Exosomes as Emerging Non-Invasive Biomarkers of Cervical Cancer: A Systematic Review and Meta-Analysis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. Information Sources
2.4. Study Selection
2.5. Data Extraction
2.6. Risk of Bias in Individual Studies
2.7. Summary Measures
2.8. Confidence in Cumulative Evidence
3. Results
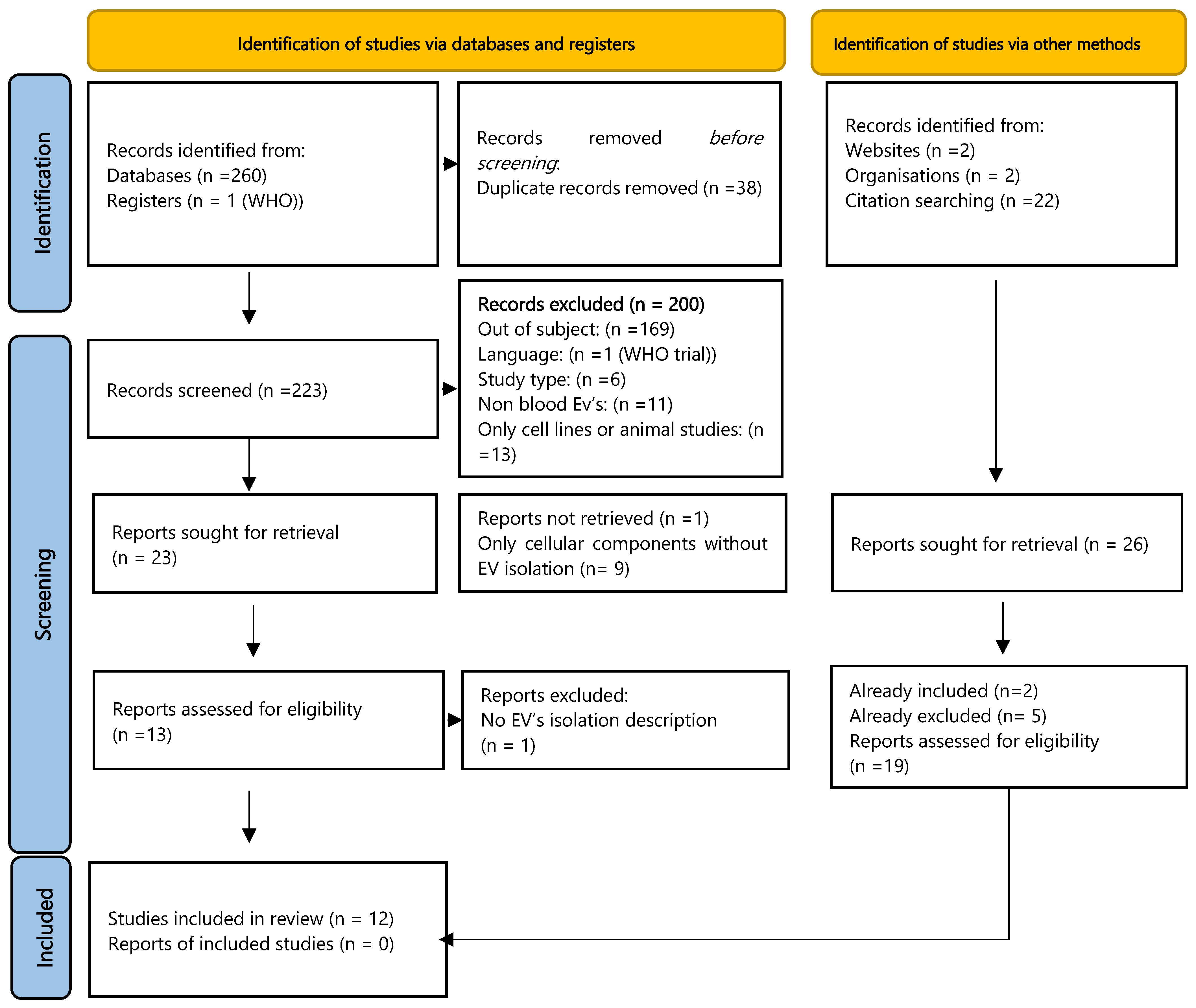
3.1. Study Selection
3.2. Study Characteristics
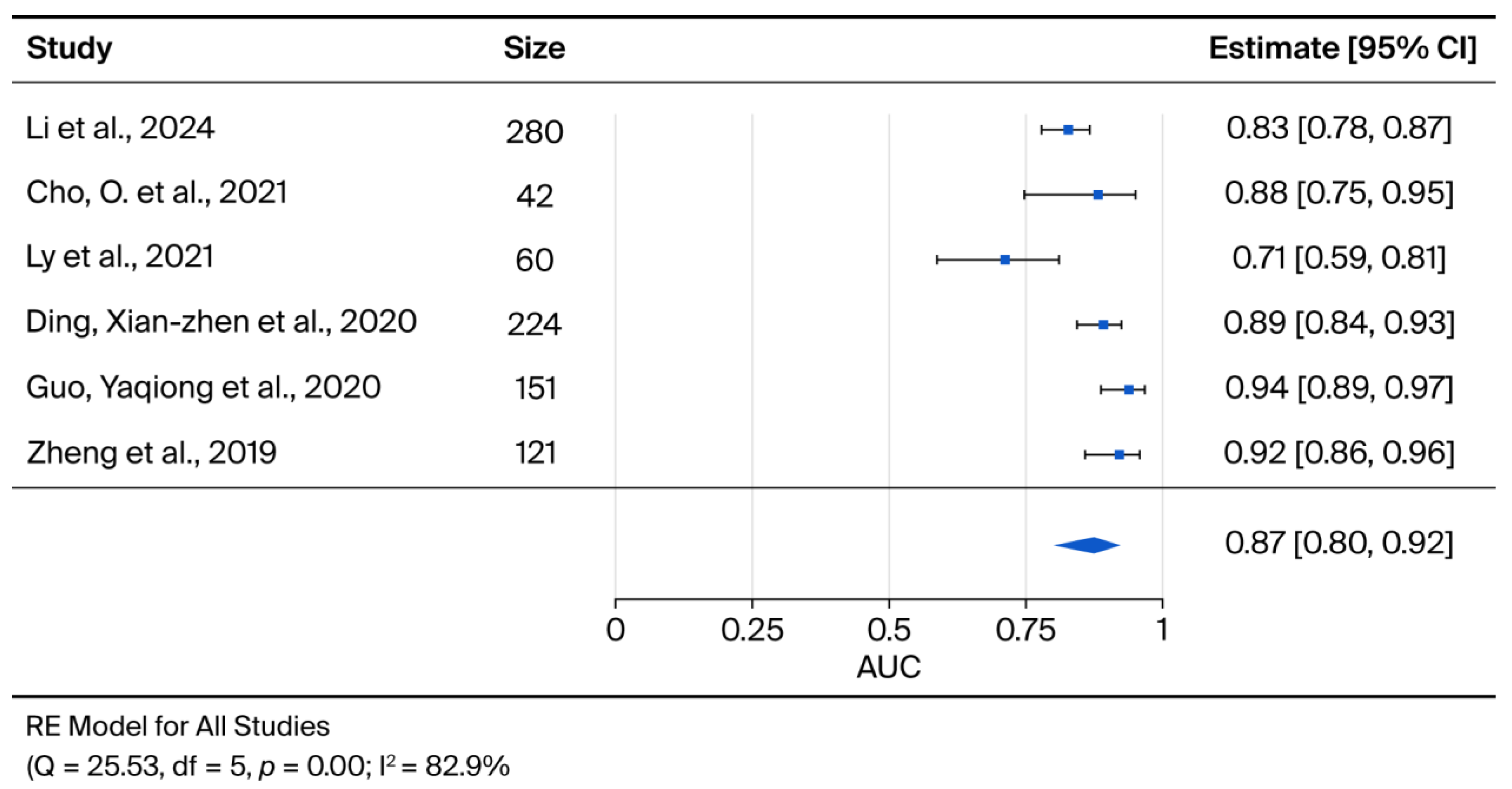
3.3. Results of Syntheses for Diagnosis
3.4. Results of Syntheses for Prognosis
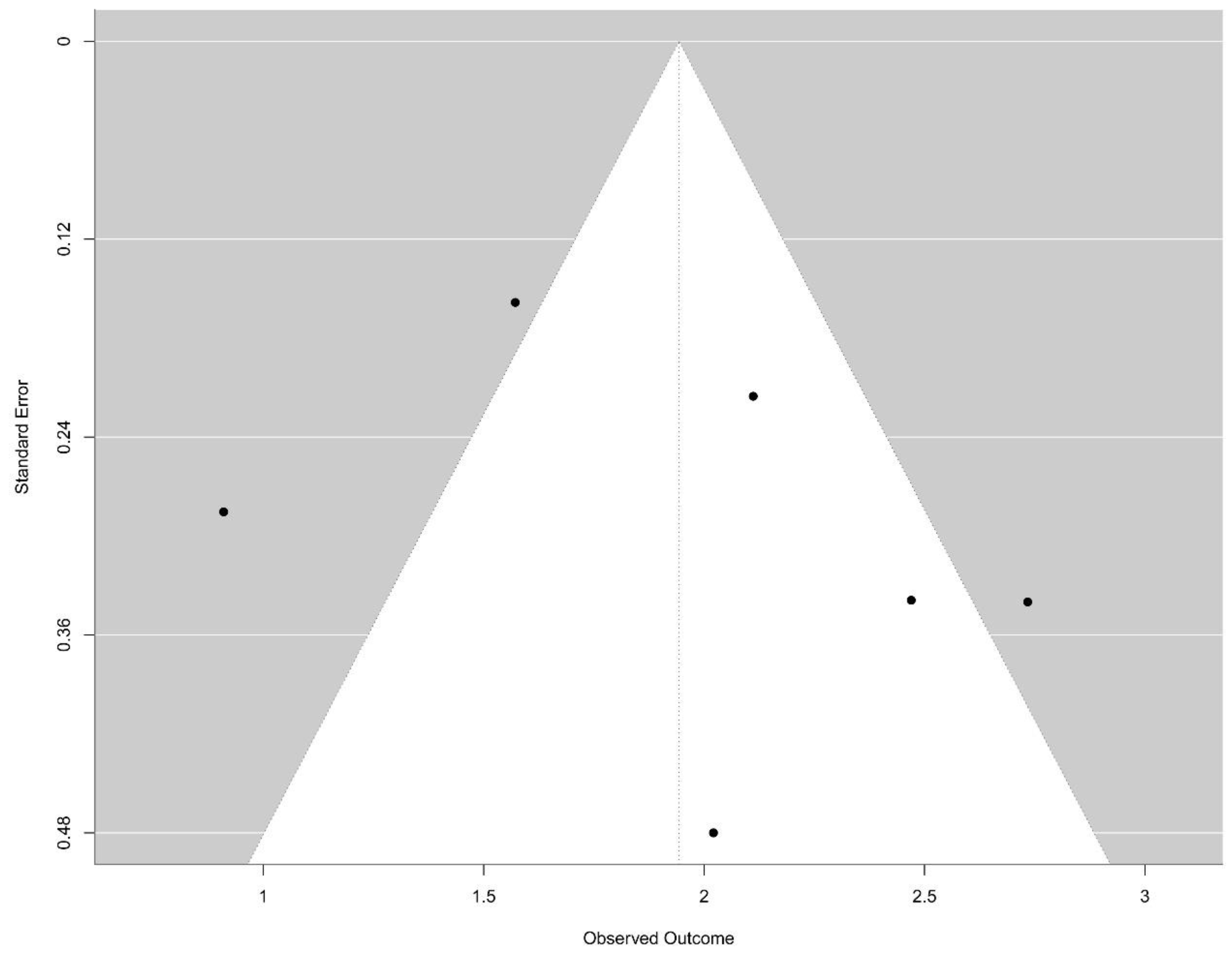
3.5. Reporting Biases
4. Discussion
4.1. Summary of Main Results
4.2. Results in the Context of Published Literature
4.3. Strengths and Weaknesses
4.4. Implications for Practice and Future Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CC | cervical cancer |
| EVs | Extracellular Vesicles |
| AUC | Area Under the Curve |
| miRNAs | microRNAs |
| lncRNAs | long non-coding RNAs |
| ctDNA | circulating tumour DNA |
References
- Dyba, T.; Randi, G.; Bray, F.; Martos, C.; Giusti, F.; Nicholson, N.; Gavin, A.; Flego, M.; Neamtiu, L.; Dimitrova, N.; et al. The European cancer burden in 2020, Incidence and mortality estimates for 40 countries and 25 major cancers. Eur. J. Cancer 2021, 157, 308–347. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjose, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018, a worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Lindquist, S.; Kjaer, S.K.; Frederiksen, K.; Ornskov, D.; Munk, C.; Waldstrom, M. Comparative analysis of HPV testing versus cytology in Danish cervical cancer screening: Insights from a large-scale implementation study. Gynecol. Oncol. 2024, 191, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Cibula, D.; Raspollini, M.R.; Planchamp, F.; Centeno, C.; Chargari, C.; Felix, A.; Fischerová, D.; Jahnn-Kuch, D.; Joly, F.; Kohler, C.; et al. ESGO/ESTRO/ESP Guidelines for the management of patients with cervical cancer-Update 2023. Int. J. Gynecol. Cancer 2023, 33, 649–666. [Google Scholar] [CrossRef]
- Jalali, M.; Lu, Y.; del Real Mata, C.; Rak, J.; Mahshid, S. Nanoscopic technologies toward molecular profiling of single extracellular vesicles for cancer liquid biopsy. Appl. Phys. Rev. 2025, 12, 011312. [Google Scholar] [CrossRef]
- Lawrence, S.R.; Shah, K.M. Prospects and Current Challenges of Extracellular Vesicle-Based Biomarkers in Cancer. Biology 2024, 13, 694. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, L.; Li, S.; Li, W.; Zheng, X.; Yang, Y.; Fu, W.; Yi, J.; Wang, J.; Du, G. The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 2021, 11, 2783–2797. [Google Scholar] [CrossRef]
- Leetanaporn, K.; Hanprasertpong, J.; Navakanitworakul, R. Molecular insights and clinical impacts of extracellular vesicles in cancer. Oncol. Rev. 2021, 15, 542. [Google Scholar] [CrossRef]
- Buglyó, B.S.G. The Role of Exosomes in Cancer Progression. Int. J. Mol. Sci. 2021, 23, 8. [Google Scholar] [CrossRef]
- Wang, K.H.; Ding, D.C. The Role and Applications of Exosomes in Gynecological Cancer: A Review. Cell Transplant. 2023, 32, 9636897231195240. [Google Scholar] [CrossRef]
- Cakir, M.O.; Selek, M.; Yilmaz, B.; Ozdogan, M.; Ashrafi, G.H. Exosomes in HPV-Associated Cancers: From Biomarkers to Engineered Therapeutics. Cancers 2025, 17, 3386. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Guyatt, G.; Oxman, A.D.; Akl, E.A.; Kunz, R.; Vist, G.; Brozek, J.; Norris, S.; Falck-Ytter, Y.; Glasziou, P.; DeBeer, H.; et al. GRADE guidelines: 1. Introduction-GRADE evidence profiles and summary of findings tables. J. Clin. Epidemiol. 2011, 64, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Mallett, S.; Takwoingi, Y.; Davenport, C.F.; Hyde, C.J.; Whiting, P.F.; Deeks, J.J.; Leeflang, M.M.; QUADAS-C Group. QUADAS-C: A Tool for Assessing Risk of Bias in Comparative Diagnostic Accuracy Studies. Ann. Intern. Med. 2021, 174, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.M.; Paik, S.; Hayes, D.F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl. Cancer Inst. 2009, 101, 1446–1452. [Google Scholar] [CrossRef]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. REporting recommendations for tumour MARKer prognostic studies (REMARK). Eur. J. Cancer 2005, 41, 1690–1696. [Google Scholar] [CrossRef]
