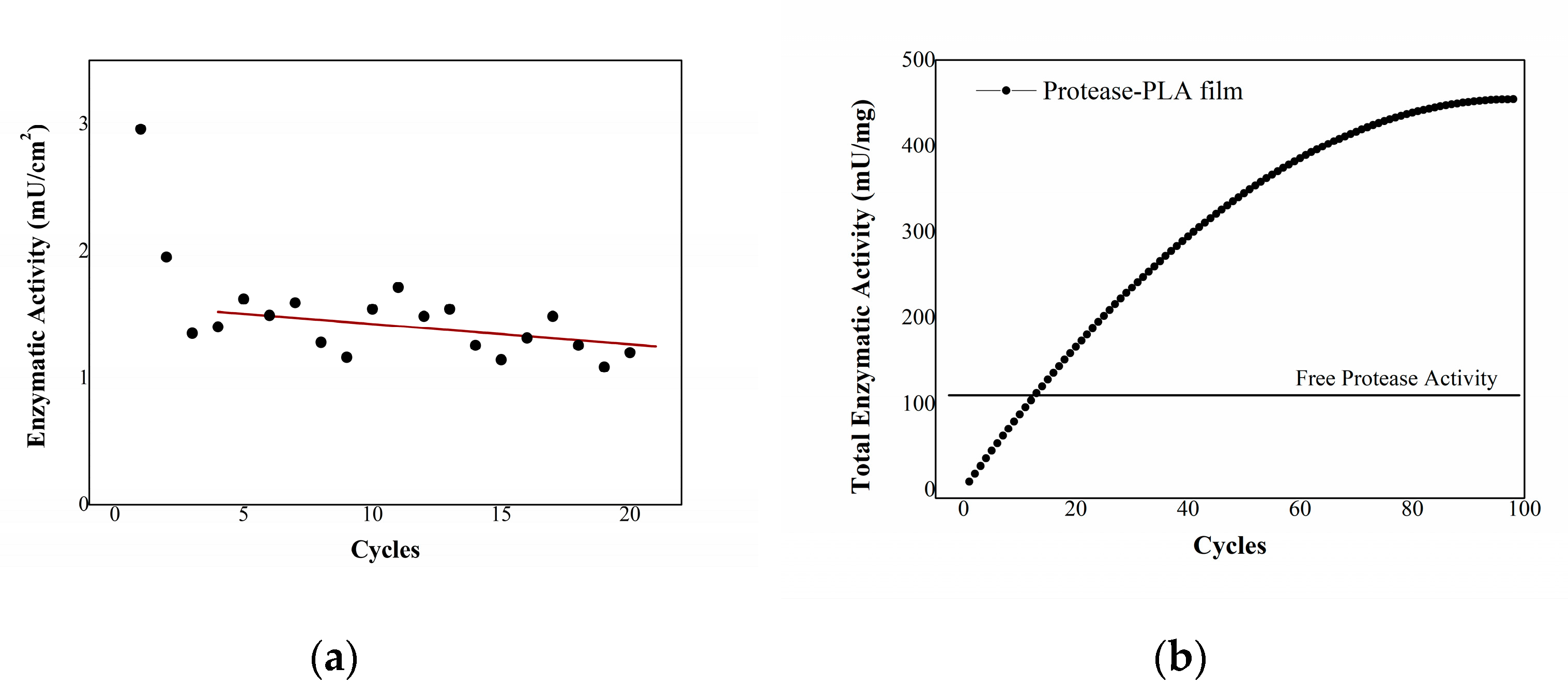
2.4. Stability of Enzymatic Activity over Time
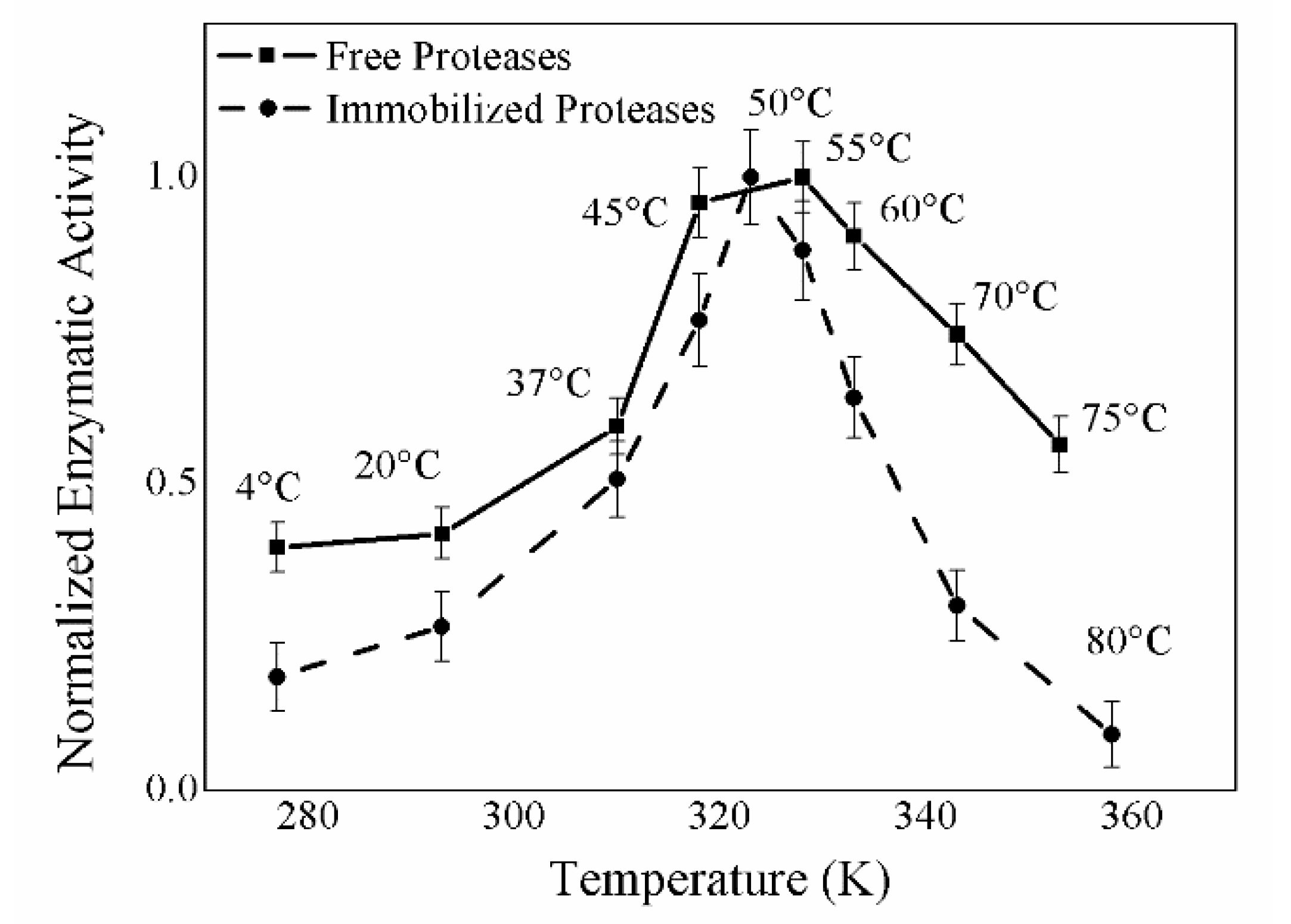
2.5. Temperature and pH Effect on Enzymatic Activity
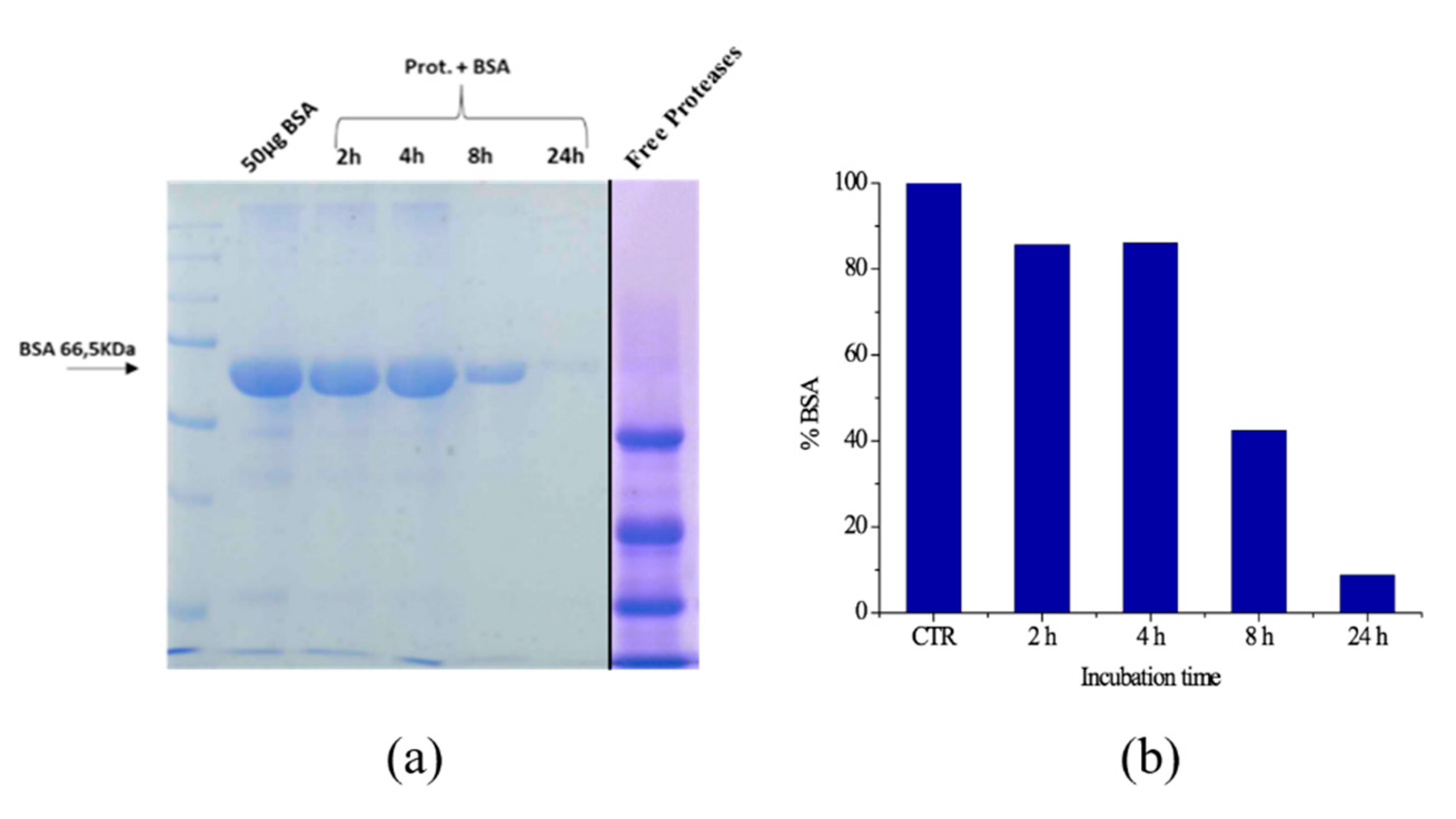
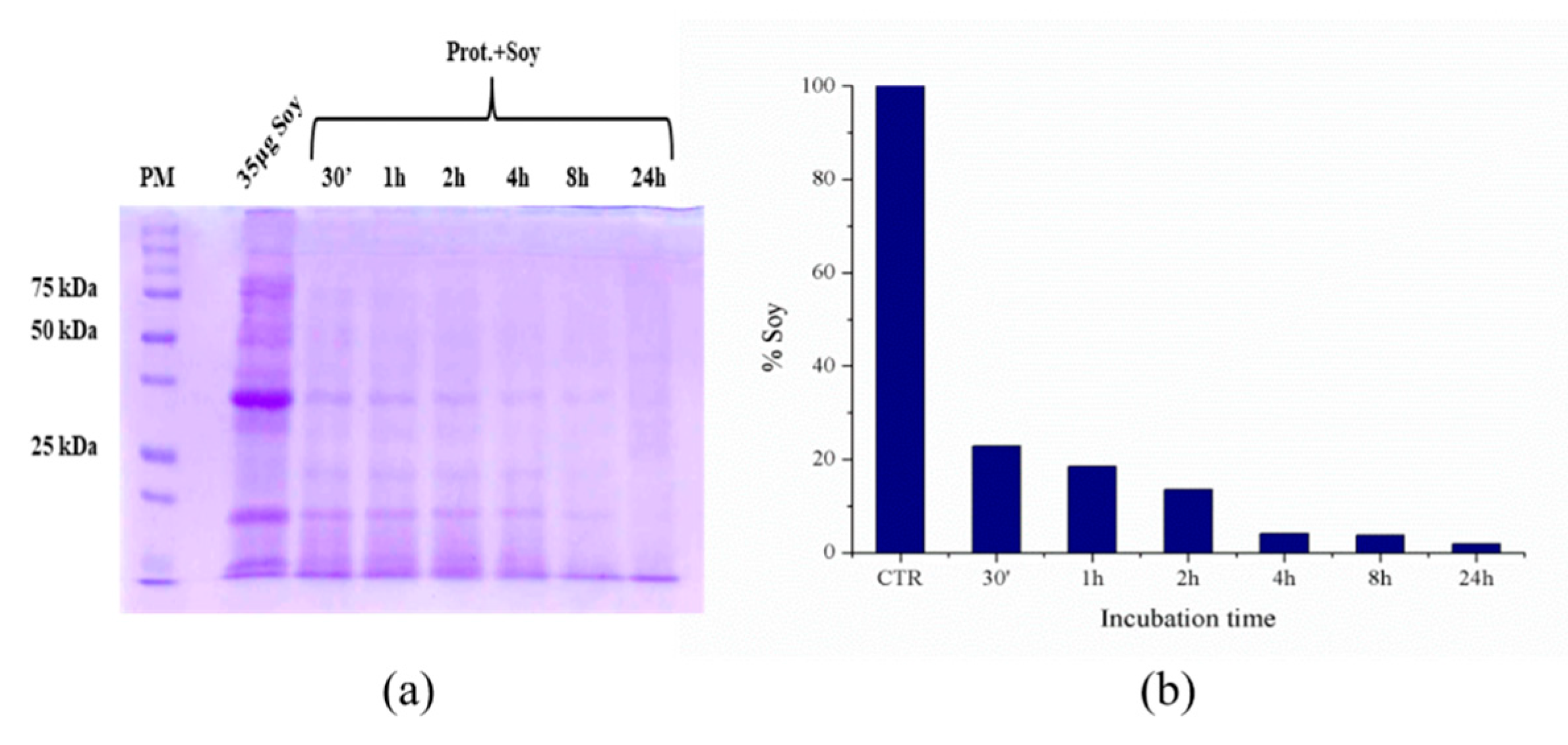
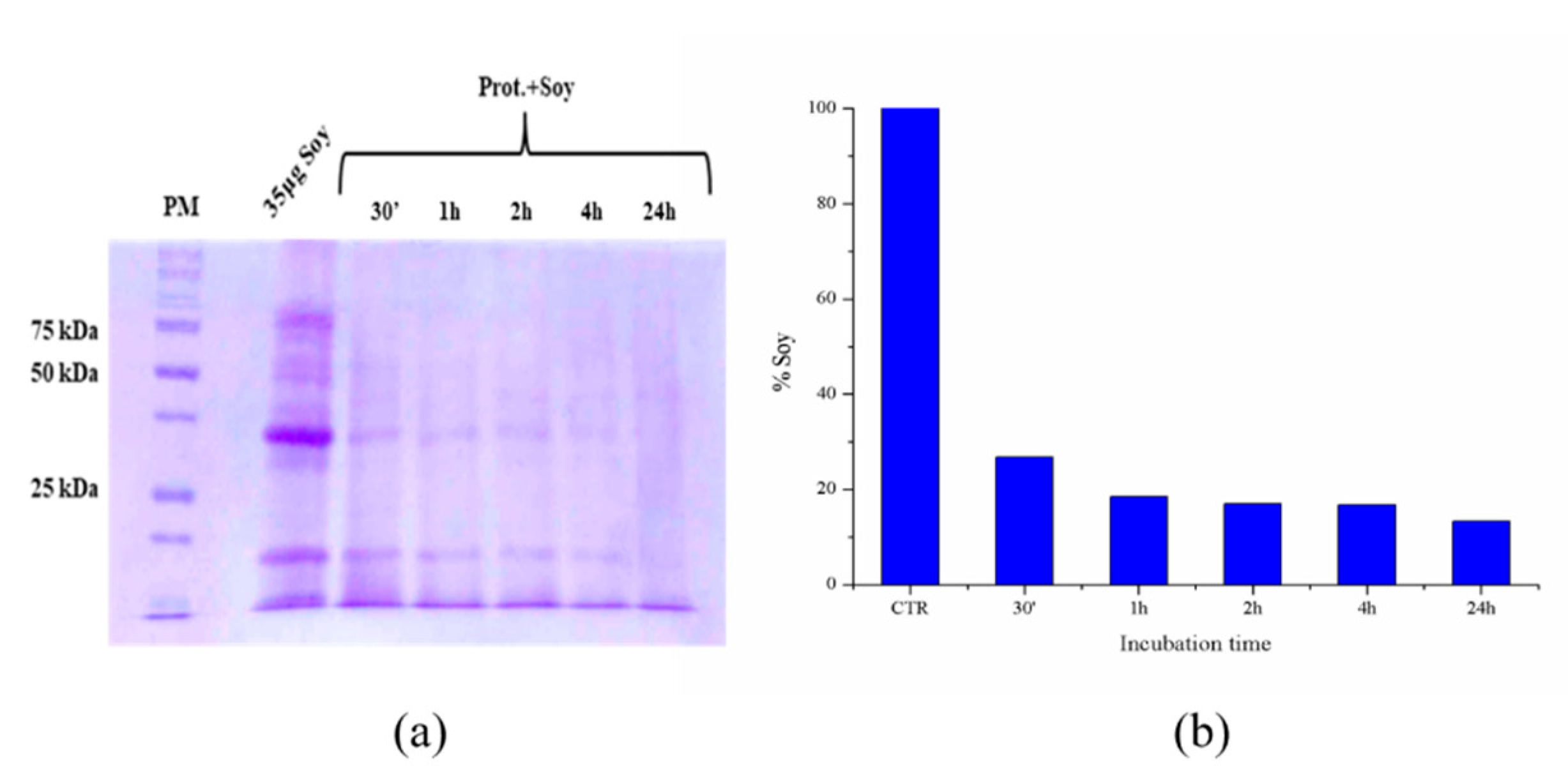
2.6. Hydrolysis of Model Biomasses
2.7. pH-Stat
2.8. Peptide Analysis in the Protein Hydrolysate
3. Materials and Methods
3.1. Materials
3.2. Enzyme Immobilization
3.3. Morphological and Chemical Characterization
3.4. Enzymatic Tests
3.5. Protein Extract from Soybean Waste
3.6. Polyacrylamide Gel Electrophoresis in Denaturing Conditions (SDS-PAGE)
3.7. Coomassie Brilliant Blue Gel Staining
3.8. pH-Stat
3.9. Peptides Extraction and LC/MS Analysis
4. Conclusions
5. Patents
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ravindran, R.; Hassan, S.; Williams, G.; Jaiswal, A. A review on bioconversion of agro-industrial wastes to industrially important enzymes. Bioengineering 2018, 5, 93. [Google Scholar] [CrossRef]
- Milczek, E.M. Commercial applications for enzyme-mediated protein conjugation: New developments in enzymatic processes to deliver functionalized proteins on the commercial scale. Chem. Rev. 2017, 118, 119–141. [Google Scholar] [CrossRef]
- Schmid, A.; Hollmann, F.J.; Park, B.; Bühler, B. The use of enzymes in the chemical industry in Europe. Curr. Opin. Biotechnol. 2002, 13, 359–366. [Google Scholar] [CrossRef]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1988, 62, 597–635. [Google Scholar] [CrossRef]
