NSUN2 Negatively Regulates TP53 mRNA Stability to Promote the Malignant Progression of Nasopharyngeal Carcinoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Cell Culture
2.3. Plasmids and Construction of Stable Cell Lines
2.4. Western Blot Analysis
2.5. Cell Counting Kit-8 (CCK-8) Assay
2.6. Colony Formation Assays
2.7. Migration and Invasion Assays
2.8. Flow Cytometry-Mediated Apoptosis Assay
2.9. RNA Extraction
2.10. Actinomycin D Experiment
2.11. RNA Binding Protein Immunoprecipitation (RIP) Assay
2.12. Dot Blot Assay
2.13. Dual-Luciferase Reporter Assay
2.14. Immunohistochemistry (IHC)
2.15. Nude Mouse Xenograft Model
2.16. Gene Expression and Clinical Correlation Analysis
2.17. Statistical Analysis
3. Results
3.1. High NSUN2 Expression in NPC and Its Association with Poor Prognosis
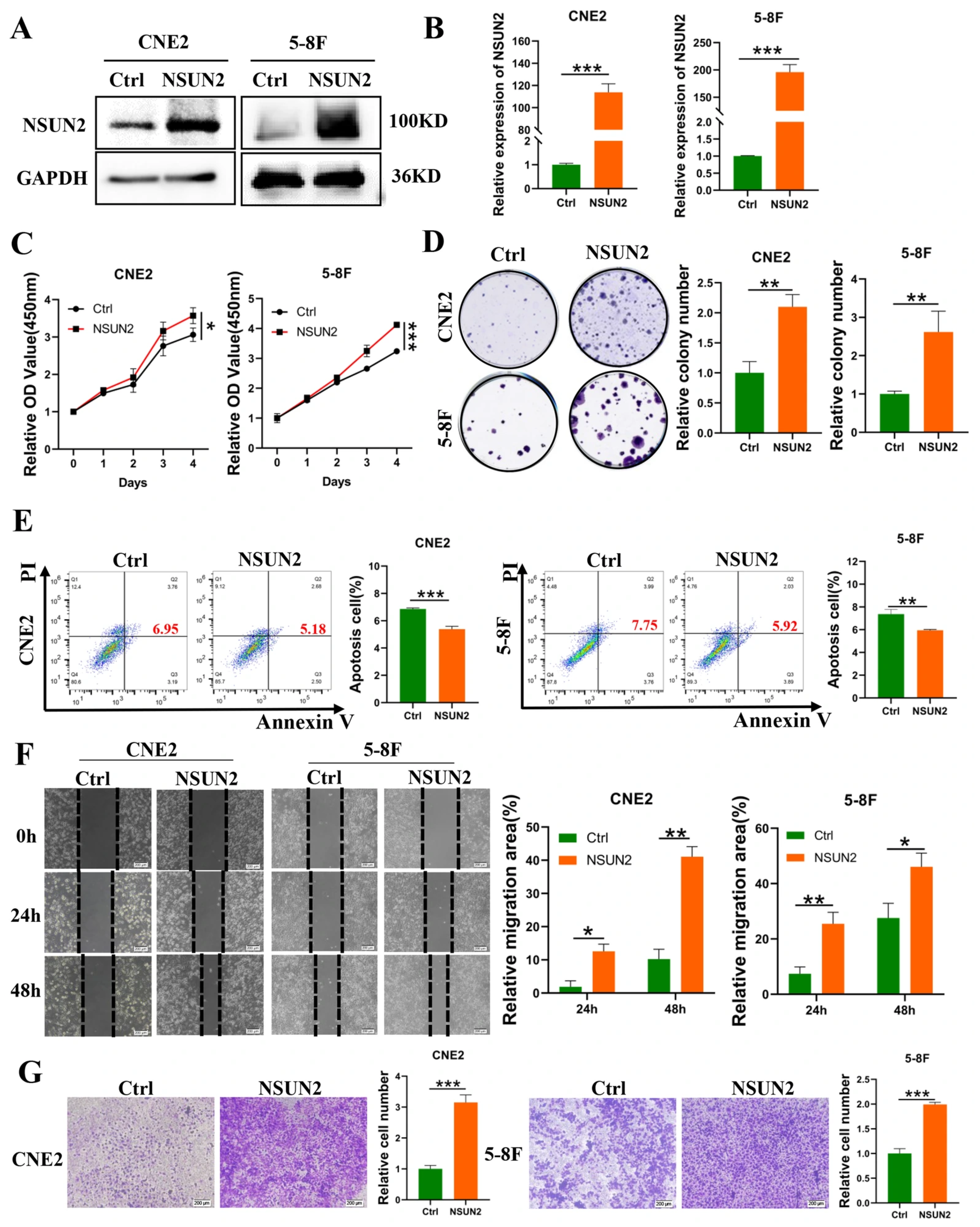
3.2. NSUN2 Promotes Proliferation, Migration, and Invasion, and Reduces NPC Cell Apoptosis
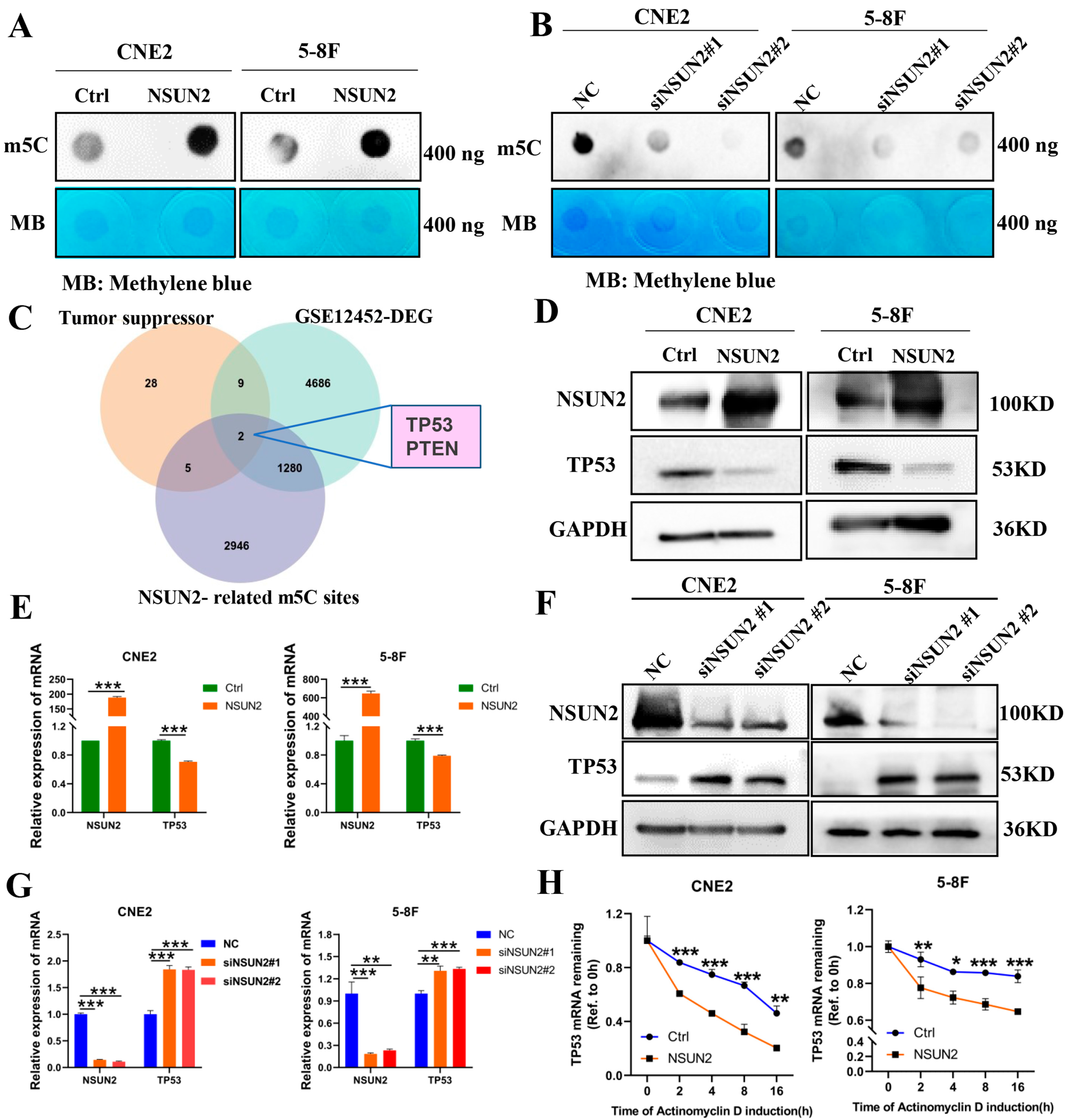
3.3. NSUN2 Negatively Regulates TP53 Expression via m5C Modification
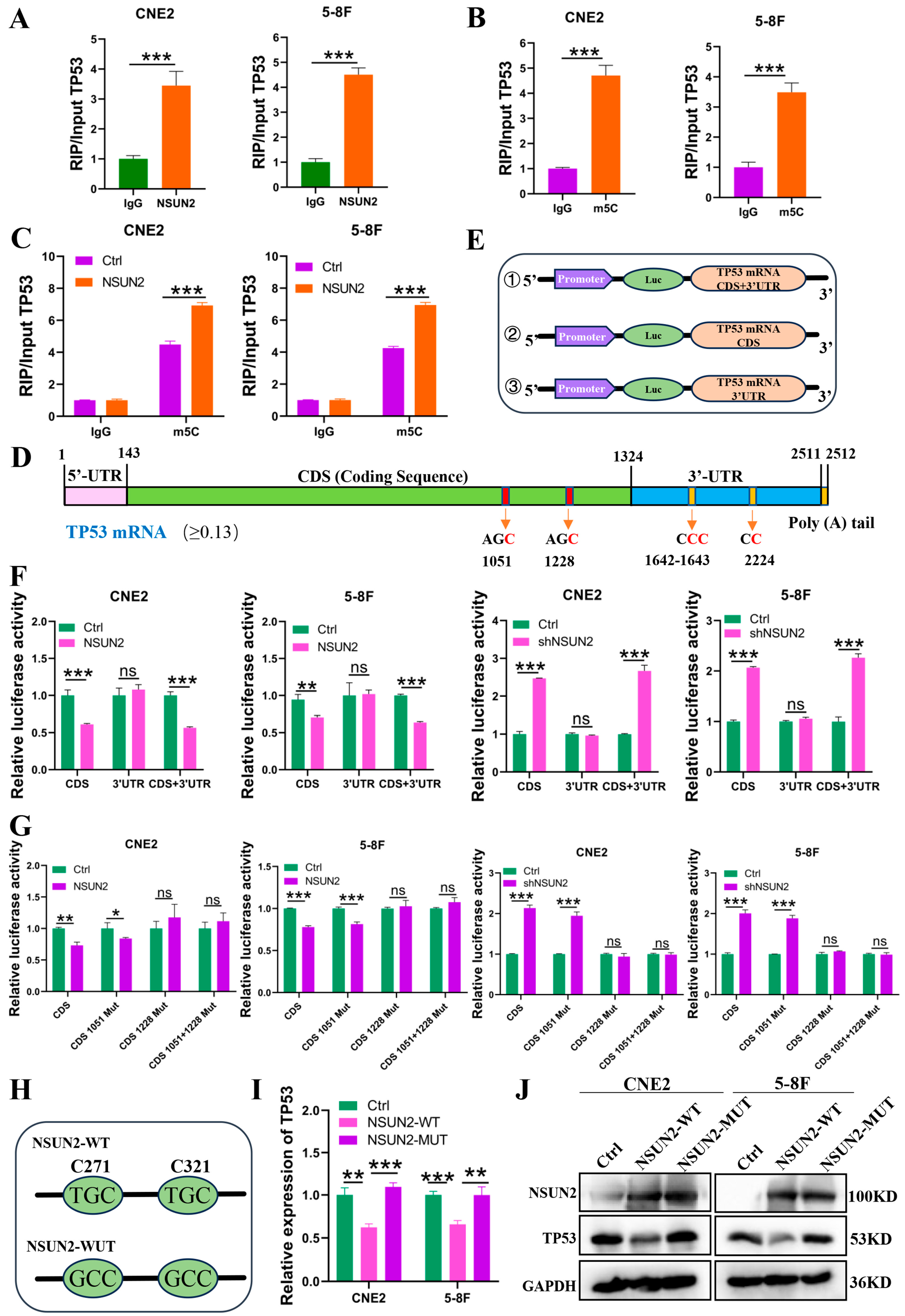
3.4. NSUN2 Increases m5C Modification at the TP53 mRNA CDS 1228 Site, Reducing TP53 Expression
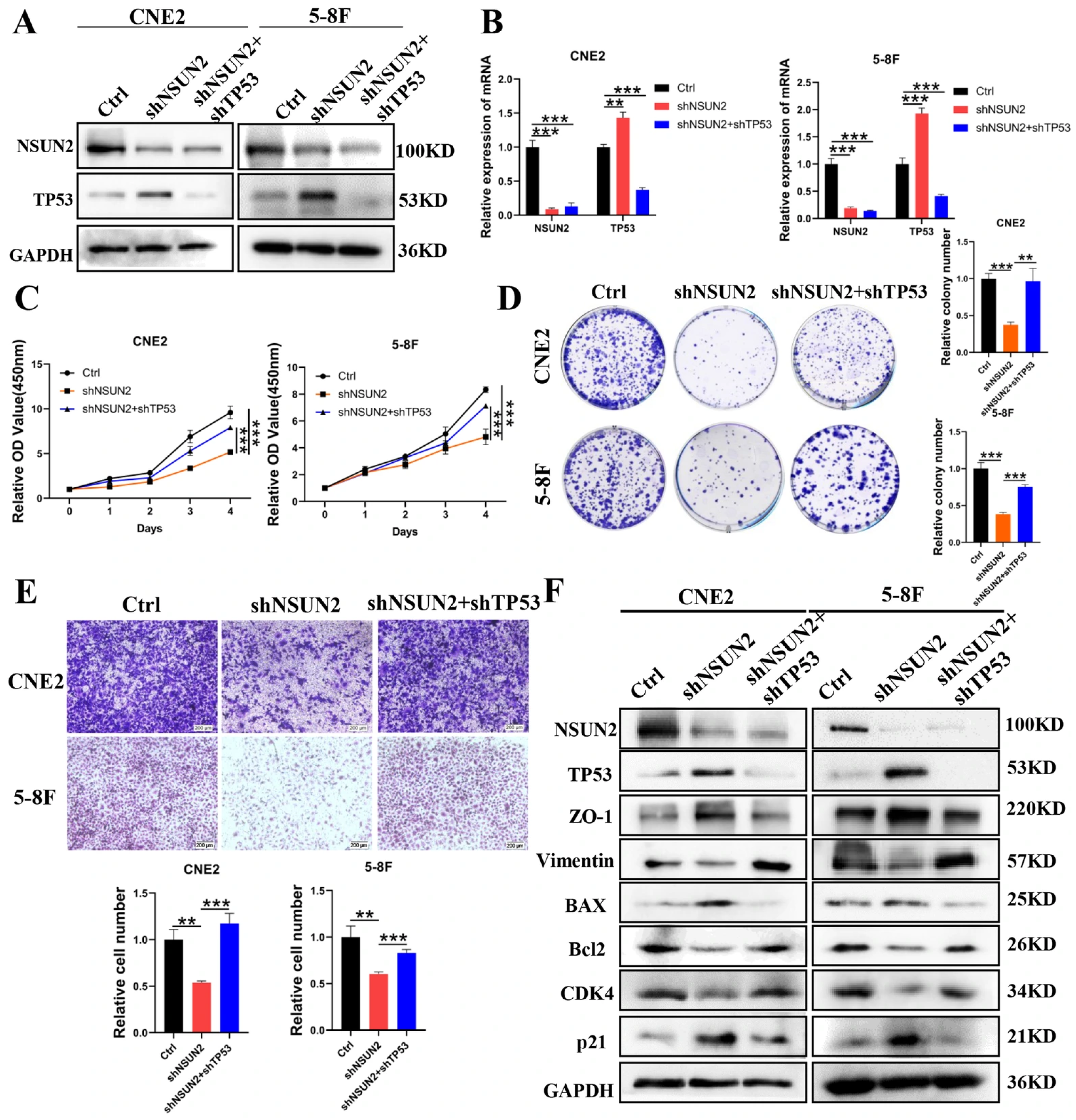
3.5. TP53 Knockdown Reverses the Suppressive Effects of NSUN2 Knockdown on NPC Cells
3.6. NSUN2 Knockdown Inhibits NPC Growth by Regulating In Vivo TP53 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.P.; Chan, A.T.C.; Le, Q.T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.C.W.; Hui, E.P.; Lo, K.W.; Lam, W.K.J.; Johnson, D.; Li, L.; Tao, Q.; Chan, K.C.A.; To, K.F.; King, A.D.; et al. Nasopharyngeal carcinoma: An evolving paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 679–695. [Google Scholar] [CrossRef]