- Hamza, T.; Schwarzer, G.; Salanti, G. crossnma: An R package to synthesize cross-design evidence and cross-format data using network meta-analysis and network meta-regression. BMC Med. Res. Methodol. 2024, 24, 169. [Google Scholar] [CrossRef]
- Cumpston, M.; Li, T.; Page, M.J.; Chandler, J.; Welch, V.A.; Higgins, J.P.; Thomas, J. Updated guidance for trusted systematic reviews: A new edition of the Cochrane Handbook for Systematic Reviews of Interventions. Cochrane Database Syst. Rev 2019, 10, Ed000142. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Ding, X.Z.; Zhang, S.Q.; Deng, X.L.; Qiang, J.H. Serum Exosomal lncRNA DLX6-AS1 Is a Promising Biomarker for Prognosis Prediction of Cervical Cancer. Technol. Cancer Res. Treat. 2021, 20, 1533033821990060. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, X.; Wang, K.; He, Y. Appraising the Value of Serum and Serum-Derived Exosomal LncRNA-EXOC7 as a Promising Biomarker in Cervical Cancer. Clin. Lab. 2020, 66, 1357–1363. [Google Scholar] [CrossRef]
- Cho, O.; Kim, D.W.; Cheong, J.Y. Screening Plasma Exosomal RNAs as Diagnostic Markers for Cervical Cancer: An Analysis of Patients Who Underwent Primary Chemoradiotherapy. Biomolecules 2021, 11, 1691. [Google Scholar] [CrossRef]
- Li, Z.; Wang, H.; Yang, R.; Jin, X.; Han, Q.; She, Z.; Ge, P. Plasma-Derived Exosomal tRF-Phe-GAA-001 and tRF-Gly-GCC-037 as Novel Diagnostic Biomarkers for Cervical Cancer. Indian J. Clin. Biochem. 2024, 40, 683–690. [Google Scholar] [CrossRef]
- Lv, A.; Tu, Z.; Huang, Y.; Lu, W.; Xie, B. Circulating exosomal miR-125a-5p as a novel biomarker for cervical cancer. Oncol. Lett. 2020, 21, 54. [Google Scholar] [CrossRef]
- Ma, G.; Song, G.; Zou, X.; Shan, X.; Liu, Q.; Xia, T.; Zhou, X.; Zhu, W. Circulating plasma microRNA signature for the diagnosis of cervical cancer. Cancer Biomark. 2019, 26, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Hou, L.; Ma, Y.; Zhou, L.; Wang, F.; Cheng, B.; Wang, W.; Lu, B.; Liu, P.; Lu, W.; et al. Exosomal let-7d-3p and miR-30d-5p as diagnostic biomarkers for non-invasive screening of cervical cancer and its precursors. Mol. Cancer 2019, 18, 76. [Google Scholar] [CrossRef] [PubMed]
- Molika, P.; Leetanaporn, K.; Rungkamoltip, P.; Roytrakul, S.; Hanprasertpong, J.; Navakanitworakul, R. Proteomic analysis of small extracellular vesicles unique to cervical cancer. Transl. Cancer Res. 2023, 12, 3113–3128. [Google Scholar] [CrossRef] [PubMed]
- Ao, K.; Yin, M.; Lyu, X.; Xiao, Y.; Chen, X.; Zhong, S.; Wen, X.; Yuan, J.; Ye, M.; Zhang, J.; et al. METTL3-mediated HSPA9 m6A modification promotes malignant transformation and inhibits cellular senescence by regulating exosomal mortalin protein in cervical cancer. Cancer Lett. 2024, 587, 216658. [Google Scholar] [CrossRef]
- Someya, M.; Hasegawa, T.; Tsuchiya, T.; Kitagawa, M.; Fukushima, Y.; Gocho, T.; Mafune, S.; Ikeuchi, Y.; Kozuka, Y.; Idogawa, M.; et al. Predictive value of an exosomal microRNA-based signature for tumor immunity in cervical cancer patients treated with chemoradiotherapy. Med. Mol. Morphol. 2023, 56, 38–45. [Google Scholar] [CrossRef]
- Cho, O.; Kim, D.W.; Cheong, J.Y. Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study. J. Clin. Med. 2021, 10, 2110. [Google Scholar] [CrossRef]
- Molika, P.; Bissanum, R.; Surachat, K.; Pattanapanyasat, K.; Hanprasertpong, J.; Chotigeat, W.; Navakanitworakul, R. Exploration of Extracellular Vesicle Long Non-Coding RNAs in Serum of Patients with Cervical Cancer. Oncology 2024, 102, 53–66. [Google Scholar] [CrossRef]