- Cesaretti, A.; Montegiove, N.; Calzoni, E.; Leonardi, L.; Emiliani, C. Protein hydrolysates: From agricultural waste biomasses to high added-value products. (Minireview) AgroLife Sci. J. 2020, 9, 79–87. [Google Scholar]
- Divakar, G.; Sunitha, M.; Vasu, P.; Ellaiah, P. Optimization of process parameters for alkaline protease production under solid-state fermentation by thermoactinomyces thalpophilus PEE 14. Indian J. Biotechnol. 2006, 5, 80–83. [Google Scholar]
- Gupta, R.; Beg, Q.; Khan, S.; Chauhan, B. An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl. Microbiol. Biotechnol. 2002, 60, 381–395. [Google Scholar] [CrossRef]
- Usharani, B.; Muthuraj, M. Production and characterization of protease enzyme from bacillus laterosporus. Afr. J. Microbiol. Res. 2010, 4, 1057–1063. [Google Scholar] [CrossRef]
- Salihi, A.; Asoodeh, A.; Aliabadian, M. Production and biochemical characterization of an alkaline protease from aspergillus oryzae CH93. Int. J. Biol. Macromol. 2017, 94, 827–835. [Google Scholar] [CrossRef]
- Sawant, R.; Nagendran, S. Protease: An enzyme with multiple industrial applications. World J. Pharm. Sci. 2014, 3, 568–579. [Google Scholar]
- Souza, P.M.D.; Bittencourt, M.L.D.A.; Caprara, C.C.; Freitas, M.D.; Almeida, R.P.C.D.; Silveira, D.; Fonseca, Y.M.; Ferreira Filho, E.X.; Pessoa, A.J.; Magalhães, P.O. A biotechnology perspective of fungal proteases. Braz. J. Microbiol. 2015, 46, 337–346. [Google Scholar] [CrossRef]