- Lee, A.W.M.; Ng, W.T.; Chan, J.Y.W.; Corry, J.; Makitie, A.; Mendenhall, W.M.; Rinaldo, A.; Rodrigo, J.P.; Saba, N.F.; Strojan, P.; et al. Management of locally recurrent nasopharyngeal carcinoma. Cancer Treat. Rev. 2019, 79, 101890. [Google Scholar] [CrossRef]
- Chan, A.T. Nasopharyngeal carcinoma. Ann. Oncol. 2010, 21 (Suppl. 7), vii308–vii312. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, Y.; Fan, R.; Gao, K.; Xie, S.; Wang, F.; Zhang, J.; Zhang, H.; He, Y.; Xie, Z.; et al. Treatment of Recurrent Nasopharyngeal Carcinoma: A Sequential Challenge. Cancers 2022, 14, 4111. [Google Scholar] [CrossRef]
- Siak, P.Y.; Khoo, A.S.; Leong, C.O.; Hoh, B.P.; Cheah, S.C. Current Status and Future Perspectives about Molecular Biomarkers of Nasopharyngeal Carcinoma. Cancers 2021, 13, 3490. [Google Scholar] [CrossRef]
- Dong, Z.; Cui, H. The Emerging Roles of RNA Modifications in Glioblastoma. Cancers 2020, 12, 736. [Google Scholar] [CrossRef]
- Xuan, J.J.; Sun, W.J.; Lin, P.H.; Zhou, K.R.; Liu, S.; Zheng, L.L.; Qu, L.H.; Yang, J.H. RMBase v2.0: Deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018, 46, D327–D334. [Google Scholar] [CrossRef]
- Bohnsack, K.E.; Hobartner, C.; Bohnsack, M.T. Eukaryotic 5-methylcytosine (m(5)C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 2019, 10, 102. [Google Scholar] [CrossRef]
- Boo, S.H.; Kim, Y.K. The emerging role of RNA modifications in the regulation of mRNA stability. Exp. Mol. Med. 2020, 52, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, A.; Sun, B.F.; Yang, Y.; Han, Y.N.; Yuan, X.; Chen, R.X.; Wei, W.S.; Liu, Y.; Gao, C.C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Y.; Xiang, Y.; Yadav, T.; Ouyang, J.; Phoon, L.; Zhu, X.; Shi, Y.; Zou, L.; Lan, L. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc. Natl. Acad. Sci. USA 2022, 119, e2116251119. [Google Scholar] [CrossRef]