- Xue, J.; Qin, S.; Ren, N.; Guo, B.; Shi, X.; Jia, E. Extracellular vesicle biomarkers in circulation for the diagnosis of gastric cancer: A systematic review and meta-analysis. Oncol. Lett. 2023, 26, 423. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, T.; Wu, Q.; Yang, X.; Hao, J.; Deng, X.; Yang, S.; Gu, C.; Wang, Z. Diagnostic performance of various liquid biopsy methods in detecting colorectal cancer: A meta-analysis. Cancer Med. 2020, 9, 5699–5707. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Y.; Tian, X.; Wang, Q.; Huang, H.; Lu, X.; Qi, M.; Cao, X.; Lei, J. Diagnostic and predictive value of liquid biopsy-derived exosome miR-21 for breast cancer: A systematic review and meta-analysis. Expert. Rev. Mol. Diagn. 2023, 23, 315–324. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Andresen, T.L.; Simonsen, J.B. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 109–116. [Google Scholar] [CrossRef]
- Andre, M.; Caobi, A.; Miles, J.S.; Vashist, A.; Ruiz, M.A.; Raymond, A.D. Diagnostic potential of exosomal extracellular vesicles in oncology. BMC Cancer 2024, 24, 322. [Google Scholar] [CrossRef]
- Herrero, C.; de la Fuente, A.; Casas-Arozamena, C.; Sebastian, V.; Prieto, M.; Arruebo, M.; Abalo, A.; Colás, E.; Moreno-Bueno, G.; Gil-Moreno, A.; et al. Extracellular Vesicles-Based Biomarkers Represent a Promising Liquid Biopsy in Endometrial Cancer. Cancers 2019, 11, 2000. [Google Scholar] [CrossRef]
- Paterson, E.; Blenkiron, C.; Danielson, K.; Henry, C. Recommendations for extracellular vesicle miRNA biomarker research in the endometrial cancer context. Transl. Oncol. 2022, 23, 101478. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Kim, H.S.; Park, S.J.; Lee, E.J.; Kim, S.I.; Song, G.; Lim, W. Inhibition of miR-214-3p Aids in Preventing Epithelial Ovarian Cancer Malignancy by Increasing the Expression of LHX6. Cancers 2019, 11, 1917. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Lei, N.; Zhou, J.; Chen, M.; Guo, R.; Qin, B.; Li, Y.; Chang, L. Extracellular vesicles in ovarian cancer chemoresistance, metastasis, and immune evasion. Cell Death Dis. 2022, 13, 64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Su, X.; Li, S.; Liu, Z.; Wang, Q.; Zeng, H. Low serum exosomal miR-484 expression predicts unfavorable prognosis in ovarian cancer. Cancer Biomark. 2020, 27, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, O.; Wormland, S.; Bittner, A.K.; Collenburg, M.; Horn, P.A.; Kimmig, R.; Kasimir-Bauer, S.; Rebmann, V. Programmed death receptor ligand-2 (PD-L2) bearing extracellular vesicles as a new biomarker to identify early triple-negative breast cancer patients at high risk for relapse. J. Cancer Res. Clin. Oncol. 2023, 149, 1159–1174. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, Y.; Li, X.; Guo, A. CCAAT enhancer binding protein delta activates vesicle associated membrane protein 3 transcription to enhance chemoresistance and extracellular PD-L1 expression in triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2024, 43, 115. [Google Scholar] [CrossRef]
- Thakur, A.; Parra, D.C.; Motallebnejad, P.; Brocchi, M.; Chen, H.J. Exosomes: Small vesicles with big roles in cancer, vaccine development, and therapeutics. Bioact. Mater. 2022, 10, 281–294. [Google Scholar]
- Ridolfi, A.; Brucale, M.; Montis, C.; Caselli, L.; Paolini, L.; Borup, A.; Boysen, A.T.; Loria, F.; van Herwijnen, M.J.C.; Kleinjan, M.; et al. AFM-Based High-Throughput Nanomechanical Screening of Single Extracellular Vesicles. Anal. Chem. 2020, 92, 10274–10282. [Google Scholar] [CrossRef]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Beeram, R.; Vepa, K.R.; Soma, V.R. Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques. Biosensors 2023, 13, 328. [Google Scholar] [CrossRef] [PubMed]
| Rule | Inclusion Criteria | Exclusion Criteria |
|---|---|---|
| Study Type | Original research or replication studies published in peer-reviewed journals. No limitations imposed regarding publication date. | In vitro or animal studies without human data, case reports, review articles, editorials and per-spectives. |
| Participants | Non pregnant with HPV related cervical cancer. Older than 18 years. Without other invasive diseases. Non hospitalised. Studies involving all stages of cervical cancer. Studies involving CIN and/or healthy controls for comparison. | Under 18 years of age. Pregnant. Other invasive diseases. Only Animal models. Only Cell lines. |