- Zambare, V.; Nilegaonkar, S.; Kanekar, P. A novel extracellular protease from pseudomonas aeruginosa MCM B-327: Enzyme production and its partial characterization. New Biotechnol. 2012, 28, 173–181. [Google Scholar] [CrossRef]
- Poldermans, B. Proteolytic enzymes. In Proteolytic enzymes in Industry: Production and Applications; Gerhartz, W., Ed.; VCH: Weinheim, Germany, 1990; pp. 108–123. [Google Scholar]
- Kumar, C.G.; Takagi, H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv. 1999, 17, 561–594. [Google Scholar] [CrossRef]
- Husain, Q. Magnetic nanoparticles as a tool for the immobilization/stabilization of hydrolases and their applications: An overview. Biointerface Res. Appl. Chem. 2016, 6, 1585–1606. [Google Scholar]
- Husain, Q. Nanocarriers immobilized proteases and their industrial applications: An overview. J. Nanosci. Nanotechnol. 2018, 18, 486–499. [Google Scholar] [CrossRef]
- Tavano, O.L. Protein hydrolysis using proteases: An important tool for food biotechnology. J. Mol. Catal. B Enzym. 2013, 90, 1–11. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Wang, J.; Xing, L. Preparation of active corn peptides from zein through double enzymes immobilized with calcium alginate–chitosan beads. Process Biochem. 2014, 49, 1682–1690. [Google Scholar] [CrossRef]