- Chen, H.; Yang, H.; Zhu, X.; Yadav, T.; Ouyang, J.; Truesdell, S.S.; Tan, J.; Wang, Y.; Duan, M.; Wei, L.; et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat. Commun. 2020, 11, 2834. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Shi, Q.; Zheng, L.; Li, Q.; Yang, L.; Zhang, Y. Gene signatures of m5C regulators may predict prognoses of patients with head and neck squamous cell carcinoma. Am. J. Transl. Res. 2020, 12, 6841–6852. [Google Scholar] [PubMed]
- Yuan, H.; Liu, J.; Zhao, L.; Wu, P.; Chen, G.; Chen, Q.; Shen, P.; Yang, T.; Fan, S.; Xiao, B.; et al. Prognostic Risk Model and Tumor Immune Environment Modulation of m5C-Related LncRNAs in Pancreatic Ductal Adenocarcinoma. Front. Immunol. 2021, 12, 800268. [Google Scholar] [CrossRef]
- Chellamuthu, A.; Gray, S.G. The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer. Cells 2020, 9, 1758. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wu, G.; Ye, Y.; Zhang, J.; Zeng, L.; Huang, X.; Zheng, Y.; Bai, R.; Zhuang, L.; Li, M.; et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene 2021, 40, 5814–5828. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Z.; Zhu, Y.; Zhu, Q.; Yang, Y.; Jin, Y.; Zhang, F.; Jiang, L.; Ye, Y.; Li, H.; et al. NOP2/Sun RNA methyltransferase 2 promotes tumor progression via its interacting partner RPL6 in gallbladder carcinoma. Cancer Sci. 2019, 110, 3510–3519. [Google Scholar] [CrossRef]
- Mei, L.; Shen, C.; Miao, R.; Wang, J.Z.; Cao, M.D.; Zhang, Y.S.; Shi, L.H.; Zhao, G.H.; Wang, M.H.; Wu, L.S.; et al. RNA methyltransferase NSUN2 promotes gastric cancer cell proliferation by repressing p57(Kip2) by an m(5)C-dependent manner. Cell Death Dis. 2020, 11, 270. [Google Scholar] [CrossRef]
- Chen, L.; Ding, J.; Wang, B.; Chen, X.; Ying, X.; Yu, Z.; Dong, P. RNA methyltransferase NSUN2 promotes hypopharyngeal squamous cell carcinoma proliferation and migration by enhancing TEAD1 expression in an m(5)C-dependent manner. Exp. Cell Res. 2021, 404, 112664. [Google Scholar] [CrossRef]
