Abstract
Although traumatic brain injury (TBI) is a global public health issue, not all injuries necessitate additional hospitalisation. Thinking, memory, attention, personality, and movement can all be negatively impacted by TBI. However, only a small proportion of nonsevere TBIs necessitate prolonged observation. Clinicians would benefit from an electroencephalography (EEG)-based computational intelligence model for outcome prediction by having access to an evidence-based analysis that would allow them to securely discharge patients who are at minimal risk of TBI-related mortality. Despite the increasing popularity of EEG-based deep learning research to create predictive models with breakthrough performance, particularly in epilepsy prediction, its use in clinical decision making for the diagnosis and prognosis of TBI has not been as widely exploited. Therefore, utilising 60s segments of unprocessed resting-state EEG data as input, we suggest a long short-term memory (LSTM) network that can distinguish between improved and unimproved outcomes in moderate TBI patients. Complex feature extraction and selection are avoided in this architecture. The experimental results show that, with a classification accuracy of 87.50 ± 0.05%, the proposed prognostic model outperforms three related works. The results suggest that the proposed methodology is an efficient and reliable strategy to assist clinicians in creating an automated tool for predicting treatment outcomes from EEG signals.
1. Introduction
Traumatic brain injury (TBI) is one of the most common and costly health and socioeconomic problems worldwide. The incidence of TBI is higher than those of complex diseases such as breast cancer, AIDS, multiple sclerosis, and Parkinson’s disease, such that TBI is considered the leading cause of mortality and disability in persons under 45 years of age. In 2019, there were approximately 223,135 TBI-related hospitalizations and 64,362 TBI-related deaths [1]. Previous studies from the United States [2] and New Zealand [3] estimate approximately 500–800 new cases of TBI per 100,000 people per year. However, there are few estimates of the burden of TBI in low- and middle-income countries (LMICs) [4]. Outcomes for patients who sustain a TBI are highly variable, making it difficult for providers to accurately predict their prognosis and long-term quality of life. The high incidence of TBI and uncertain outcomes in these cases highlight the need to develop better tools to support difficult decision making.
In clinical practice, TBI is classified as mild, moderate, or severe based on clinical guidelines. The Glasgow Coma Scales (GCS) is a standard neurologic scoring system used to assess the severity of patients with severe injuries admitted to the emergency department [5,6] The GCS is a 15-point behavioral observation scale that defines severity based on eye, verbal, and motor responses [7]. Individuals with a score of 3–8 are classified as severe, 9–13 as moderate, and those between 14 and 15 as mild TBI [8,9]. Mild TBIs are defined as temporary changes in consciousness resulting from an external force applied to the skull and account for more than 70% of reported brain injuries, including concussions [10]. In most people, symptoms resolve completely within 3 months of a mild TBI, despite evidence of chronic pathologic changes in brain tissue [10]. Unlike mild traumatic brain injury, severe TBI is associated with persistent loss of consciousness (>24 h) and a mortality rate of 24%, with 43% of surviving patients having chronic physical and emotional disabilities [11,12].
Previous studies of moderate TBI have found that about 60% of patients had an intracranial head injury on admission as detected by computed tomography (CT) [13,14], 20–84% were admitted to an intensive care unit (ICU) [13,15], and about 15% underwent surgery for a mass lesion or skull fracture [14], and the mortality rate was low (0.9–8%) [14,15]. Moreover, the vast majority of patients were only moderately or not at all disabled, indicating independence in daily life (74–85%) [15,16], and many even recovered well, indicating no disability (55–75%) [13,14,15]. Although the prognosis is better in moderate than in severe TBI, early and accurate prognosis using a predictive model is critical so that patients’ families can make an informed decision about whether to discontinue life support or continue supportive care.
Despite advances in prognostication management of TBI (i.e., inpatient rehabilitation services), about 50% of people with moderate TBI will either experience further deterioration in their daily lives or die within five years after their injury. According to the TBI Model System (TBIMS) National Database [17], 22% of TBI patients died, 30% deteriorated, 22% remained in the same condition, and only 26% of survivors experienced full neurological recovery. Even years after recovery, hardly any formerly employed patients return to full-time employment. Despite regaining full consciousness, patients with moderate TBI continue to have difficulty reintegrating into society [18]. In Malaysia, TBI due to road traffic accidents is the fourth leading cause of death among young people (15–40 years; 16.8%) and children (0–14 years; 3.0%) [19], and the sixth leading cause (7.20%) of hospitalization in Malaysian government and private hospitals, due to injuries and other consequences of external causes [20].
TBI is a disruption of brain function caused by a blow to the head [21]. TBI negatively leads to a broad spectrum of structural and functional brain injuries, especially electrophysiologic changes, because of both missing and interrupted brain circuits and alterations to surviving structures. The prediction of outcome for patients with TBI is a complicated but important task during early hospitalization. Some clinical predictors have been proposed and predictive models have been developed [22,23,24,25]. Most studies have used traditional regression techniques to determine these characteristics [26,27]. Severity, diagnosis, and outcome level, as measured by the Glasgow Outcome Scale (GOS) [28], were defined as the most relevant clinical indicators for predicting the outcome of TBI, despite a poor implementation in clinical practice. In this context, great efforts have been made in recent years to implement and develop artificial intelligence tools. Clinicians would benefit significantly from an electroencephalography (EEG)-based artificial intelligence model for outcome prediction, as this would give them access to evidence-based analysis that would allow them to safely discharge patients who are at minimal risk of TBI-related mortality.
The machine learning (ML) technique, particularly supervised ML, has also been used to predict clinical outcomes after TBI. According to research by Haveman et al. [29], random forest (RF), which uses clinical variables and quantitative EEG predictors to predict moderate to severe TBI outcomes, has an area under the receiver operating characteristic curve (AUC) of 0.94–0.81. van den Brink et al. [30] used linear discriminant analysis (LDA) to predict each patient’s chances of recovery, based on spectral amplitude and connectivity features, with an accuracy of 75%. In our previous work [31,32], a prediction model for the recovery of TBI patients was developed based on the RUSBoost model to cope with the highly imbalanced EEG dataset. The RUSBoost model performed best in discriminating between two outcomes, with an AUC of 0.97–0.95. Noor and Ibrahim [33], in a review, supported the use of quantitative EEG predictors and ML models to predict the recovery outcomes of moderate to severe TBI patient groups. The review in [33] showed that the performance of traditional ML algorithms, including support vector machine (SVM), RF, and boosting algorithms (i.e., Adaboost, RUSBoost), when combined with signal processing techniques (short time Fourier transform (STFT), power spectral density (PSD), coherence, and connectivity), requires sufficient hand-crafted features to train ML classifiers to achieve satisfactory classification accuracy.