| Exposure | Blood extracellular vesicles (EVs) Various types of EVs (e.g., exosomes, microvesicles, apoptotic bodies). EV-associated biomarkers (e.g., miRNA, nucleic acids, HPV DNA). | Without blood EVs. Examination of only cellular components without EV isolation. Documentation of EVs in cervical cancer, but without assessment of their biomarker potential. |
| Comparators | Healthy women. Cervical intraepitelial neoplasia. EV biomarkers across different stages of cervical cancer. | Absence of a clear comparison group or baseline. Comparison of only different EV isolation methods without clinical relevance. |
| Outcomes | Blood biomarker (EVs: miRNA, mRNA, lncRNAs HPVDNA or HPVRNA, ctDNA). Diagnostic biomarker (e.g., screening and/or early detection). Prognosis biomarker. | Other fluids: urine or cervicovaginal fluid. Absence of clear reporting of diagnostic or prognostic metrics. Focusing solely on therapeutic applications of EVs without biomarker assessment. Only qualitative outcomes without quantitative data. |
| Study (Author/ Year) | Country | Study Type | Sample Type | EVs Isolation Method | EVs Characterization Method (EVs Parameters) | EVs quantification Method | Type of Biomarker | Techniques for Biomarker Identification and Quantification | Specific Biomarker | Expression Level | Cases (N) | Healthy Controls (N) | CIN Controls (N) | Sensitivity (%) | Specificity (%) | AUC (IC 95%) | OR (IC 95%) | p Value | Included in the Meta-Analyses |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ding, Xian-zhen et al., 2020 [21] | China | Prospective cohort study | Serum | Total Exosome Isolation Kit (Invitrogen, CA, USA) | Western Blot (CD63, CD9, GM130) | Not performed | lncRNA | RNA extraction: miRNeasy Micro Kit (QIAGEN, Hilden, DE) RNA quality: Agilent 2100 Bioanalyzer RNA analysis: RT-qPCR (Taqman probes, Thermo Waltham, MA, USA) | lncRNA DLX6-AS1 | Upregulated (significantly higher) | 114 | 110 | 60 | 78.1% (CC vs. Healthy); 75.4% (CC vs. CIN) | 88.2% (CC vs. Healthy); 71.8% (CC vs. CIN) | 0.892 (95% CI: 0.844–0.929) for CC vs. Healthy; 0.831 (95% CI: 0.775–0.877) for CC vs. CIN | OS: Univariate analysis: HR = 3.54 (95% CI: 1.852–6.422, p = 0.008); Multivariate analysis: HR = 3.38 (95% CI: 1.742–6.178, p = 0.009) | Univariated: p: 0.008; multivariated: p: 0.009 | yes |
| Guo Yaqiong 2020 [22] | China | Retrospective cross-sectional study | Serum | exoRNeasy Serum/ Plasma Midi Kit (QIAGEN, Hilden, DE) | Western Blot (CD9, CD63, GAPDH) | Not performed | lncRNA | RNA extraction: miRNeasy Kit | lncRNA-EXOC7 | Higher level | 101 (52 pre-operative/newly diagnosed cases, 30 post-operative cases, and 19 recurrence cases) | 50 | Not included | Not provided | Not provided | 0.9388 for serum and 0.8982 for exosomal lncRNA-EXOC7 | Not provided | 0.041 for FIGO stage correlation with serum expression of lncRNA-EXOC7 | yes |
| Cho O. et al., 2021 [23] | South Korea | Retrospective cohort study with a nested case-control element | Plasma | Exo2D (ExosomePlus, Seoul, Republic of Korea). | Not provided | Not performed | miRNA, mRNA, snoRNA | RNA extraction: RNeasy Serum/Plasma Kit (Qiagen)RNA quantification: Quant-IT Ribogreen (Invitrogen Carlsbad, CA, USA) RNA quality: Agilent 2100 Bioanalyzer RNA analysis: small RNA sequencing (Ilumina, San Diego, CA, USA) | 13 miRNAs; 43 piRNAs; 28 lncRNAs, and 67 mRNAs →15 selected RNAs: miRNA: miR-142-3p; mRNAs: CXCL5, KIF2A, RGS18, ARL6IP5, and DAPP1; lncRNA: LINC00989 (via targeting RGS18); snoRNAs: SNORD17, SCARNA12, SNORA6, SNORA12, SCRNA1, SNORD97, SNORD62, and SNORD38A | Upregulated: RGS18. Downregulated: SNORA12; SNORD97 | 30 | 12 | Not included | RGS18 + SNORA12 + SNORD97: 96.7%; RGS18: 96.7%; SNORA12 + SNORD97: 73.3%. | RGS18 + SNORA12 + SNORD97: 100%; RGS18: 91.7%; SNORA12 + SNORD97: 100%. | RGS18 + SNORA12 + SNORD97: 0.992; RGS18: 0.964; SNORA12 + SNORD97: 0.883 | Not provided | p < 0.01 | yes |
