MRI-Based Radiomics Ensemble Model for Predicting Radiation Necrosis in Brain Metastasis Patients Treated with Stereotactic Radiosurgery and Immunotherapy
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participation
2.2. Radiosurgery Technique
2.3. Response Assessment and Necrosis Definition
2.4. Image Processing and Radiomic Feature Extraction
2.5. Feature Dimensionality Reduction
2.6. Model Development and Validation
2.7. Model Interpretation
2.8. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. Model Evaluation
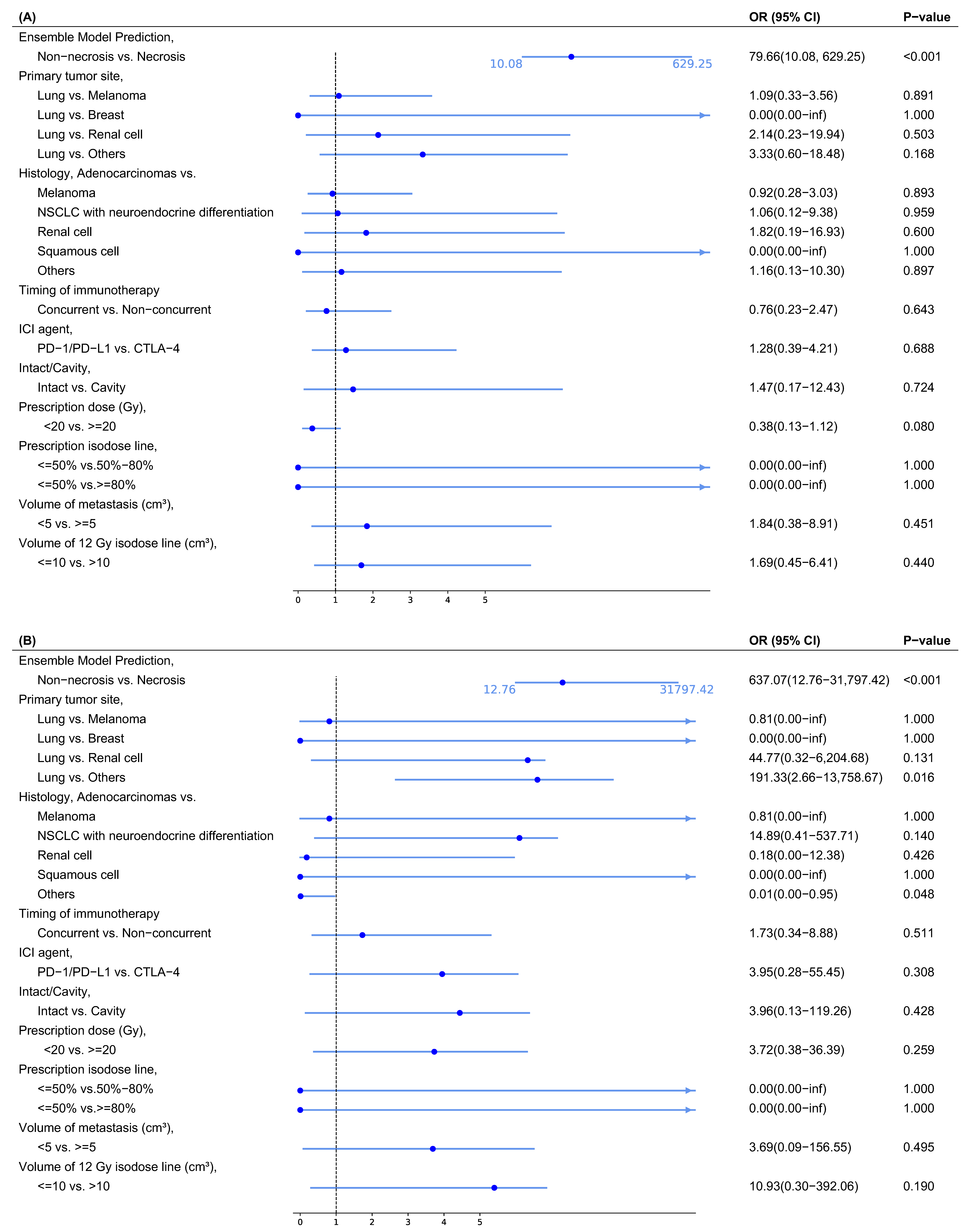
3.3. Model Association with Radiation Necrosis
3.4. Feature Importance and Clinical Relevance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lassman, A.B.; DeAngelis, L.M. Brain metastases. Neurol. Clin. 2003, 21, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.H.; Kotecha, R.; Chao, S.T.; Ahluwalia, M.S.; Sahgal, A.; Chang, E.L. Current approaches to the management of brain metastases. Nat. Rev. Clin. Oncol. 2020, 17, 279–299. [Google Scholar] [PubMed]
- Vaios, E.J.; Winter, S.F.; Shih, H.A.; Dietrich, J.; Peters, K.B.; Floyd, S.R.; Kirkpatrick, J.P.; Reitman, Z.J. Novel Mechanisms and Future Opportunities for the Management of Radiation Necrosis in Patients Treated for Brain Metastases in the Era of Immunotherapy. Cancers 2023, 15, 2432. [Google Scholar] [CrossRef]
- Griffith, B.; Jain, R. Perfusion Imaging in Neuro-Oncology: Basic Techniques and Clinical Applications. Radiol. Clin. N. Am. 2015, 53, 497–511. [Google Scholar]
- Lee, D.; Riestenberg, R.A.; Haskell-Mendoza, A.; Bloch, O. Brain Metastasis Recurrence Versus Radiation Necrosis: Evaluation and Treatment. Neurosurg. Clin. N. Am. 2020, 31, 575–587. [Google Scholar] [CrossRef]
- Salari, E.; Elsamaloty, H.; Ray, A.; Hadziahmetovic, M.; Parsai, E.I. Differentiating Radiation Necrosis and Metastatic Progression in Brain Tumors Using Radiomics and Machine Learning. Am. J. Clin. Oncol. 2023, 46, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Parekh, V.S.; Lee, E.; Chen, X.; Redmond, K.J.; Pillai, J.J.; Peng, L.; Jacobs, M.A.; Kleinberg, L.R. A Multidimensional Connectomics- and Radiomics-Based Advanced Machine-Learning Framework to Distinguish Radiation Necrosis from True Progression in Brain Metastases. Cancers 2023, 15, 4113. [Google Scholar]