Recent developments in ML and the availability of huge EEG datasets have driven the development of deep learning architectures, especially for the analysis of EEG signals. The robust and reliable classification of EEG signals is a critical step in making the use of EEG more widely applicable and less dependent on experienced experts. EEG is an electrophysiological monitoring method that records the electrical activity of the brain. The method is usually noninvasive, as the electrodes are placed on the patient’s scalp; it has a high temporal range, and it is relatively inexpensive [21]. Because of these advantages, EEG has become widely used in neuroscience, including brain–computer interface (BCI) [34], sleep analysis [35,36,37], and seizure detection [38,39,40,41]. As pointed out in [42], despite the exponential increase in the number of publications using EEG and DL, EEG is not as widely used for TBI diagnosis and prognosis.
To address this gap, we propose a binary EEG-based deep learning approach (i.e., long short-term memory (LSTM)) that can discriminate between improved and unimproved outcomes in patients with moderate TBI by using raw resting EEG data as input to the model. Our research aims to develop an easy-to-use deep learning prognostic model (LSTM-based network) as a tool for the early prediction of outcomes in moderate TBI, supporting clinical decision making (clinical care of patients). The existing knowledge gap is a significant barrier to improving clinical care for patients with moderate TBI. To our knowledge, the potential of deep learning LSTM-based EEG networks specifically for predicting outcomes in moderate TBI has not been explored. Despite the high temporal resolution of EEG, modern techniques ignore the temporal dependence of the signal. The LSTM network [43] was developed in order to solve the problem of long-term dependence of the recurrent neural network (RNN), which is primarily used to predict time series data, and is therefore perfectly suited to discovering the temporal aspects of EEG. To evaluate the performance of the proposed prediction model, it is compared with three other similar works on predicting TBI outcomes from EEG signals.
2. Methodology
2.1. Participants
EEG recordings for this study were obtained from 14 male nonsurgical moderate TBI patients recruited from Universiti Sains Malaysia Hospital in Kubang Kerian, Kelantan. The study procedure was approved by the Universiti Sains Malaysia Human Research Ethics Committee under approval number USM/JEPeM/1511045. Inclusion criteria were patients aged 18 to 65 years with moderate TBI (Glasgow Coma Scale (GCS) of 9 to 13) due to traffic accidents [44]. The initial impact involved the left or right brain, as confirmed by CT scan in the emergency department. Exclusion criteria included injuries resulting in severe scalp and skull abnormalities, bone fractures, and illicit drug use that could interfere with research compliance. Each participant signed an informed consent form before the start of the experiment. The consent form briefly described the purpose of the project, and side effects and inconveniences associated with the procedures.
2.2. Patients’ Outcome Assessment
Clinicians assessed patient recovery by telephone call between 4 weeks, 6 months, and 1 year after injury. The GOS [45] was used as the primary outcome, and the results were divided into good (GOS score of 5) and poor outcomes (GOS score of 1–4) approximately 12 months after injury. In this study, an expert (i.e., a neurosurgeon) from our team evaluated the neurological outcomes of patients with moderate TBI using the GOS scores (see Table 1) corresponding to the specific degree of improvement of each patient.

Table 1.
Description of GOS score for outcome judgment.
2.3. EEG Data Acquisition
Stimulus-free EEG was performed entirely in the EEG laboratory. The patients were instructed to relax in a quiet, semidark environment, sit comfortably, and close their eyes while their resting-state EEG data were recorded for approximately 5 min and 50 s. Real-time EEG data were acquired throughout using a 64-channel EEG cap (ANT Neuro, Enschede, The Netherlands). All 64 gel sensors were attached to the EEG head helmet, referenced to the connected mastoids (M1–M2), and grounded at 10% anterior Fz (see Figure 1). Impedance was kept below 10 kΩ. A SynAmps programmable direct current (DC) broadband amplifier with gain up to 2500 and accuracy of 0.033/bit was used to record brain signals at 55 millivolt (mV) at DC 70 Hz. The EEG signals were digitized to 1 kHz using 16-bit AC converters (AC)–DC. The CPz channel was configured as an electrooculography (EOG) channel to capture gaze and blink artifacts. Therefore, only 63 EEG channels were used to record brain signals and served as input data for our prediction model.
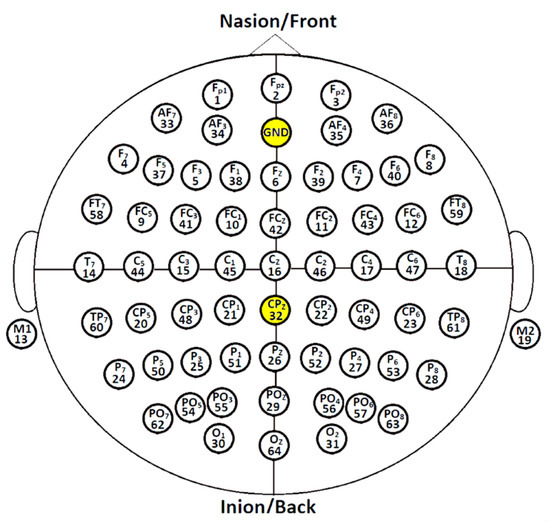
Figure 1.
Topographical placement of 64 gel-based electrodes using the international system 10–20 configuration.
2.4. EEG Dataset Preparation
Continuous EEG data were collected from patients with moderate TBI who underwent follow-up. The first measurement (4 to 10 weeks after the accident) provided eighteen moderate TBI data. The second measurement (i.e., six months after the accident) provided eleven EEG data. The third measurement (one year after the accident) contributed to three EEG data points. Patients who did not participate in the follow-up EEG measurements within the specified time period were disqualified. The raw, unfiltered EEG data were exported for future studies. In total, 32 EEG recordings were obtained from the 14 patients.