| Li et al., 2024 [24] | China | Retrospective cross-sectional study | Plasma | Ultracentrifugation | - Transmission electron microscopy (TEM) - Nanoparticle tracking analysis (NTA): qNano (Izon Science Ltd., New Zealand) - Western blot (CD9, GM130 and TSG 101) | Not performed | tRNA | RNA extraction: Trizol RNA analysis: RT-qPCR | tRF-Phe-GAA-001; tRF-Gly-GCC-037 | Downregulated | 140 (plasma) | 140 (plasma) | Not included | For plasma exosomal tRF-Phe-GAA-001: 72.1%; tRF-Gly-GCC-037: 83.6%; Combined: 92.9%; For early stage: tRF-Phe-GAA-001: 80.2%% tRF-Gly-GCC-037: 81.5% | For plasma exosomal tRF-Phe-GAA-001: 85.7%; tRF-Gly-GCC-037: 69.3%; Combined: 83.6%; For early stage: tRF-Phe-GAA-001: 81.4% tRF-Gly-GCC-037: 69.3% | For plasma exosomal tRF-Phe-GAA-001: 0.8682; tRF-Gly-GCC-037: 0.8283; Combined: 0.9337 For early-stage CC plasma samples: tRF-Phe-GAA-001: 0.8963; tRF-Gly-GCC-037: 0.8103; Combined: 0.9432 | Not provided | p < 0.0001 for both. | yes |
| Lv et al., 2021 [25] | China | Cross-sectional | Plasma | ExoEasy Maxi kit (Qiagen) | - Transmission electron microscopy (TEM) - Nanoparticle Tracking Analysis (NTA): ZetaView PMX | Not performed | miRNA | RNA extraction: Discovery cohort: miRNeasy Serum/Plasma Kit (Qiagen Hilden, DE, USA) Validation cohort: miRNeasy kit RNA analysis: Discovery cohort: miRNA sequencing (Illumina; San Diego, CA, USA); Validation cohort: RT-qPCR (syber green, giagen) | miR-125a-5p | Downregulated | Discovery cohort: 6 (cervical cancer) Validation cohort: 38 (cervical cancer) | Discovery cohort: 6 Validation cohort: 22 | Not included | 59.1% (at a cut-off value of 2.537 for miR-125a-5p) | 84.2% (at a cut-off value of 2.537 for miR-125a-5p) | 0.7129 (95% CI, 0.561–0.865) | Not provided | p < 0.001 for initial miRNA differential expression and p < 0.001 for difference in miR-125a-5p levels between cases and controls. | yes |
| Ma et al., 2019 [26] | China | Retrospective cross-sectional study | Plasma | ExoQuick exosome precipitation solution (System Biosciences, Palo Alto, CA, USA) | Not performed | Not performed | miRNA | RNA extraction: MirVana Paris Kit (Ambion, Austin, TX, USA)/Trizol RNA quantification: Quant-IT Ribogreen (Invitrogen, Carlsbad, CA, USA) RNA quality: Agilent 2100 Bioanalyzer RNA analysis: RT-qPCR (SYBR Green, TaKaRa, Kusatsu, Japan) | miR-146a-5p, miR-151a-3p, miR-2110, miR-21-5p (a four-miRNA panel). The expression of miR-21-5p was also increased in plasma exosomes of CC patients compared with those in NCs, albeit not reaching statistical significance. | Upregulated | 24 (cervical cancer samples) All samples used for exosome analysis belonged to validation set. | 24. All samples used for exosome analysis belonged to validation set. | Not included | Not provided | Not provided | Not provided for exossome samples | Not provided | miR-146a-5p: p: 0.041; miR-151a-3p p: 0.08; miR-2110: p.032; miR-21-5p: p: 0.079 | no |
| Zheng et al., 2019 [27] | China | Retrospective cohort study with a nested case-control element | Plasma | ExoQuick Exosome Precipitation Solution (SBI Cat #:100356EXOQ20A-1, Mountain View, CA, USA) mixture with RNase A | Not provided | Not performed | miRNA | RNA extraction: Discovery cohort: miRNeasy Micro Kit Validation cohort: miRNeasy Micro Kit RNA quantification/quality: Discovery cohort: Agilent 2100 Bioanalyzer-Santa Clara, CA, USA; Qubit Validation cohort: Agilent 2100 Bioanalyzer; Qubit RNA analysis: Discovery cohort: miRNA sequencing (Ilumina, San Diego, CA, USA) Validation cohort: ddPCR | Eight miRNAs (let-7a-3p, let-7d-3p, miR-30d-5p, miR-144-5p, miR-182-5p, miR-183-5p, miR-215-5p, and miR-4443). Best predictors: let-7d-3p, miR-30d-5p | Down-regulated in CIN II+ compared to CIN I | Discovery cohort: 34 Validation cohort: 63 CC | Discovery cohort: 23 Validation cohort: 50 | Discovery cohort: 5 CIN I, 59 CIN II-III | Not provided | Not provided | 0.922 (for miRNA panel) | Not provided | Not provided | yes |
| Molika et al., 2023 [28] | Thailand | Retrospective cohort study with a nested case-control element | Serum | Differential ultracentrifugation (dUC) combined with size-exclusion chromatography (SEC) | Transmission electron microscopy (TEM) and Western blot (CD9, CD63, CD81, Cytocrome C) | Not performed | Protein | LC-MS/MS (Liquid Chromatography with tandem Mass Spectrometry) | ERI3, COX5A, SGSM3 (identified as uniquely expressed proteins in sEVs from CC patients; validation pending) | Up regulated (high level at CC) | 90 | 30 | Not included | Not provided | Not provided | Not provided | Not provided | several | no |