- Florez, E.; Nichols, T.; Parker, E.E.; Lirette, S.T.; Howard, C.M.; Fatemi, A. Multiparametric Magnetic Resonance Imaging in the Assessment of Primary Brain Tumors Through Radiomic Features: A Metric for Guided Radiation Treatment Planning. Cureus 2018, 10, e3426. [Google Scholar] [CrossRef]
- Chen, X.; Parekh, V.S.; Peng, L.; Chan, M.D.; Redmond, K.J.; Soike, M.; McTyre, E.; Lin, D.; Jacobs, M.A.; Kleinberg, L.R. Multiparametric radiomic tissue signature and machine learning for distinguishing radiation necrosis from tumor progression after stereotactic radiosurgery. Neuro-Oncol. Adv. 2021, 3, vdab150. [Google Scholar]
- Lyu, Q.; Namjoshi, S.V.; McTyre, E.; Topaloglu, U.; Barcus, R.; Chan, M.D.; Cramer, C.K.; Debinski, W.; Gurcan, M.N.; Lesser, G.J.; et al. A transformer-based deep-learning approach for classifying brain metastases into primary organ sites using clinical whole-brain MRI images. Patterns 2022, 3, 100613. [Google Scholar] [CrossRef]
- Acquitter, C.; Piram, L.; Sabatini, U.; Gilhodes, J.; Moyal Cohen-Jonathan, E.; Ken, S.; Lemasson, B. Radiomics-Based Detection of Radionecrosis Using Harmonized Multiparametric MRI. Cancers 2022, 14, 286. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yang, Z.; Zeng, Z.; Ren, H.; Jiang, M.; Hu, Y.; Xu, Y.; Zhang, H.; Ma, K.; Long, L. MRI-based radiomics models for the early prediction of radiation-induced temporal lobe injury in nasopharyngeal carcinoma. Front. Neurol. 2023, 14, 1135978. [Google Scholar]
- Zhou, X.; Yu, Y.; Feng, Y.; Ding, G.; Liu, P.; Liu, L.; Ren, W.; Zhu, Y.; Cao, W. Attention mechanism based multi-sequence MRI fusion improves prediction of response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Radiat. Oncol. 2023, 18, 175. [Google Scholar]
- Jain, P.; Santhanalakshmi, S. Early detection of brain tumor and survival prediction using deep learning and an ensemble learning from radiomics images. In Proceedings of the 2022 IEEE 3rd Global Conference for Advancement in Technology (GCAT), Bangalore, India, 7–9 October 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–9. [Google Scholar]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. An ensemble learning approach for brain cancer detection exploiting radiomic features. Comput. Methods Programs Biomed. 2020, 185, 105134. [Google Scholar]