2.5. EEG Data Processing and Input EEG Signal Representation
To eliminate transient artifacts due to contamination at the beginning of data collection, the first 60 s of data points were removed. It was found that patients were less comfortable during the initial phase of recording. The remaining data were neither re-referenced nor further filtered or processed to remove artifacts due to line noise, eye blinks and movements, or electromyogram (EMG) contamination. Resting-state EEG data were acquired in raw format and converted to Matlab (.mat) format for further processing and analysis using EEGLAB [48]. The EEG signals were downsampled from 1000 Hz to 100 Hz using the EEGLAB function pop_resample() (using the integer factor D of 10), because processing signals with excessive temporal resolution resulted in an additional and perhaps not useful computational burden [49,50,51]. Therefore, the following 60-second sets (60 one-second segments) of synchronous segment were extracted from each of the 63-channel time series comprising each subject’s raw resting-state EEG dataset. The input signals were arranged as a matrix M (i.e., channel amplitude versus time) [52], as shown in Equation (1):
where k is the number of data samples and j is the number of EEG channels. In this experiment, and k = (60 s × 100 data points/s = 6000 data points). Therefore, a 63 × 6000 matrix represented one EEG of each subject. After the data processing phase, each column of the matrix (i.e., 63 × 6000) was fed as an input feature to the LSTM module, which consisted of LSTM cells as shown in Figure 2.
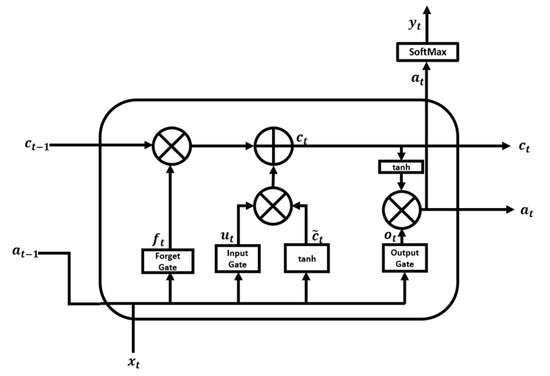
Figure 2.
LSTM cell structure.
2.6. Long Short-Term Memory
LSTM [43] is a variation of recurrent neural network (RNN) that overcomes the difficulties of gradient vanishing and exploding. An RNN uses past knowledge to predict output and captures a temporal correlation between the previous state and the current input during training. Due to its short memory, an RNN cannot recover past information for long time series. During backpropagation, the gradient is reduced to such a minimal value that no parameters are appreciably changed, so that RNNs cannot learn from the data. Details on RNN networks can be found in Salehinejad et al. [53]. In the present work, LSTM networks were used to address the vanishing gradient problem and allow our model to learn long sequences. The distinguishing feature of LSTMs that makes them a better choice than RNNs is their internal architecture, which is shown in Figure 2.
An LSTM cell consists of four gates: the forget gate , the input gate , the cell candidate gate , and the output gate . The LSTM essentially consists of four stages. The first stage is the forget gate layer, represented by Equation (2). In this stage, the sigmoid layer decides which information to forget.
where is the input vector of the LSTM unit, is the previously hidden state vector that can be considered as the output vector of the previous LSTM unit, denotes the weight matrices, denotes the bias vector parameters for the forget layer that need to be optimized during model training, and is a sigmoid function that returns values (y-axis) between 0 and 1. Next, the LSTM unit chooses between 0 and 1 to forget.
The second stage is the input gate of Equation (3) and the tanh layer of Equation (4). The sigmoid layer selects which values to update, and the tanh layer generates a new candidate value for , a cell input activation vector. Finally, the values from the two layers are combined and appended to the cell state. The sigmoid function is used as the activation function for gate input, and the hyperbolic tanh function is used as the activation function for block input and output.
The third step is to create a new cell state by updating the previous state, as shown in Equation (5). First, the information selected for deletion by the forget gate is discarded, and then the information to be added is appended.
The final step is to decide which value to output, using Equation (6) with the output gate layer. The output gate layer determines which part of the cell state should be exported via sigmoid for the input data, and then the generated value is multiplied by the cell state via the tanh layer, as indicated in Equation (7).
The architecture of the LSTM network used in this work is shown in Figure 3. The proposed LSTM network was fed with a 63 × 6000 matrix representing 60 s of raw EEG recording (i.e., 1 s recording = 100 data points). For each time step (i.e., 1 s), 100 data points were passed to the LSTM.
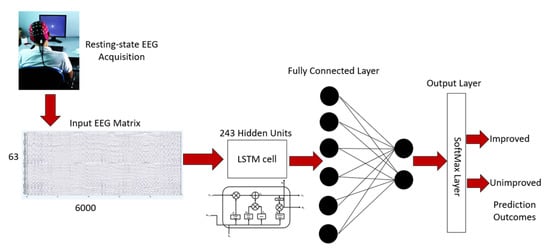
Figure 3.
Moderate TBI outcome prediction architecture.
The LSTM was trained for 60 time steps (i.e., 60 s of EEG recordings). The LSTM was set to contain 243 hidden units followed by a dropout layer, a fully connected layer, and a classification layer, namely SoftMax, to classify the 2 classes of output as shown in Figure 3. The number of neurons in the SoftMax layer represented the number of classes (i.e., improved vs. unimproved). In addition, the model was regularized by applying a dropout layer to avoid overfitting the model by a factor of 50% [54].
Five hyperparameters were kept constant throughout the training of the proposed LSTM architecture. The parameters and their values are listed in Table 2. The learning rate was set to 0.001 followed by a minibatch size of 3, L2 regularization was set to 0.0005 to prevent overfitting, and there were 30 training repetitions per epoch. The training iteration was set to a modest value to avoid overfitting the network with a higher training iteration. On the other hand, if each iteration was too short, there might not be enough training repetitions to properly feed the network with training data. Adaptive Moment Estimation (Adam) was used as an optimizer because it was appropriately suited for a learning rate of 0.001 for better accuracy. The NVIDIA GeForce GTX 1060 6GB GPU was used to train the LSTM model.

Table 2.
Network training parameters and values.
2.7. Data Augmentation Approach for Imbalanced Dataset
A data augmentation technique was applied to the training set to minimize overfitting [55]. Our dataset included EEGs from eight patients with unimproved and 24 patients with improved group outcomes. One underrepresented class was a positive class (i.e., unimproved) and was of primary interest from a learning standpoint. The other class, which was abundant, was designated a negative class (i.e., improved), and these two groups constituted the entire dataset. The ratio between the two classes is referred to as the imbalanced ratio (IR). The dataset had an imbalanced ratio of 1:3 (unimproved: improved outcomes). The performance of the classifier would be affected by the class imbalance. During training, learning algorithms that are insensitive to the class imbalance could classify all samples into the majority class to minimize the error rate [56].