- Zhu, W.; Wan, F.; Xu, W.; Liu, Z.; Wang, J.; Zhang, H.; Huang, S.; Ye, D. Positive epigenetic regulation loop between AR and NSUN2 promotes prostate cancer progression. Clin. Transl. Med. 2022, 12, e1028. [Google Scholar] [CrossRef]
- Chen, Y.; Zuo, X.; Wei, Q.; Xu, J.; Liu, X.; Liu, S.; Wang, H.; Luo, Q.; Wang, Y.; Yang, Y.; et al. Upregulation of LRRC8A by m(5)C modification-mediated mRNA stability suppresses apoptosis and facilitates tumorigenesis in cervical cancer. Int. J. Biol. Sci. 2023, 19, 691–704. [Google Scholar] [CrossRef]
- Sun, Z.; Xue, S.; Zhang, M.; Xu, H.; Hu, X.; Chen, S.; Liu, Y.; Guo, M.; Cui, H. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 2020, 39, 6906–6919. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Zhang, J.; Zhang, X. NSun2 promotes cell migration through methylating autotaxin mRNA. J. Biol. Chem. 2020, 295, 18134–18147. [Google Scholar] [CrossRef]
- Chen, T.; Xu, Z.G.; Luo, J.; Manne, R.K.; Wang, Z.; Hsu, C.C.; Pan, B.S.; Cai, Z.; Tsai, P.J.; Tsai, Y.S.; et al. NSUN2 is a glucose sensor suppressing cGAS/STING to maintain tumorigenesis and immunotherapy resistance. Cell Metab. 2023, 35, 1782–1798. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Zhang, J.; Zhou, T.; Rao, S.S.; Li, Q.; Xiao, L.Y.; Wei, S.T.; Zhang, H.F. Epigenetically upregulated NSUN2 confers ferroptosis resistance in endometrial cancer via m(5)C modification of SLC7A11 mRNA. Redox Biol. 2024, 69, 102975. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, C.; Tong, X.; Chen, S.; Hu, X.; Pan, B.; Sun, X.; Chen, Z.; Shi, X.; Hu, Y.; et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021, 12, 842. [Google Scholar] [CrossRef]
- Lu, L.; Zhu, G.; Zeng, H.; Xu, Q.; Holzmann, K. High tRNA Transferase NSUN2 Gene Expression is Associated with Poor Prognosis in Head and Neck Squamous Carcinoma. Cancer Investig. 2018, 36, 246–253. [Google Scholar] [CrossRef]