| Ao, K. et al., 2024 [29] | China | Cross-sectional | Plasma | Discovery cohort: ExoQuick test Validation cohort: Ultracentrifugation (UC) | Transmission electron microscopy (TEM) (morphology, size), Nanoparticle Tracking Analysis (NTA) (size) | Not performed | Protein | Discovery cohort: Label-free Quantitative LC/MS Proteomic analysis Validation cohort: Enzyme-linked immunosorbent assay (ELISA) | Mortalin (HSPA9) | Up regulated (high level at CC) | 19 (prior to stage IIa) | 15 (individuals with cervicitis as normal controls) | 15 (patients with precancerous lesions—HSIL or CINII/III) | Not provided | p < 0.01 | no | |||
| Study (Author/Year) | Country | Study Type | Sample Type | EVs Isolation Method | EVs Characterization Method | EVs Quantification Method | Type of Biomarker | Techniques for Biomarker Identification and Quantification | Specific Biomarker | Expression Level | Cases (N) | Controls (N) | Sensitivity (%) | Specificity (%) | AUC (IC 95%) | OR (IC 95%) | p Value | Included in the Meta-Analyses |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Guo, Yaqiong 2020 [22] | China | Retrospective cross-sectional study | Serum | exoRNeasy Serum/Plasma Midi Kit (QIAGEN, Hilden, Germany) | Western Blot (CD9, CD63, GAPDH) | Not performed | lncRNA | RNA extraction: miRNeasy Kit | lncRNA-EXOC7 | Higher level | 101 (52 pre-operative/newly diagnosed cases, 30 post-operative cases, and 19 recurrence cases) | 50 | Not provided | Not provided | 0.9333 | Not provided | Exosomal level of lncRNA-EXOC7 was correlated with the FIGO stage (p = 0.010) | no |
| Cho O. et al., 2021 [31] | South Korea | Prospective cohort study | Plasma | Exo2D (ExosomePlus, Seoul, Republic of Korea). | Not provided | Not performed | miRNA | RNA extraction: RNeasy Serum/Plasma Kit (Qiagen, Hilden, Germany) RNA quantification: Quant-IT Ribogreen (Invitrogen, Carlsbad, CA, USA) RNA quality: Agilent 2100 Bioanalyzer, Santa Clara, CA, USA. RNA analysis: small RNA sequencing (Ilumina, San Diego, CA, USA) | miR-1228-5p, miR-146a-3p, miR-33a-5p, miR-3200-3p, miR-501-3p, and miR-6815-5p | Upregulated | 28 (cases: two weeks after the initiation of CCRT) | Before and after CCRT | Not provided | Not provided | Not provided | Not provided | Not provided | no |
| Molika, Piyatida et al. 2024 [32] | Thailand | Prospective Cohort Study (with cross-sectional elements) | Serum | Differential ultracentrifugation | - Transmission electron microscopy (TEM) - Western blot (CD9, CD63, Alix, albumin, and cytochrome C) | Not performed | lncRNA | RNA extraction: ExoRNeasy Kit (Qiagen, Hilden, Germany) RNA quantification: Agilent 2100 Bioanalyzer RNA quality: Agilent 2100 Bioanalyzer Santa Clara, CA, USA RNA analysis: small RNA sequencing (Ilumina, San Diego, CA, USA) | LINC00941, LINC00482, LINC01116, LINC02274, LINC01489, LINC01910, LINC02454, DSG2-AS1, HCG15, TCEAL3-AS1, LINC01960, LINC01398, LINC02251, RNASEH1-AS1, and LINC01271 | Upregulated | 60 (Cervical cancer patients, staged I–III) | 27 healthy | Not provided | Not provided | Not provided | Not provided | p < 0.05 | no |
| Someya et al., 2023 [30] | Japan | Prospective Cohort Study (with cross-sectional elements) | Plasma | exoRNeasy Serum/Plasma Kit (QIAGEN, Hilden, Germany) | Not performed | Not performed | miRNA | RNA quality: Agilent 2100 Bioanalyzer, Santa Clara, CA, USA RNA analysis: mir-RNA sequencing (Ilumina, San Diego, CA, USA) | Nine-miRNA signature (miR-148a-5p, miR-1915-3p, miR-3960, miR-183-5p, miR-196b-5p, miR-200c-3p, miR-182-5p, miR-374a-5p, miR-431-5p) | Down expression at recurrence: miR-148a-5p; miR-1915-3p; miR-3960; UP expression: miR-183-5p; miR-183-5p; miR-200c-3p; miR-182-5p; miR-374a-5p miR-431-5p | 45 | Recurrence (19: 15 deaths) vs. non recurrence (26) | Not provided | Not provided | miR-148a-5p: 0.713 miR-1915-3p: 0.682 miR-3960: 0.806;miR-183-5p: 0.680; miR-183-5p 0.686; miR-200c-3p 0.721; miR-182-5p 0.709; miR-374a-5p 0.729; miR-431-5p: 0.729 | miR-148a-5p: 4.7 miR-1915-3p: 4.1 miR-3960: 22.4;miR-183-5p: 3.8; miR-183-5p: 5.9; miR-200c-3p: 11.9; miR-182-5p 5.9; miR-374a-5p 5.9; miR-431-5p: 6.3 | miR-148a-5p: 0.007 miR-1915-3p: 0.031 miR-3960: >0.001; miR-183-5p: 0.027; miR-183-5p: 0.028; miR-200c-3p: 0.006; miR-182-5p: 0.007; miR-374a-5p: 0.003; miR-431-5p: 0.003 | no |