- Dholey, M.; Santosham, R.J.M.; Ray, S.; Das, J.; Chatterjee, S.; Ahmed, R.; Mukherjee, J. Ensemble Methods with [18F]FDG-PET/CT Radiomics in Breast Cancer Response Prediction; Springer Nature: Cham, Switzerland, 2023; pp. 369–379. [Google Scholar]
- Jain, M.; Rai, C.; Jain, J. A Novel Method for Differential Prognosis of Brain Degenerative Diseases Using Radiomics-Based Textural Analysis and Ensemble Learning Classifiers. Comput. Math. Methods Med. 2021, 2021, 7965677. [Google Scholar] [CrossRef]
- Li, Z. Extracting spatial effects from machine learning model using local interpretation method: An example of SHAP and XGBoost. Comput. Environ. Urban Syst. 2022, 96, 101845. [Google Scholar]
- Van den Broeck, G.; Lykov, A.; Schleich, M.; Suciu, D. On the tractability of SHAP explanations. J. Artif. Intell. Res. 2022, 74, 851–886. [Google Scholar]
- Garreau, D.; Luxburg, U. Explaining the explainer: A first theoretical analysis of LIME. In Proceedings of the International Conference on Artificial Intelligence and Statistics: PMLR, Valencia, Spain, 2–4 May 2020; pp. 1287–1296. [Google Scholar]
- Slack, D.; Hilgard, S.; Jia, E.; Singh, S.; Lakkaraju, H. Fooling lime and shap: Adversarial attacks on post hoc explanation methods. In Proceedings of the AAAI/ACM Conference on AI, Ethics, and Society, New York, NY, USA, 7–9 February 2020; pp. 180–186. [Google Scholar]
- Visani, G.; Bagli, E.; Chesani, F.; Poluzzi, A.; Capuzzo, D. Statistical stability indices for LIME: Obtaining reliable explanations for machine learning models. J. Oper. Res. Soc. 2022, 73, 91–101. [Google Scholar]
- Ruben, J.D.; Dally, M.; Bailey, M.; Smith, R.; McLean, C.A.; Fedele, P. Cerebral radiation necrosis: Incidence, outcomes, and risk factors with emphasis on radiation parameters and chemotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 499–508. [Google Scholar] [PubMed]
- Korytko, T.; Radivoyevitch, T.; Colussi, V.; Wessels, B.W.; Pillai, K.; Maciunas, R.J.; Einstein, D.B. 12 Gy gamma knife radiosurgical volume is a predictor for radiation necrosis in non-AVM intracranial tumors. Int. J. Radiat. Oncol. Biol. Phys. 2006, 64, 419–424. [Google Scholar]