To overcome the problem of the unbalanced dataset, a data augmentation method (a random oversampling method) was performed, in which the positive class (unimproved) was randomly replicated to match the negative class (improved). Finally, the amounts of unimproved and improved data in the training set were balanced. Other oversampling methods, such as the synthetic minority oversampling technique (SMOTE) and the adaptive synthetic approach (ADASYN) [57,58], work by generating synthetic samples of the majority class instead of oversampling with replacement. Both of these algorithms generate artificial instances based on features rather than the data space [59]. In the case of EEG data, both oversampling methods are best suited for EEG data features [60]. Since the actual EEG signal was used as input, it was not possible to generate synthetic EEG epochs. Therefore, the negative class was duplicated to be numerically equal to the majority class.
2.8. Training Procedure and Performance Evaluation for Imbalanced Dataset
Although deep learning has achieved great success in many research applications, there are very few approaches for classifying imbalanced EEG data, as mentioned earlier. Directly applying deep learning models to an imbalanced dataset usually results in poor classification performance [61]. To address this problem, we propose to extend the LSTM network algorithm with a bootstrapping method that incorporates oversampling with decision fusion to improve the performance of LSTM in predicting TBI outcomes with imbalanced dataset distributions. To evaluate the performance of the proposed model for TBI outcomes, the bootstrap method was performed with 3-fold cross-validation to test its robustness for the classification of unseen data.
Due to the limited size of the dataset, we applied an augmentation technique (i.e., the bootstrap method [62]) to the training data to maximize classification accuracy and ensure minimal overfitting. A bootstrap method is a resampling method that generates bootstrap samples to quantify the uncertainties associated with an ML method despite a small dataset. It can estimate the performance of a proposed ML architecture for a small dataset by providing a percentile confidence interval (CI) of the performance measures (i.e., classification accuracy and precision). Bootstrap resampling allows the user to mimic the process of acquiring a new dataset to estimate the performance of the proposed design without creating new samples.
In this work, bootstrap resampling was performed for all 48 subjects after the EEG dataset was balanced using random oversampling, and the resampling sample size was fixed at 48. Efron [63] suggested that a minimum of 50 to 1000 iterations of the bootstrap sample set could provide valid percentile intervals to achieve the optimal performance of the proposed architectures. Therefore, to obtain the optimal parameters of the proposed LSTM network, 300 iterations of the resampled bootstrap sample were used.
In k-fold cross-validation, the EEG data segments were divided into k-folds, trained with , and tested with the rest. The average result value obtained by repeating this process k times served as the verification result of the model. For each bootstrap sample, 3-fold cross-validation was performed, and the accuracy of the cross-validation was recorded. The mean, standard deviation (SD), and 95% CI were determined from the 300 cross-validation accuracies recorded.
In this context, widely used classification performance metrics for imbalanced datasets, according to Kaur et al. [64], were assigned in this work, represented by Equations (8)–(13) based on training and testing data.
3. Results and Discussion
Table 3 recapitulates our experimental results. To validate the performance of the proposed prediction model, it was compared with three similar works on predicting TBI outcomes from the literature. We used reliable performance metrics such as accuracy, sensitivity, specificity, G-mean, and F1 score, and included the percentage of error in the classification results. The proposed raw LSTM model was reliable with an average validation accuracy of 87.50% ± 0.05 and had a minimum classification error of 12.50%, outperforming previous studies in terms of accuracy, as shown in Table 4. The results showed a sensitivity of 91.65%, a specificity of 87.50%, and an F1 score of 87.50%. These results indicate that our proposed approach is good at predicting and discriminating between unimproved and improved outcomes in moderate TBI. The G-mean value of 87.50% showed balanced classification performance for both the negative class (i.e., improved outcomes) and the positive class (i.e., unimproved outcomes). The highest percentage of G-mean showed good performance in classifying the positive class, even when the negative class samples were correctly classified. The percentage of G-mean is important to avoid overfitting the negative class and underfitting the positive class [57,65].

Table 3.
Performance metrics computed from the results of the proposed model (raw-LSTM) for predicting moderate TBI outcomes.
We believe that our proposed model is better than other models for several reasons. First, it can handle imbalanced data well using random oversampling. Second, the LSTM block efficiently extracts relevant features during feature refinement. Finally, bootstrapping is helpful because the prediction outcomes of the trained deep learning model using a sample set of bootstraps always have a Gaussian distribution, and the 95% CI is necessary to determine the accuracy and stability of our proposed prediction model.
The limitation of a small dataset has been overcome by incorporating oversampling and bootstrapping approaches to improve the model performance of the imbalanced EEG dataset. In neuroinformatics applications, a limited dataset often becomes a problem when unexpected constraints, such as a small patient population, arise. A small dataset could lead to optimistic biases in classifier evaluation, resulting in inaccurate performance estimations. Data augmentation methods normally used in image classification are not appropriate for classifying EEG data.
Augmentation of time-series EEG data from subjects with moderate TBI may introduce random noise that can increase the chance of classification errors [66]. Li et al. [67] believed that direct geometric transformation and introduction of noise may destroy the feature in the time domain, which may have a negative effect on data augmentation. Therefore, they performed a short-time Fourier transform (STFT) to transform time-series EEG signals into spectral images; this is known as amplitude perturbation data augmentation. Lee et al. [68] demonstrated the effectiveness of the data augmentation method using the borderline synthetic minority oversampling technique (borderline-SMOTE) on EEG data from the P300 task. The results showed that the proposed methods can increase the robustness of decision boundaries to improve the classification accuracy of P300 based on the brain–computer interface (BCI). We believe that the data augmentation method (bootstrapping and oversampling) introduced to the LSTM network has the advantages of interpretability and lower computational cost.