- Zhu, X.; Li, Y.; Yang, G.; Lv, M.; Zhang, L. Covalent immobilization of alkaline proteinase on amino-functionalized magnetic nanoparticles and application in soy protein hydrolysis. Biotechnol. Progress 2019, 35, e2756. [Google Scholar] [CrossRef]
- Ohyama, T.; Minagawa, R.; Ishikawa, S.; Yamamoto, M.; Hung, N.V.P.; Ohtake, N.; Sueyoshi, K.; Sato, T.; Nagumo, Y.; Takahashi, Y. Soybean seed production and nitrogen nutrition. In A Comprehensive Survey of International Soybean Research-Genetics, Physiology, Agronomy and Nitrogen Relationships; James, E.B., Ed.; InTech: London, UK, 2013; pp. 115–157. [Google Scholar] [CrossRef][Green Version]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef]
- Haddad, T.; Noel, S.; Liberelle, B.; El Ayoubi, R.; Ajji, A.; De Crescenzo, G. Fabrication and surface modification of poly lactic acid (PLA) scaffolds with epidermal growth factor for neural tissue engineering. Biomatter 2016, 6, e1231276. [Google Scholar] [CrossRef]
- Gialanella, S.; Girardi, F.; Ischia, G.; Lonardelli, I.; Mattarelli, M.; Montagna, M. On the goethite to hematite phase transformation. J. Therm. Anal. Calorim. 2010, 102, 867–873. [Google Scholar] [CrossRef]
- Barelli, L.; Bidini, G.; Di Michele, A.; Gammaitoni, L.; Mattarelli, M.; Mondi, F.; Sisani, E. Development and validation of a Ni-based catalyst for carbon dioxide dry reforming of methane process coupled to solid oxide fuel cells. Int. J. Hydrogen Energy 2019, 44, 16582–16593. [Google Scholar] [CrossRef]