- Kim, J.M.; Miller, J.A.; Kotecha, R.; Xiao, R.; Juloori, A.; Ward, M.C.; Ahluwalia, M.S.; Mohammadi, A.M.; Peereboom, D.M.; Murphy, E.S.; et al. The risk of radiation necrosis following stereotactic radiosurgery with concurrent systemic therapies. J. Neuro-Oncol. 2017, 133, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.H.; Marcrom, S.R.; Patel, M.P.; Popple, R.A.; Travis, R.L.; McDonald, A.M.; Riley, K.O.; Markert, J.M.; Willey, C.D.; Bredel, M.; et al. Understanding the Effect of Prescription Isodose in Single-Fraction Stereotactic Radiosurgery on Plan Quality and Clinical Outcomes for Solid Brain Metastases. Neurosurgery 2023, 93, 1313–1318. [Google Scholar] [CrossRef]
- Barillaro, A.; Caroprese, M.; Cella, L.; Viggiano, A.; Buccelli, F.; Daponte, C.; Feoli, C.; Oliviero, C.; Clemente, S.; Farella, A.; et al. Stereotactic Radiation Therapy for Brain Metastases: Factors Affecting Outcomes and Radiation Necrosis. Cancers 2023, 15, 2094. [Google Scholar] [CrossRef] [PubMed]
- Helis, C.A.; Hughes, R.T.; Glenn, C.W.; Lanier, C.M.; Masters, A.H.; Dohm, A.; Ahmed, T.; Ruiz, J.; Triozzi, P.; Gondal, H.; et al. Predictors of Adverse Radiation Effect in Brain Metastasis Patients Treated with Stereotactic Radiosurgery and Immune Checkpoint Inhibitor Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 295–303. [Google Scholar] [CrossRef]
- Minniti, G.; Clarke, E.; Lanzetta, G.; Osti, M.F.; Trasimeni, G.; Bozzao, A.; Enrici, R.M. Stereotactic radiosurgery for brain metastases: Analysis of outcome and risk of brain radionecrosis. Radiat. Oncol. 2011, 6, 48. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Gorgolewski, K.; Burns, C.D.; Madison, C.; Clark, D.; Halchenko, Y.O.; Waskom, M.L.; Ghosh, S.S. Nipype: A flexible, lightweight and extensible neuroimaging data processing framework in python. Front. Neuroinform. 2011, 5, 12318. [Google Scholar] [CrossRef]
- Shinohara, R.T.; Sweeney, E.M.; Goldsmith, J.; Shiee, N.; Mateen, F.J.; Calabresi, P.A.; Jarso, S.; Pham, D.L.; Reich, D.S.; Crainiceanu, C.M. Statistical normalization techniques for magnetic resonance imaging. NeuroImage Clin. 2014, 6, 9–19. [Google Scholar] [CrossRef]
- Depeursinge, A.; Andrearczyk, V.; Whybra, P.; van Griethuysen, J.; Müller, H.; Schaer, R.; Vallières, M.; Zwanenburg, A. Standardised convolutional filtering for radiomics. arXiv 2020, arXiv:2006.05470. [Google Scholar]