The results of the comparison between our proposed method and other models for predicting TBI outcomes are shown in Table 4. Chennu et al. [69] developed a prediction model using a support vector machine (SVM) as a classifier to discriminate between positive and negative recovery outcomes. Schorr et al. [70] determined the predictive power using a receiver operating characteristic curve (ROC) calculated from a multivariate autoregression analysis of difference coherence between multiple patient groups (i.e., minimally unconscious syndrome versus unresponsive wakefulness syndrome; unimproved versus improved; traumatic versus nontraumatic). The developed prognostic model yielded an accuracy of 78.03%, suggesting that coherence analysis may be a useful prognostic tool for predicting recovery outcomes in the future. Lee et al. [71] developed a logistic regression model to calculate the percentage of correct or incorrect predictions according to a dichotomized output of the model.

Table 4.
Comparison with previous studies.
Table 4.
Comparison with previous studies.
| Architecture | Accuracy ± SD | [CI] |
|---|---|---|
| Support Vector Machine (SVM); | 81.98 ± 5.13 | [80.69, 83.27] |
| Chennu et al. [69] | ||
| Multivariate Auto Regression (MVAR); | 78.03 ± 21.07 | [73.29, 82.77] |
| Schorr et al. [70] | ||
| Logistic Regression (LR); | 49.97 ± 2.51 | [49.56, 50.37] |
| Lee et al. [71] | ||
| Proposed Raw-LSTM | 87.50 ± 0.05 | [87.12, 88.34] |
Table 4 shows that the SVM performed comparably (i.e., with an accuracy of 81.98%). However, to ensure such high performance, preprocessing and feature extraction must be carefully performed to ensure that high quality and discriminative features can be extracted. On the other hand, the MVAR model has a relatively modest classification accuracy of 78.03%, and the logistic regression model shows the lowest classification accuracy of 49.97%. Compared to these works, our proposed approach (i.e., raw-LSTM) shows competitive performance and provides the highest accuracy of 87.50% with the lowest error rate of 12.50%.
To the best of our knowledge, no existing binary classification approach uses deep learning to predict moderate TBI outcomes. Therefore, the proposed approach using LSTM would be beneficial to overcome the limitations of ML approaches (i.e., SVM, MVAR, and logistic regression). These MLs are very sensitive to noise and artifacts, which can easily affect the training of the classifier and lead to incorrect classification. The conventional ML model also ignores the dependency of EEG channels and assumes that individual features are not correlated. Therefore, more effort must be expended to eliminate the noise and artifacts in order to successfully train the classifier. The main advantage of an LSTM-based network is its ability to detect temporal correlations between channels, which can ensure robust LSTM training. This is the first case of a high-performing, less time-consuming EEG-based platform for automatic classification of TBI outcomes that relies only on easily obtained raw resting-state EEG data.
Furthermore, our results provide preliminary evidence of the importance of developing a prognostic model for initial clinical management in the moderate TBI patient group. This is because patients with moderate TBI, who are in a dangerous situation and at high risk of neurological deterioration (ND), are usually associated with a poor prognosis [9,15,72]. Despite the possibility of deterioration and the need for critical care, a substantial proportion of patients with moderate TBI are treated in nontrauma centers.
It should be noted here that, according to Watanitanon et al. [57], some TBI patients with a GCS of 13, who are typically considered to be at lower risk, are in fact still at risk for serious adverse outcomes, and that there are many patients with moderate TBI who may also benefit from treatment for severe TBI if their condition worsens [73]. We believe that the high discriminative ability of our proposed model demonstrates its potential for classifying the outcomes of moderate TBI by risk of ND using easily obtained, task-free raw EEG data. This was not explored in the current work but could be explored in future work. ND usually leads to a fatal outcome or severe disability and is a prognostic factor associated with an unfavorable outcome. If patients with moderate TBI who are at high risk for ND can be identified in advance, then they can be provided with a high level of monitoring and treatment, including neurologic assessment and intensive care.
4. Conclusions
A novel deep learning approach for predicting moderate TBI outcomes based on EEG signal recordings was proposed. The prediction model uses an LSTM-based network to accurately discriminate EEG signals for unimproved and improved recovery outcomes. The proposed model works directly with raw EEG signals, without requiring feature extraction. The use of oversampling methods to overcome the imbalanced EEG dataset and the inclusion of a bootstrapping method with 300 iterations improved the system’s performance, resulting in the highest accuracy of 87.50%. To test the classification accuracy of the system and ensure its robustness and stability, 3-fold cross-validation was used. A comparison between our proposed approach and previous studies using ML techniques shows that the proposed system outperforms previous work in terms of classification accuracy, even though it uses raw EEG signals. The proposed system takes advantage of an LSTM-based network, which is ideal for processing sequential data points such as EEG time series data. This also proves that deep learning networks form a robust classifier for EEG signals that outperforms traditional learning techniques. The present model also provides a solid foundation for future work on predicting TBI outcomes, and its high performance suggests that this method has potential for clinical application. Future work may focus on improving LSTM networks with an attention mechanism in a transformer LSTM, using an encoder–decoder LSTM model for predicting moderate TBI outcomes. Finally, the application of novel deep learning feature fusion techniques to predict TBI outcomes based on EEG signals may be worth mentioning as one of the directions for future work.
Author Contributions
N.S.E.M.N., C.Q.L. and H.I. contributed to the main idea and the methodology of the research. N.S.E.M.N. designed the experiment, performed the simulations and wrote the original manuscript. H.I. contributed significantly in improving the technical and grammatical content of the manuscript. C.Q.L., H.I. and J.M.A. contributed in terms of supervision and project funding. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Higher Education (MoHE), Malaysia, via the Trans-disciplinary Research Grant Scheme (TRGS) with grant number TRGS/1/2015/USM/01/6/2 and in part by MoHE through Skim Latihan Akademik Bumiputera (SLAB).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Human Research Ethics Committee, Universiti Sains Malaysia (USM), with an approval number USM/JEPeM/1511045.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Resting-state electroencephalography (EEG) signals with eyes closed supporting the results of this study were restricted by the Universiti Sains Malaysia (USM) Human Research Ethics Committee to protect patient privacy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/traumaticbraininjury/get_the_facts.html (accessed on 5 July 2022).