- Caponi, S.; Liguori, L.; Giugliarelli, A.; Matterelli, M.; Morresi, A.; Sassi, P.; Urbanelli, L.; Musio, C. Raman micro-spectroscopy: A powerful tool for the monitoring of dynamic supramolecular changes in living cells. Biophys. Chem. 2013, 182, 58–63. [Google Scholar] [CrossRef]
- Mattana, S.; Mattarelli, M.; Urbanelli, L.; Sagini, K.; Emiliani, C.; Dalla Serra, M.; Fioretto, D.; Caponi, S. Non-contact mechanical and chemical analysis of single living cells by microspectroscopic techniques. Light Sci. Appl. 2018, 7, 17139. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.T.K.; Koenig, J.L. Raman studies of silane coupling agents. Mater. Sci. Eng. 1975, 20, 145. [Google Scholar] [CrossRef]
- Hiraoui, M.; Guendouz, M.; Lorrain, N.; Moadhen, A.; Haji, L.; Oueslati, M. Spectroscopy studies of functionalized oxidized porous silicon surface for biosensing applications. Mater. Chem. Phys. 2011, 128, 151–156. [Google Scholar] [CrossRef]
- Cazzolli, G.; Caponi, S.; Defant, A.; Gambi, C.M.C.; Marchetti, S.; Mattarelli, M.M.; Rossi, B.; Rossi, F.; Viliani, G. Aggregation processes in micellar solutions: A Raman study. J. Raman Spectrosc. 2012, 43, 1877–1883. [Google Scholar] [CrossRef]
- Sassi, P.; Caponi, S.; Ricci, M.; Morresi, A.; Oldenhof, H.; Wolkers, W.F.; Fioretto, D. Infrared versus light scattering techniques to monitor the gel to liquid crystal phase transition in lipid membranes. J. Raman Spectrosc. 2015, 46, 644–651. [Google Scholar] [CrossRef]