- Zheng, L.M.; Li, M.N.; Wei, J.X.; Chen, S.P.; Xue, C.N.; Duan, Y.M.; Tang, F.Q.; Li, G.Y.; Xiong, W.; She, K.L.; et al. NOP2/Sun RNA methyltransferase 2 is a potential pan-cancer prognostic biomarker and is related to immunity. PLoS ONE 2023, 18, e0292212. [Google Scholar] [CrossRef] [PubMed]
- Voskarides, K.; Giannopoulou, N. The Role of TP53 in Adaptation and Evolution. Cells 2023, 12, 512. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Hassin, O.; Oren, M. Drugging p53 in cancer: One protein, many targets. Nat. Rev. Drug Discov. 2023, 22, 127–144. [Google Scholar] [CrossRef]
- Ghosh, M.; Saha, S.; Bettke, J.; Nagar, R.; Parrales, A.; Iwakuma, T.; van der Velden, A.W.M.; Martinez, L.A. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell 2021, 39, 494–508.e495. [Google Scholar] [CrossRef]
- Ghosh, M.; Saha, S.; Li, J.; Montrose, D.C.; Martinez, L.A. p53 engages the cGAS/STING cytosolic DNA sensing pathway for tumor suppression. Mol. Cell 2023, 83, 266–280.e266. [Google Scholar] [CrossRef]
- Muller, M.; Wilder, S.; Bannasch, D.; Israeli, D.; Lehlbach, K.; Li-Weber, M.; Friedman, S.L.; Galle, P.R.; Stremmel, W.; Oren, M.; et al. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J. Exp. Med. 1998, 188, 2033–2045. [Google Scholar] [CrossRef]
- Menendez, D.; Snipe, J.; Marzec, J.; Innes, C.L.; Polack, F.P.; Caballero, M.T.; Schurman, S.H.; Kleeberger, S.R.; Resnick, M.A. p53-responsive TLR8 SNP enhances human innate immune response to respiratory syncytial virus. J. Clin. Investig. 2019, 129, 4875–4884. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Ma, S.; Zheng, Z.; Wang, X.; Chen, S.; Chang, T.; Liang, Z.; Jiang, Y.; Xu, S.; Liu, R. Multi-omics analysis of expression and prognostic value of NSUN members in prostate cancer. Front. Oncol. 