- Rutland-Brown, W.; Langlois, J.A.; Thomas, K.E.; Xi, Y.L. Incidence of traumatic brain injury in the United States, 2003. J. Head Trauma Rehabil. 2006, 21, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Feigin, V.; Theadom, A.; Barker-Collo, S.; Starkey, N.; McPherson, K.; Kahan, M.; Dowell, A.; Brown, P.; Parag, V.; Kydd, R.; et al. Incidence of traumatic brain injury in New Zealand: A population-based study. Lancet Neurol. 2013, 12, 53–64. (In English) [Google Scholar] [CrossRef] [PubMed]
- Dewan, M.; Rattani, A.; Gupta, S.; Baticulon, R.; Hung, Y.-C.; Punchak, M.; Agrawal, A.; Adeleye, A.; Shrime, M.; Rubiano, A.; et al. Estimating the global incidence of traumatic brain injury. J. Neurosurg. 2019, 130, 1080–1097. [Google Scholar] [CrossRef]
- Teasdale, G.; Jennett, B. Assessment of coma and impaired consciousness. A practical scale. Lancet 1974, 2, 81–84. (In English) [Google Scholar] [CrossRef]
- Teasdale, G.; Murray, G.; Parker, L.; Jennett, B. Adding up the Glasgow Coma Score. Acta Neurochir. Suppl. (Wien) 1979, 28, 13–16. [Google Scholar]
- Gomez, D.; Byrne, J.; Alali, A.; Xiong, W.; Hoeft, C.; Neal, M.; Subacius, H.; Nathens, A. Inclusion of highest glasgow coma scale motor component score in mortality risk adjustment for benchmarking of trauma center performance. J. Am. Coll. Surg. 2017, 225, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Mena, J.H.; Sanchez, A.I.; Rubiano, A.M.; Peitzman, A.B.; Sperry, J.L.; Gutierrez, M.I.; Puyana, J.C. Effect of the modified Glasgow Coma Scale score criteria for mild traumatic brain injury on mortality prediction: Comparing classic and modified Glasgow Coma Scale score model scores of 13. J. Trauma 2011, 71, 1185–1193. (In English) [Google Scholar] [CrossRef]
- Watanitanon, A.; Lyons, V.H.; Lele, A.V.; Krishnamoorthy, V.; Chaikittisilpa, N.; Chandee, T.; Vavilala, M.S. Clinical Epidemiology of Adults With Moderate Traumatic Brain Injury. Crit. Care Med. 2018, 46, 781–787. (In English) [Google Scholar] [CrossRef]
- Cassidy, J.D.; Carroll, L.J.; Peloso, P.M.; Borg, J.; von Holst, H.; Holm, L.; Kraus, J.; Coronado, V.G.; WHO Collaborating Centre Task Force on Mild Traumatic Brain Injury. Incidence, risk factors and prevention of mild traumatic brain injury: Results of the WHO Collaborating Centre Task Force on Mild Traumatic Brain Injury. J. Rehabil. Med. 2004, 36, 28–60. [Google Scholar] [CrossRef]
- Selassie, A.W.; Zaloshnja, E.; Langlois, J.A.; Miller, T.; Jones, P.; Steiner, C. Incidence of long-term disability following traumatic brain injury hospitalization, United States, 2003. J. Head Trauma Rehabil. 2008, 23, 123–131. [Google Scholar] [CrossRef]
- Steppacher, I.; Kaps, M.; Kissler, J. Against the odds: A case study of recovery from coma after devastating prognosis. Ann. Clin. Transl. Neurol. 2016, 3, 61–65. [Google Scholar] [CrossRef]
- Andriessen, T.M.J.C.; Horn, J.; Franschman, G.; van der Naalt, J.; Haitsma, I.; Jacobs, B.; Steyerberg, E.W.; Vos, P.E. Epidemiology, severity classification, and outcome of moderate and severe traumatic brain injury: A prospective multicenter study. J. Neurotrauma 2011, 28, 2019–2031. [Google Scholar] [CrossRef] [PubMed]
- Fearnside, M.; Mcdougall, P. Moderate Head Injury: A system of neurotrauma care. Aust. N. Z. J. Surg. 1998, 68, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, A.; Servadei, F.; Marchesini, G.; Stein, S.C.; Vandelli, A. Early predictors of unfavourable outcome in subjects with moderate head injury in the emergency department. J. Neurol. Neurosurg. Psychiatry 2008, 79, 567. [Google Scholar] [CrossRef]
- Compagnone, C.; d’Avella, D.; Servadei, F.; Angileri, F.F.; Brambilla, G.; Conti, C.; Cristofori, L.; Delfini, R.; Denaro, L.; Ducati, A.; et al. Patients with moderate head injury: A prospective multicenter study of 315 patients. Neurosurgery 2009, 64, 690–697. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Health and Human Services. Centers for Disease Control and Prevention. Moderate to Severe Traumatic Injury Is a Lifelong Condition. Available online: https://www.cdc.gov/traumaticbraininjury/pdf/moderate_to_severe_tbi_lifelong-a.pdf (accessed on 11 November 2022).
- Einarsen, C.E.; Naalt, J.V.; Jacobs, B.; Follestad, T.; Moen, K.G.; Vik, A.; Håberg, A.K.; Skandsen, T. Moderate traumatic brain injury: Clinical characteristics and a prognostic model of 12-month outcome. World Neurosurg. 2018, 114, e1199–e1210. [Google Scholar] [CrossRef] [PubMed]
- Department of Statistics Malaysia. Statistics on Causes of Death, Malaysia. 2021. Available online: https://www.dosm.gov.my/v1/index.php?r=column/cthemeByCat&cat=401&bul_id=R3VrRUhwSXZDN2k4SGN6akRhTStwQT09&menu_id=L0pheU43NWJwRWVSZklWdzQ4TlhUUT09 (accessed on 4 July 2022).
- Ministry of Health Malaysia Planning Division. Ministry of Health Malaysia, Kementerian Kesihatan Malaysia (KKM) Health Facts; Ministry of Health Malaysia Planning Division: Putrajaya, Malaysia, 2021.