- Karami, Z.; Akbari-adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Chen, H.M.; Muramoto, K.; Yamauchi, F. Structural analysis of antioxidative peptides from Soybean. beta.-Conglycinin. J. Agric. Food Chem. 1995, 43, 574–578. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Malmo, C.; Vilasi, S.; Iannuzzi, C.; Tacchi, S.; Cametti, C.; Irace, G.; Sirangelo, I. Tetracycline inhibits W7FW14F apomyoglobin fibril extension and keeps the amyloid protein in a pre-fibrillar, highly cytotoxic state. FASEB J. 2005, 19, 346. [Google Scholar] [CrossRef]
- Riga, A.; Zhang, J.; Collis, J. Characterization of drawn and undrawn poly-L-lactide films by differential scanning calorimetry. J. Therm. Anal. Calorim. 2004, 75, 257–268. [Google Scholar] [CrossRef]
- Scarponi, F.; Mattana, S.; Corezzi, S.; Caponi, S.; Comez, L.; Sassi, P.; Morresi, A.; Paolantoni, M.; Urbanelli, L.; Emiliani, C.; et al. High-performance versatile setup for simultaneous Brillouin-Raman microspectroscopy. Phys. Rev. X 2017, 7, 031015. [Google Scholar] [CrossRef]
- Anson, M.L. The estimation of pepsin, trypsin, papain, and cathepsin with hemoglobin. J. Gen. Physiol. 1938, 22, 79. [Google Scholar] [CrossRef]
- Folin, O.; Ciocalteau, V. On tyrosine and tryptophane determinations in proteins. J. Biol. Chem. 1927, 73, 627–660. [Google Scholar] [CrossRef]