2022, 12, 965571. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wei, Y.; Liu, Y.; Wei, J.; Zhou, X.; Duan, Y.; Chen, S.; Xue, C.; Zhan, Y.; Zheng, L.; et al. BRD7 inhibits enhancer activity and expression of BIRC2 to suppress tumor growth and metastasis in nasopharyngeal carcinoma. Cell Death Dis. 2023, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wen, Q.; Xu, L.; Xie, G.; Li, J.; Luo, J.; Chu, S.; Shi, L.; Huang, D.; Li, J.; et al. Activation of Akt/mTOR pathway is associated with poor prognosis of nasopharyngeal carcinoma. PLoS ONE 2014, 9, e106098. [Google Scholar] [CrossRef]
- Chen, L.; Liu, S.; Tao, Y. Regulating tumor suppressor genes: Post-translational modifications. Signal Transduct. Target. Ther. 2020, 5, 90. [Google Scholar] [CrossRef]
- Weinberg, R.A. Tumor suppressor genes. Science 1991, 254, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, Z.; Zhang, C.; Zhang, L.; Chen, H.; Xiao, N.; Bai, L.; Liu, H.; Wan, J. 5-Methylcytosine transferase NSUN2 drives NRF2-mediated ferroptosis resistance in non-small cell lung cancer. J. Biol. Chem. 2024, 300, 106793. [Google Scholar] [CrossRef]
- Li, P.; Huang, D. NSUN2-mediated RNA methylation: Molecular mechanisms and clinical relevance in cancer. Cell. Signal. 2024, 123, 111375. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, J.; Feng, L.; Li, O.; Huang, L.; Zhou, S.; Xu, Y.; An, K.; Zhang, Y.; Chen, R.; et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol. Cancer 2023, 22, 81. [Google Scholar] [CrossRef]
- Pan, A.; Xue, Y.; Ruan, X.; Dong, W.; Wang, D.; Liu, Y.; Liu, L.; Lin, Y.; E, T.; Lin, H.; et al. m5C modification of LINC00324 promotes angiogenesis in glioma through CBX3/VEGFR2 pathway. Int. J. Biol. Macromol. 2024, 257, 128409. [Google Scholar] [CrossRef]