- Whybra, P.; Zwanenburg, A.; Andrearczyk, V.; Schaer, R.; Apte, A.P.; Ayotte, A.; Baheti, B.; Bakas, S.; Bettinelli, A.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Convolutional Filters for Reproducible Radiomics and Enhanced Clinical Insights. Radiology 2024, 310, e231319. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature selection: A data perspective. ACM Comput. Surv. (CSUR) 2017, 50, 1–45. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Arlot, S.; Celisse, A. A survey of cross-validation procedures for model selection. Statist. Surv. 2010, 4, 40–79. [Google Scholar] [CrossRef]
- Fitzgerald, M.; Saville, B.R.; Lewis, R.J. Decision curve analysis. JAMA 2015, 313, 409–410. [Google Scholar] [CrossRef]
- Nachar, N. The Mann-Whitney U: A test for assessing whether two independent samples come from the same distribution. Tutor. Quant. Methods Psychol. 2008, 4, 13–20. [Google Scholar] [CrossRef]
- Greenwood, P.E.; Nikulin, M.S. A Guide to Chi-Squared Testing; John Wiley & Sons: Hoboken, NJ, USA, 1996. [Google Scholar]
- Peng, C.-Y.J.; Lee, K.L.; Ingersoll, G.M. An introduction to logistic regression analysis and reporting. J. Educ. Res. 2002, 96, 3–14. [Google Scholar] [CrossRef]
- Sperandei, S. Understanding logistic regression analysis. Biochem. Medica 2014, 24, 12–18. [Google Scholar] [CrossRef]

| Training Cohort (n = 125) | Validation Cohort (n = 84) | ||||||
|---|---|---|---|---|---|---|---|
| Necrosis (n = 10) | Non-Necrosis (n = 115) | p-Value | Necrosis (n = 5) | Non-Necrosis (n = 79) | p-Value | ||
| Gender | 0.465 | 0.739 | |||||
| Female | 6 (60) | 49 (43) | 3 (60) | 33 (42) | |||
| Male | 4 (40) | 66 (57) | 2 (40) | 46 (58) | |||
| Age | 0.041 | 0.449 | |||||
| <=60 | 7 (70) | 43 (37) | 2 (40) | 27 (34) | |||
| >60 | 3 (30) | 72 (63) | 3 (60) | 52 (66) | |||
| Death | 0.737 | 0.572 | |||||
| Dead | 5 (50) | 70 (61) | 2 (40) | 50 (63) | |||
| Not Dead | 5 (50) | 45 (39) | 3 (60) | 29 (37) | |||
| Primary tumor site | 0.418 | 0.390 | |||||
| Lung | 5 (50) | 61 (53) | 2 (40) | 44 (56) | |||
| Melanoma | 3 (30) | 40 (35) | 2 (40) | 29 (37) | |||
| Breast | 0 (0) | 4 (3) | 0 (0) | 0 (0) | |||
| Renal cell | 0 (0) | 4 (3) | 1 (20) | 3 (4) | |||