- Jang, H.M.; Cho, H.U.; Park, S.K.; Ha, J.H.; Park, J.M. Influence of thermophilic aerobic digestion as a sludge pre-treatment and solids retention time of mesophilic anaerobic digestion on the methane production, sludge digestion and microbial communities in a sequential digestion process. Water Res. 2014, 48, 1–14. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Adler-Nissen, J. Limited enzymic degradation of proteins: A new approach in the industrial application of hydrolases. J. Chem. Technol. Biotechnol. 1982, 32, 138–156. [Google Scholar] [CrossRef]
- Jacobsen, C.F.; LBonis, J.; Linderstrem-Lang, K.; Ottesen, M. Methods of Biochemical Analysis; Glick, D., Ed.; Interscience Publisher Inc.: New York, NY, USA, 1957; Volume 4, pp. 187–202. [Google Scholar]
- Corte, L.; Dell’Abate, M.T.; Magini, A.; Migliore, M.; Felici, B.; Roscini, L.; Sardella, R.; Tancini, B.; Emiliani, C.; Cardinali, G.; et al. Assessment of safety and efficiency of nitrogen organic fertilizers from animal-based protein hydrolysates—A laboratory multidisciplinary approach. J. Sci. Food Agric. 2014, 94, 235–245. [Google Scholar] [CrossRef]




| Formulations | First Heating | |||||||
|---|---|---|---|---|---|---|---|---|
| Tg (°C) | ΔHcc1 (J/g) | Tcc1 (°C) | ΔHcc2 (J/g) | Tcc2 (°C) | ΔHm (J/g) | Tm (°C) | Xm (%) | |
| PLA | 62.3 | 2.9 | 94.2 | 1.5 | 160.5 | 40.7 | 180.3 | 39.1 |
| activated PLA | - | - | - | - | - | 43.3 | 150.4/177.2 | 46.5 |