- Grit, J.L.; McGee, L.E.; Tovar, E.A.; Essenburg, C.J.; Wolfrum, E.; Beddows, I.; Williams, K.; Sheridan, R.T.C.; Schipper, J.L.; Adams, M.; et al. p53 modulates kinase inhibitor resistance and lineage plasticity in NF1-related MPNSTs. Oncogene 2024, 43, 1411–1430. [Google Scholar] [CrossRef]
- Ma, Z.; Guo, D.; Wang, Q.; Liu, P.; Xiao, Y.; Wu, P.; Wang, Y.; Chen, B.; Liu, Z.; Liu, Q. Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics 2019, 9, 2967–2983. [Google Scholar] [CrossRef] [PubMed]



| Targets | Sequence |
|---|---|
| siRNA | |
| siNSUN2#1 | 5′-GGAGAACAAGCTGTTCGAG-3′ |
| siNSUN2#2 | 5′-GAGATCCTCTTCTATGATC-3′ |
| shRNA | |
| shNSUN2 | F: GATCCGAAGCATCGTGCTGAAGTACTCGAGTACTTCAGCACGATGCTTCTTTTTG R: AATTCAAAAAGAAGCATCGTGCTGAAGTACTCGAGTACTTCAGCACGATGCTTCG |
| shTP53 | F: GATCCGACTCCAGTGGTAATCTACCTCGAGGTAGATTACCACTGGAGTCTTTTTG R: AATTCAAAAAGACTCCAGTGGTAATCTACCTCGAGGTAGATTACCACTGGAGTCG |
| TP53 related primer | |
| TP53-CDS | F: CGCCGTGTAATTCTAGGAAACTACTTCCTGAAAACAA R: CCGCCCCGACTCTAGTTCTGTCTTGAACATGAGTTT |
| TP53-3′UTR | F: CGCCGTGTAATTCTAGAGGACTTCCATTTGCTTT R: CCGCCCCGACTCTAGATATAAAAATGGGATATAAAAAGGG |
| TP53-CDS + 3′UTR | F: GCCGTGTAATTCTAGAAGAGAATCTCCGCAAGAAA R: CCGCCCCGACTCTAGATATAAAAATGGGATATAAAAAGGG |
| TP53 CDS 1051 Mut | F: GCTGCCCCCAGGGAGTACTAAGCGAGCACTGCCCAA R: CAGTGCTCGCTTAGTACTCCCTGGGGGCAGCTCGTG |
| TP53 CDS 1228 Mut | F: GGAGCCAGGGGGGAGTAGGGCTCACTCCAGCCACCT R: GCTGGAGTGAGCCCTACTCCCCCCTGGCTCCTTCCC |
| TP53 CDS 1051 + 1228 Mut | F: GCTGCCCCCAGGGAGTACTAAGCGAGCACTGCCCAA R: GCTGGAGTGAGCCCTACTCCCCCCTGGCTCCTTCCC |
| Primer | |
| NSUN2 | F: 5′-AAGAAAGATGGCGTGTGTGG-3′ R: 5′-TATTCAGCAGCACATTCCGC-3′ |
| TP53 | F: 5′-CTCAGATAGCGATGGTCTGG-3′ R: 5′-CTGTCATCCAAATACTCCACAC-3′ |
| GAPDH | F: 5′-CAACGGATTTGGTCGTATTGG-3′ R: 5′-TGACGGTGCCATGGAATTT-3′ |
| 18S RNA | F: 5′-TCTTAGCTGAGTGTCCCGCG-3′ R: 5′-ATCATGGCCTCAGTTCCGAA-3′ |
| Variable Features | NSUN2 Expression | p | |