- Schmitt, S.; Dichter, M.A. Electrophysiologic Recordings in Traumatic Brain Injury, 1st ed.; Elsevier B.V.: Amsterdam, The Netherlands, 2015; pp. 319–339. [Google Scholar]
- Walker, W.C.; Stromberg, K.A.; Marwitz, J.H.; Sima, A.P.; Agyemang, A.A.; Graham, K.M.; Harrison-Felix, C.; Hoffman, J.M.; Brown, A.W.; Kreutzer, J.S.; et al. Predicting long-term global outcome after traumatic brain injury: Development of a practical prognostic tool using the traumatic brain injury model systems national database. J. Neurotrauma 2018, 35, 1587–1595. [Google Scholar] [CrossRef]
- Maas, A.I.R.; Marmarou, A.; Murray, G.D.; Teasdale, S.G.M.; Steyerberg, E.W. Prognosis and clinical trial design in traumatic brain injury: The IMPACT study. J. Neurotrauma 2007, 24, 232–238. [Google Scholar] [CrossRef]
- Han, J.; King, N.K.K.; Neilson, S.J.; Gandhi, M.P.; Ng, I. External validation of the CRASH and IMPACT prognostic models in severe traumatic brain injury. J. Neurotrauma 2014, 31, 1146–1152. (In English) [Google Scholar] [CrossRef]
- MRC CRASH Trial Collaborators; Perel, P.; Arango, M.; Clayton, T.; Edwards, P.; Komolafe, E.; Poccock, S.; Roberts, I.; Shakur, H.; Steyerberg, E.; et al. Predicting outcome after traumatic brain injury: Practical prognostic models based on large cohort of international patients. BMJ 2008, 336, 425–429. [Google Scholar] [CrossRef]
- Lingsma, H.F.; Roozenbeek, B.; Steyerberg, E.W.; Murray, G.D.; Maas, A.I.R. Early prognosis in traumatic brain injury: From prophecies to predictions. Lancet Neurol. 2010, 9, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Noor, N.S.E.M.; Ibrahim, H. Predicting outcomes in patients with traumatic brain injury using machine learning models. In Intelligent Manufacturing and Mechatronics, Melaka, Malaysia; Springer: Berlin/Heidelberg, Germany, 2019; pp. 12–20. [Google Scholar]
- Weir, J.; Steyerberg, E.W.; Butcher, I.; Lu, J.; Lingsma, H.F.; McHugh, G.S.; Roozenbeek, B.; Maas, A.I.R.; Murray, G.D. Does the extended glasgow outcome scale add value to the conventional glasgow outcome scale? J. Neurotrauma 2012, 29, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Haveman, M.E.; Putten, M.J.A.M.V.; Hom, H.W.; Eertman-Meyer, C.J.; Beishuizen, A.; Tjepkema-Cloostermans, M.C. Predicting outcome in patients with moderate to severe traumatic brain injury using electroencephalography. Crit. Care 2019, 23, 401. [Google Scholar] [CrossRef] [PubMed]
- van den Brink, R.L.; Nieuwenhuis, S.; van Boxtel, G.J.M.; Luijtelaar, G.v.; Eilander, H.J.; Wijnen, V.J.M. Task-free spectral EEG dynamics track and predict patient recovery from severe acquired brain injury. NeuroImage Clin. 2018, 17, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Noor, N.S.E.M.; Ibrahim, H.; Lah, M.H.C.; Abdullah, J.M. Improving Outcome Prediction for Traumatic Brain Injury from Imbalanced Datasets Using RUSBoosted Trees on Electroencephalography Spectral Power. IEEE Access 2021, 9, 121608–121631. [Google Scholar] [CrossRef]
- Noor, N.S.E.M.; Ibrahim, H.; Lah, M.H.C.; Abdullah, J.M. Prediction of Recovery from Traumatic Brain Injury with EEG Power Spectrum in Combination of Independent Component Analysis and RUSBoost Model. Biomedinformatics 2022, 2, 106–123. [Google Scholar] [CrossRef]
- Noor, N.S.E.M.; Ibrahim, H. Machine learning algorithms and quantitative electroencephalography predictors for outcome prediction in traumatic brain injury: A systematic review. IEEE Access 2020, 8, 102075–102092. [Google Scholar] [CrossRef]
- He, Y.; Eguren, D.; Azorín, J.M.; Grossman, R.G.; Luu, T.P.; Contreras-Vidal, J.L. Brain–machine interfaces for controlling lower-limb powered robotic systems. J. Neural Eng. 2018, 15, 021004. [Google Scholar] [CrossRef]
- Chen, R.; Parhi, K.K. Seizure Prediction using Convolutional Neural Networks and Sequence Transformer Networks. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 6483–6486. [Google Scholar]
- Jana, R.; Mukherjee, I. Deep learning based efficient epileptic seizure prediction with EEG channel optimization. Biomed. Signal Process. Control 2021, 68, 102767. [Google Scholar] [CrossRef]
- Attia, T.P.; Viana, P.F.; Nasseri, M.; Richardson, M.P.; Brinkmann, B.H. Seizure forecasting from subcutaneous EEG using long short term memory neural networks: Algorithm development and optimization. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 3599–3602. [Google Scholar]
- Sekkal, R.N.; Bereksi-Reguig, F.; Ruiz-Fernandez, D.; Dib, N.; Sekkal, S. Automatic sleep stage classification: From classical machine learning methods to deep learning. Biomed. Signal Process. Control 2022, 77, 103751. [Google Scholar] [CrossRef]
- Zhao, D.; Jiang, R.; Feng, M.; Yang, J.; Wang, Y.; Hou, X.; Wang, X. A deep learning algorithm based on 1D CNN-LSTM for automatic sleep staging. Technol. Health Care 2022, 30, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Khalili, E.; Asl, B.M. Automatic sleep stage classification using temporal convolutional neural network and new data augmentation technique from raw single-channel EEG. Comput. Methods Programs Biomed. 2021, 204, 106063. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; You, B.; Liu, Y.; Dai, Y. Patient-specific method of sleep electroencephalography using wavelet packet transform and Bi-LSTM for epileptic seizure prediction. Biomed. Signal Process. Control 2021, 70, 102963. [Google Scholar] [CrossRef]
- Gong, S.; Xing, K.; Cichocki, A.; Li, J. Deep learning in EEG: Advance of the last ten-year critical period. IEEE Trans. Cogn. Dev. Syst. 2021. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Levin, H.S.; Diaz-Arrastia, R.R. Diagnosis, prognosis, and clinical management of mild traumatic brain injury. Lancet Neurol. 2015, 14, 506–517. (In English) [Google Scholar] [CrossRef]
- McMillan, T.; Wilson, L.; Ponsford, J.; Levin, H.; Teasdale, G.; Bond, M. The glasgow outcome scale—40 years of application and refinement. Nat. Rev. Neurol. 2016, 12, 477. [Google Scholar] [CrossRef]