| Study/ Cases (N)/ Histology | Age a CC (Years) | FIGO Staging (N) | Healthy Controls (N) | CIN Controls (N) | HPV Infection Cases | EV Biomarker | Main Findings |
|---|---|---|---|---|---|---|---|
| [21] N: 114 (Histology not provided) | <50:56 ≥50:58 | I–II: 79 III–IV: 35 | 110 | 60 | Negative: 17 Positive: 97 | lncRNA DLX6-AS1 | Serum exosomal lncRNA DLX6-AS1 levels were significantly higher in CC patients than in CIN and healthy controls (both p < 0.001); Higher serum exosomal lncRNA DLX6-AS1 levels were observed in CIN patients compared with controls (p = 0.011). |
| [22] b N: 101 (Histology not provided) | <50:51 ≥50:50 | IB–IIA: 38 IIB–IIIA: 27 IIIB–IV: 36 | 50 | Not included | Not provided | lncRNA-EXOC7 (serum and exosomal) | Newly diagnosed CC cases presented a 3.18-fold higher serum level of lncRNA-EXOC7 and a 2.93-fold higher exosomal level of lncRNA-EXOC7 than healthy subjects. ROC curve analysis showed that the AUCs of serum and exosomal lncRNA-EXOC7 in distinguishing CC patients from healthy controls were 0.9338 and 0.8982, respectively. Exosomal level was also elevated in recurrent cases (p < 0.0001, AUC = 0.9123). |
| [23] N: 30: SCC: 22; AC: 4; ACC 2; UC: 2 | 49.2 ± 11.6 | 12 | Not included | Not provided | miRNA: miR-142-3p; mRNAs: CXCL5, KIF2A, RGS18, APL6IP5, DAPP1; snoRNAs: SNORD17, SCARNA12, SNORA6, SNORA12, SCRNA1, SNORD97, SNORD62, SNORD38A | SNORA12: the most relative to ALC2 among other snoRNAs, which showed that snoRNA was more suitable for cancer diagnose than the other three RNA classes (e.g., snRNA, tRNA, and yRNA) because it had an additional four RNAs that were differentially expressed in the cancer group. | |
| [24] N: 140 SCC: 96; AC: 44 | <55:72 ≥55:68 | I–IIA:81 IIB–IV: 59 | 140 | Not included | Positive: 129 Negative: 11 | tRF-Phe-GAA-001; tRF-Gly-GCC-037 | Significant decrease in the levels of exosomal tRF-Phe-GAA-001 and tRF-Gly-GCC-037 compared to healthy controls (p < 0.0001 for both). combination, the AUC rose to 0.9337, with a sensitivity of 92.9% and specificity of 83.6%. |
| [25] N: 38 (SCC:28; AC: 10) | <50:6 ≥50:32 | IA–IIA: 29 IIB–IVB: 9 Missing: 8 | 22 | Not included | Positive: 30 Negative: 8 | miR-125a-5p | Level of plasma exosomal miR-125a-5p was a potential marker for differentiating between non-cervical and cervical cancer, with an ROC area under the curve value of 0.7129 [95% confidence interval (CI), 0.561–0.865] |
| [26] N: 97 (combined phase) (SCC: 76; AC: 21) | ≥60:12 <60:85 | I: 74 II: 23 | 87 | Not included | Not provided | miR-146a-5p, miR-151a-3p, miR-2110, miR-21-5p | The expression of miR-21-5p was increased in plasma exosomes of CC patients compared with those in NCs, albeit not reaching statistical significance. |
| [27] N: 34 Cases defined as CIN II + (59 CINII-III + 34 CC: SCC: 21 and AC:13) | 50 ± 24 | Not provided | 23 | 5 | Not provided | Eight miRNAs (let-7a-3p, let-7d-3p, miR-30d-5p, miR-144-5p, miR-182-5p, miR-183-5p, miR-215-5p, and miR-4443). Best predictors (plasma and tissue): let-7d-3p and miR-30d-5p | AUC for miRNA panel, discriminate CIN II+ from Healthy/CIN I: 0.922. Individual AUC DEmiRs: let-7a-3p: 0.811; let-7d-3d: 0.888; mir-30d-5p: 0.915; mir-215-5p: 0.817; miR-144-5p: 0.882; miR-182-5p: 0.773; miR-183-5p: 0.814; miR-443: 0.817. |
| [28] N: 90 (Histology not provided) | 30–65 | Not provided | 30 | Not included | Not provided | ERI3, COX5A, SGSM3 | ERI3, COX5A, and SGSM3 are uniquely expressed proteins in EVs derived from patients with CC. |
| [29] N: 19 SCC | 43.6 (24–70) | IA1:5 IB1:4 IIA: 3 IIB: 1 Missing: 6 | 15 | 15: CINII: 4 CINIII: 4 | HPV 16: 12 HPV 18: 1 HPV 31: 2 HPV 58: 2 Negative: 2 | Mortalin (or HSPA9) | The CC group boasted a higher content of mortalin in comparison to the HSIL or Control (p < 0.01). Promising target for clinical interventions as well as an early warning indicator. |