|---|---|---|---|
| Low Expression (16) | High Expression (60) | ||
| Age (year) | |||
| ≤53 | 7 (43.75%) | 32 (53.33%) | 0.4956 |
| >53 | 9 (56.25%) | 28 (46.67%) | |
| Gender | |||
| Male (n = 58) | 11 (68.75%) | 47 (78.33%) | 0.4231 |
| Female (n = 18) | 5 (31.25%) | 13 (21.67%) | |
| Clinical stages (n [%]) | |||
| Ⅰ–Ⅱ | 12 (75%) | 7 (11.67%) | *** p < 0.0001 |
| Ⅲ–Ⅳ | 4 (25%) | 53 (88.33%) | |
| Tumor size (n [%]) | |||
| T1–2 | 13 (81.25%) | 18 (30%) | *** p < 0.001 |
| T3–4 | 3 (18.75%) | 42 (70%) | |
| Lymph node metastasis (n [%]) | |||
| no | 8 (50%) | 3 (5%) | *** p < 0.0001 |
| yes | 8 (50%) | 57 (95%) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, L.; Wei, J.; Li, X.; Li, M.; Xue, C.; Chen, S.; Wei, Q.; Fan, S.; Xiong, W.; Zhou, M.; et al. NSUN2 Negatively Regulates TP53 mRNA Stability to Promote the Malignant Progression of Nasopharyngeal Carcinoma. Cancers 2025, 17, 3950. https://doi.org/10.3390/cancers17243950
Zheng L, Wei J, Li X, Li M, Xue C, Chen S, Wei Q, Fan S, Xiong W, Zhou M, et al. NSUN2 Negatively Regulates TP53 mRNA Stability to Promote the Malignant Progression of Nasopharyngeal Carcinoma. Cancers. 2025; 17(24):3950. https://doi.org/10.3390/cancers17243950
Chicago/Turabian StyleZheng, Lemei, Jianxia Wei, Xiaolong Li, Mengna Li, Changning Xue, Shipeng Chen, Qingqing Wei, Songqing Fan, Wei Xiong, Ming Zhou, and et al. 2025. "NSUN2 Negatively Regulates TP53 mRNA Stability to Promote the Malignant Progression of Nasopharyngeal Carcinoma" Cancers 17, no. 24: 3950. https://doi.org/10.3390/cancers17243950
APA StyleZheng, L., Wei, J., Li, X., Li, M., Xue, C., Chen, S., Wei, Q., Fan, S., Xiong, W., Zhou, M., & Deng, H. (2025). NSUN2 Negatively Regulates TP53 mRNA Stability to Promote the Malignant Progression of Nasopharyngeal Carcinoma. Cancers, 17(24), 3950. https://doi.org/10.3390/cancers17243950