- Jennett, B.; Snoek, J.; Bond, M.R.; Brooks, N. Disability after severe head injury: Observations on the use of the glasgow outcome scale. J. Neurol. Neurosurg. Psychiatry 1981, 44, 285–293. [Google Scholar] [CrossRef]
- Jennett, B.; Bond, M. Assessment of outcome after severe brain damage: A practical scale. Lancet 1975, 305, 480–484. [Google Scholar] [CrossRef]
- Delorme, A.; Makeig, S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 2004, 134, 9–21. [Google Scholar] [CrossRef]
- Alhussein, M.; Muhammad, G.; Hossain, M.S. EEG pathology detection based on deep learning. IEEE Access 2019, 7, 27781–27788. [Google Scholar] [CrossRef]
- Altıntop, Ç.G.; Latifoğlu, F.; Akın, A.K.; Çetin, B. A novel approach for detection of consciousness level in comatose patients from EEG signals with 1-D convolutional neural network. Biocybern. Biomed. Eng. 2022, 42, 16–26. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Q.; Ibrahim, H.; Abdullah, M.Z.; Abdullah, J.M.; Suandi, S.A.; Azman, A. Arrangements of Resting State Electroencephalography as the Input to Convolutional Neural Network for Biometric Identification. Comput. Intell. Neurosci. 2019, 2019, 7895924. [Google Scholar] [CrossRef]
- Salehinejad, H.; Sankar, S.; Barfett, J.; Colak, E.; Valaee, S. Recent advances in recurrent neural networks. arXiv 2017, arXiv:1801.01078. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Salamon, J.; Bello, J.P. Deep convolutional neural networks and data augmentation for environmental sound classification. IEEE Signal Process. Lett. 2017, 24, 279–283. [Google Scholar] [CrossRef]
- Ahlawat, K.; Chug, A.; Singh, A.P. Benchmarking framework for class imbalance problem using novel sampling approach for big data. Int. J. Syst. Assur. Eng. Manag. 2019, 10, 824–835. [Google Scholar] [CrossRef]
- Longadge, R.; Dongre, S. Class imbalance problem in data mining review. arXiv 2013, arXiv:1305.1707. [Google Scholar] [CrossRef]
- Alhudhaif, A. A novel multi-class imbalanced EEG signals classification based on the adaptive synthetic sampling (ADASYN) approach. PeerJ Comput. Sci. 2021, 7, e523. [Google Scholar] [CrossRef]
- Chawla, N.V.; Lazarevic, A.; Hall, L.O.; Bowyer, K.W. SMOTEBoost: Improving prediction of the minority class in boosting. In Proceedings of the European Conference on Principles of Data Mining and Knowledge Discovery, Cavtat-Dubrovnik, Croatia, 22–26 September 2003; Springer: Berlin/Heidelberg, Germany, 2003; pp. 107–119. [Google Scholar]
- Krishnamoorthy, V.; Shoorangiz, R.; Weddell, S.J.; Beckert, L.; Jones, R.D. Deep Learning with Convolutional Neural Network for detecting microsleep states from EEG: A comparison between the oversampling technique and cost-based learning. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 4152–4155. [Google Scholar]
- Buda, M.; Maki, A.; Mazurowski, M.A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018, 106, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Zoubir, A.M.; Boashash, B. The bootstrap and its application in signal processing. IEEE Signal Process. Mag. 1998, 15, 56–76. [Google Scholar] [CrossRef]
- Efron, B. The bootstrap and modern statistics. J. Am. Stat. Assoc. 2000, 95, 1293–1296. [Google Scholar] [CrossRef]
- Kaur, H.; Pannu, H.S.; Malhi, A.K. A systematic review on imbalanced data challenges in machine learning: Applications and solutions. ACM Comput. Surv. (CSUR) 2019, 52, 1–36. [Google Scholar] [CrossRef]
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of imbalanced data: A review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- He, C.; Liu, J.; Zhu, Y.; Du, W. Data Augmentation for Deep Neural Networks Model in EEG Classification Task: A Review. Front. Hum. Neurosci. 2021, 15, 765525. (In English) [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X.-R.; Zhang, B.; Lei, M.-Y.; Cui, W.-G.; Guo, Y.-Z. A channel-projection mixed-scale convolutional neural network for motor imagery EEG decoding. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1170–1180. [Google Scholar] [CrossRef]
- Lee, T.; Kim, M.; Kim, S.-P. Data augmentation effects using borderline-SMOTE on classification of a P300-based BCI. In Proceedings of the 2020 8th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Republic of Korea, 26–28 February 2020; pp. 1–4. [Google Scholar]
- Chennu, S.; Annen, J.; Wannez, S.; Thibaut, A.; Chatelle, C.; Cassol, H.; Martens, G.; Schnakers, C.; Gosseries, O.; Menon, D.; et al. Brain networks predict metabolism, diagnosis and prognosis at the bedside in disorders of consciousness. Brain 2017, 140, 2120–2132. [Google Scholar] [CrossRef]
- Schorr, B.; Schlee, W.; Arndt, M.; Bender, A. Coherence in resting-state EEG as a predictor for the recovery from unresponsive wakefulness syndrome. J. Neurol. 2016, 263, 937–953. [Google Scholar] [CrossRef]
- Lee, H.; Mizrahi, M.A.; Hartings, J.A.; Sharma, S.; Pahren, L.; Ngwenya, L.B.; Moseley, B.D.; Privitera, M.; Tortella, F.C.; Foreman, B. Continuous electroencephalography after moderate to severe traumatic brain injury. Crit. Care Med. 2019, 47, 574–582. (In English) [Google Scholar] [CrossRef]
- Chen, M.; Li, Z.; Yan, Z.; Ge, S.; Zhang, Y.; Yang, H.; Zhao, L.; Liu, L.; Zhang, X.; Cai, Y.; et al. Predicting neurological deterioration after moderate traumatic brain injury: Development and validation of a prediction model based on data collected on admission. J. Neurotrauma 2022, 39, 371–378. (In English) [Google Scholar] [CrossRef] [PubMed]
- Cnossen, M.C.; Polinder, S.; Andriessen, T.M.; van der Naalt, J.; Haitsma, I.; Horn, J.; Franschman, G.; Vos, P.E.; Steyerberg, E.W.; Lings, H. Causes and Consequences of Treatment Variation in Moderate and Severe Traumatic Brain Injury: A Multicenter Study. Crit. Care Med. 2017, 45, 660–669. (In English) [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).