| Study/Cases (N)/Histology | Age CC (Years) | FIGO Staging (N) | Controls (N) | EV Biomarker | Main Findings |
|---|---|---|---|---|---|
| [30] N: 45 SCC: 44 AC: 1 | 44–79 (56) | I: 2 IIa + IIb: 9 IIIa + IIIb: 17 IVa + IVb: 17 | Recurrence-free (26) and recurrence (19) | Lower expression recurrence: Higher expression: miR-183-5p, miR-200c-3p, miR-182-5p, miR-374a-5p, miR-431-5p | miRNA-based risk score, along with tumor size and infiltrating FOXP3+ T cells, were significant factors in predicting disease-specific survival. (p < 0.001). |
| [31] N:28 SCC: 22 AC: 4 ACC: 1; UC 1 | 50.0 (42.5; 56.0) | IB: 4 IIB: 1 IIIC1: 13 IIIC2: 6 IVA: 1 IVB: 3 | Before and after two weeks of treatment | miR-1228-5p, miR-146a-3p, miR-33a-5p, miR-3200-3p, miR-501-3p, and miR-6815-5p | log2FC of miRNAs contained within plasma exosomes are associated with early progression and metastasis in patients with cervical cancer. miR-16-1-3p (or 15a-3p) may be a potential upstream regulator important for the suppression of metastasis by modulation of the tumor microenvironment. |
| [32] N: 60 Not provided | Not provided | 27 healthy controls | LINC00941, LINC01910, LINC02454 | The EV lncRNAs upregulated in stage III patients compared with those in HC are involved in multiple biological processes, including HPV infection, MAPK signaling pathway, and focal adhesion. High expression of LINC00941, LINC01910, LINC02454, and DSG2-AS1 were associated with poor OS of patients with CC. |
| Outcomes a | Study Design | Risk of Bias in Individual Studies | Publication Bias | Inconsistency | Indirectness | Imprecision | Confidence in Evidence | Recommendation |
|---|---|---|---|---|---|---|---|---|
| Diagnosis and/or Prognosis | 12 observational studies. | High b | No bias detected c | High d | High e | High f | ⊕ Very low. | No recommendation can be provided based on existing data. |
| Risk of Bias | Applicability Concerns | ||||||
|---|---|---|---|---|---|---|---|
| Study (Author/Year) | Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard |
| Ao, K. et al., 2024 [29] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Cho O.et al., 2021 [23] | ☹ | ☹ | ☺ | ☺ | ☹ | ? | ☺ |
| Ding.Xian-zhen et al., 2020 [21] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Guo Ya-qiong, 2020 [22] | ☹ | ? | ☺ | ☺ | ☹ | ? | ☺ |
| Li et al., 2024 [24] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Lv et al., 2021 [25] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Ma et al., 2019 [26] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Molika et al., 2023 [28] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Zheng et al., 2019 [27] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Cho O. et al., 2021 [31] | ☹ | ? | ☺ | ☺ | ☹ | ? | ☺ |
| Molika, Piyatida et al., 2024 [32] | ☹ | ☺ | ☺ | ☺ | ☹ | ? | ☺ |
| Someya et al., 2023, [30] | ☹ | ? | ☺ | ☺ | ☹ | ? | ☺ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, F.; Caramelo, F.A.; Tomaz, J.M.P.; Santana, M.M.; Nobre, R.J.; Almeida, L.P.; Figueiredo-Dias, M. Exosomes as Emerging Non-Invasive Biomarkers of Cervical Cancer: A Systematic Review and Meta-Analysis. Cancers 2025, 17, 3945. https://doi.org/10.3390/cancers17243945
Santos F, Caramelo FA, Tomaz JMP, Santana MM, Nobre RJ, Almeida LP, Figueiredo-Dias M. Exosomes as Emerging Non-Invasive Biomarkers of Cervical Cancer: A Systematic Review and Meta-Analysis. Cancers. 2025; 17(24):3945. https://doi.org/10.3390/cancers17243945
Chicago/Turabian StyleSantos, Fernanda, Francisco A. Caramelo, Jorge M. P. Tomaz, Magda M. Santana, Rui J. Nobre, Luis P. Almeida, and Margarida Figueiredo-Dias. 2025. "Exosomes as Emerging Non-Invasive Biomarkers of Cervical Cancer: A Systematic Review and Meta-Analysis" Cancers 17, no. 24: 3945. https://doi.org/10.3390/cancers17243945
APA StyleSantos, F., Caramelo, F. A., Tomaz, J. M. P., Santana, M. M., Nobre, R. J., Almeida, L. P., & Figueiredo-Dias, M. (2025). Exosomes as Emerging Non-Invasive Biomarkers of Cervical Cancer: A Systematic Review and Meta-Analysis. Cancers, 17(24), 3945. https://doi.org/10.3390/cancers17243945