| activated PLA with immobilized protease | - | - | - | - | - | 42.2 | 149.6/177.6 | 45.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calzoni, E.; Cesaretti, A.; Tacchi, S.; Caponi, S.; Pellegrino, R.M.; Luzi, F.; Cottone, F.; Fioretto, D.; Emiliani, C.; Di Michele, A. Covalent Immobilization of Proteases on Polylactic Acid for Proteins Hydrolysis and Waste Biomass Protein Content Valorization. Catalysts 2021, 11, 167. https://doi.org/10.3390/catal11020167
Calzoni E, Cesaretti A, Tacchi S, Caponi S, Pellegrino RM, Luzi F, Cottone F, Fioretto D, Emiliani C, Di Michele A. Covalent Immobilization of Proteases on Polylactic Acid for Proteins Hydrolysis and Waste Biomass Protein Content Valorization. Catalysts. 2021; 11(2):167. https://doi.org/10.3390/catal11020167
Chicago/Turabian StyleCalzoni, Eleonora, Alessio Cesaretti, Silvia Tacchi, Silvia Caponi, Roberto Maria Pellegrino, Francesca Luzi, Francesco Cottone, Daniele Fioretto, Carla Emiliani, and Alessandro Di Michele. 2021. "Covalent Immobilization of Proteases on Polylactic Acid for Proteins Hydrolysis and Waste Biomass Protein Content Valorization" Catalysts 11, no. 2: 167. https://doi.org/10.3390/catal11020167
APA StyleCalzoni, E., Cesaretti, A., Tacchi, S., Caponi, S., Pellegrino, R. M., Luzi, F., Cottone, F., Fioretto, D., Emiliani, C., & Di Michele, A. (2021). Covalent Immobilization of Proteases on Polylactic Acid for Proteins Hydrolysis and Waste Biomass Protein Content Valorization. Catalysts, 11(2), 167. https://doi.org/10.3390/catal11020167