| Other | 2 (20) | 6 (5) | 0 (0) | 3 (4) | |||
| Histology | 0.820 | 0.357 | |||||
| Adenocarcinomas | 6 (60) | 53 (46) | 1 (20) | 36 (46) | |||
| NSCLC with neuroendocrine differentiation | 0 (0) | 9 (8) | 1 (20) | 3 (4) | |||
| Melanoma | 3 (30) | 40 (35) | 2 (40) | 29 (37) | |||
| Renal cell carcinoma | 0 (0) | 3 (3) | 1 (20) | 4 (5) | |||
| Squamous cell carcinoma | 0 (0) | 4 (3) | 0 (0) | 2 (3) | |||
| Other | 1 (10) | 6 (5) | 0 (0) | 5 (6) | |||
| Laterality | 0.766 | 0.008 | |||||
| Left | 6 (60) | 61 (53) | 0 (0) | 35 (44) | |||
| Right | 4 (40) | 49 (43) | 4 (80) | 43 (54) | |||
| Midline | 0 (0) | 5 (4) | 1 (20) | 1 (1) | |||
| Location | 0.984 | 0.095 | |||||
| Frontal | 5 (50) | 45 (39) | 0 (0) | 28 (35) | |||
| Parietal | 1 (10) | 19 (17) | 0 (0) | 14 (18) | |||
| Cerebellar | 1 (10) | 13 (11) | 1 (20) | 12 (15) | |||
| Temporal | 1 (10) | 10 (9) | 3 (60) | 8 (10) | |||
| Occipital | 1 (10) | 11 (10) | 1 (20) | 11 (14) | |||
| Basal ganglia | 0 (0) | 3 (3) | 0 (0) | 2 (3) | |||
| Brainstem | 0 (0) | 6 (5) | 0 (0) | 1 (1) | |||
| Periventricular | 0 (0) | 2 (2) | 0 (0) | 3 (4) | |||
| Other | 1 (10) | 6 (5) | 0 (0) | 0 (0) | |||
| Timing of immunotherapy | 0.917 | 1.000 | |||||
| Concurrent | 8 (80) | 84 (73) | 3 (60) | 47 (59) | |||
| Non-concurrent | 2 (20) | 31 (27) | 2 (40) | 32 (41) | |||
| ICI agent | 1.000 | 0.842 | |||||
| PD−1/PD−L1 | 8 (80) | 92 (80) | 3 (60) | 59 (75) | |||
| CTLA−4 | 2 (20) | 23 (20) | 2 (40) | 20 (25) | |||
| Intact/Cavity | 1.000 | 0.571 | |||||
| Intact | 10 (100) | 109 (95) | 4 (80) | 76 (96) | |||
| Cavity | 0 (0) | 6 (5) | 1 (20) | 3 (4) | |||
| Prescription dose (Gy) | 0.554 | 0.252 | |||||
| <20 | 4 (40) | 24 (21) | 2 (40) | 15 (19) | |||
| >=20 | 6 (60) | 91 (79) | 3 (60) | 64 (81) | |||
| Prescription isodose line | 0.160 | 0.336 | |||||
| <=50% | 10 (100) | 91 (79) | 5 (100) | 66 (84) | |||
| 50–80% | 0 (0) | 15 (13) | 0 (0) | 7 (8) | |||
| >=80% | 0 (0) | 9 (8) | 0 (0) | 6 (8) | |||
| Volume of metastasis (cm3) | 0.106 | 0.009 | |||||
| <5 | 10 (100) | 104 (90) | 3 (60) | 75 (95) | |||
| >=5 | 0 (0) | 11 (10) | 2 (40) | 4 (5) | |||
| V12 (cm3) | 0.066 | 0.013 | |||||
| <=10 | 9 (90) | 97 (84) | 3 (60) | 72 (91) | |||
| >10 | 1 (10) | 18 (16) | 2 (40) | 7 (9) | |||
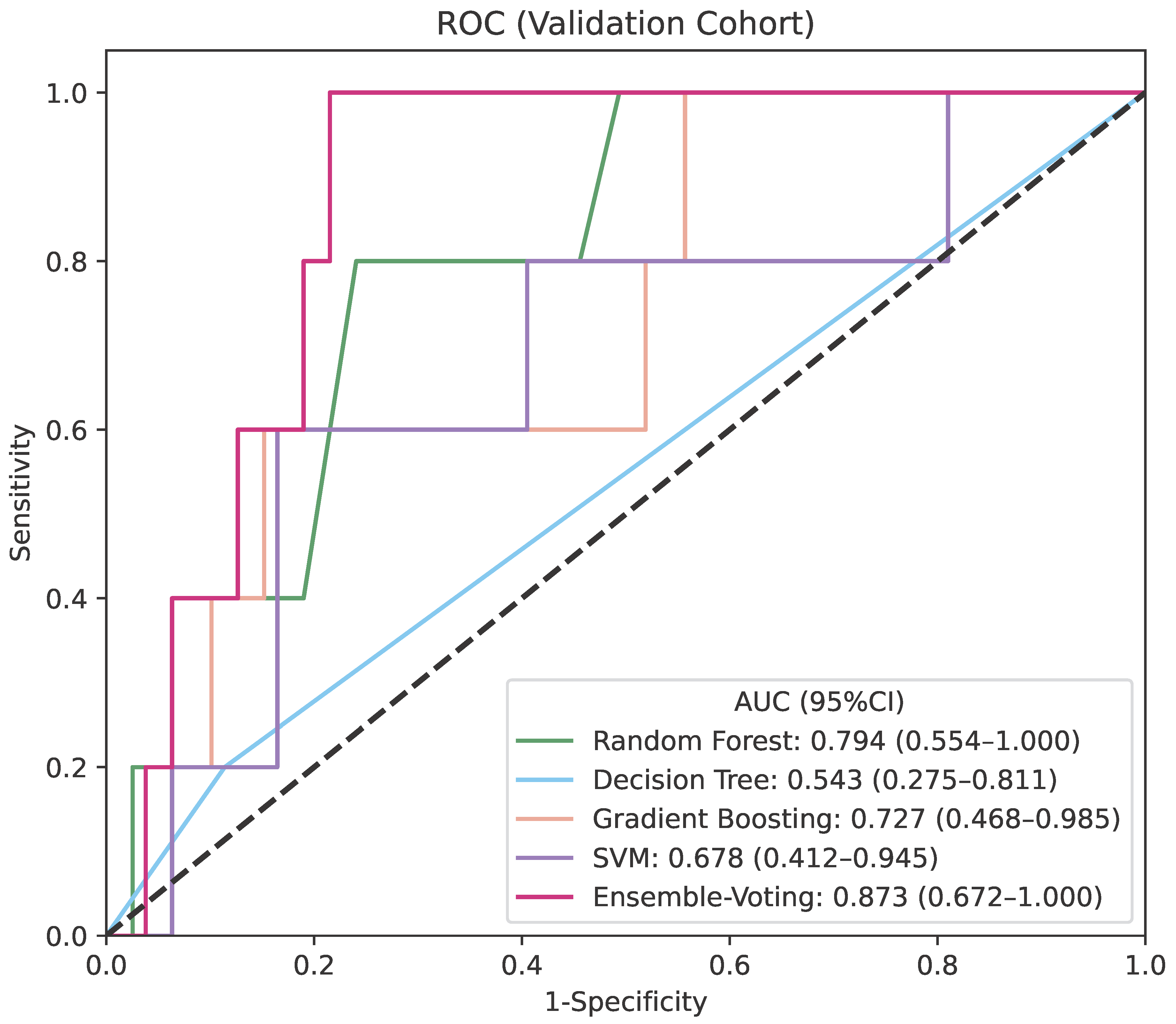
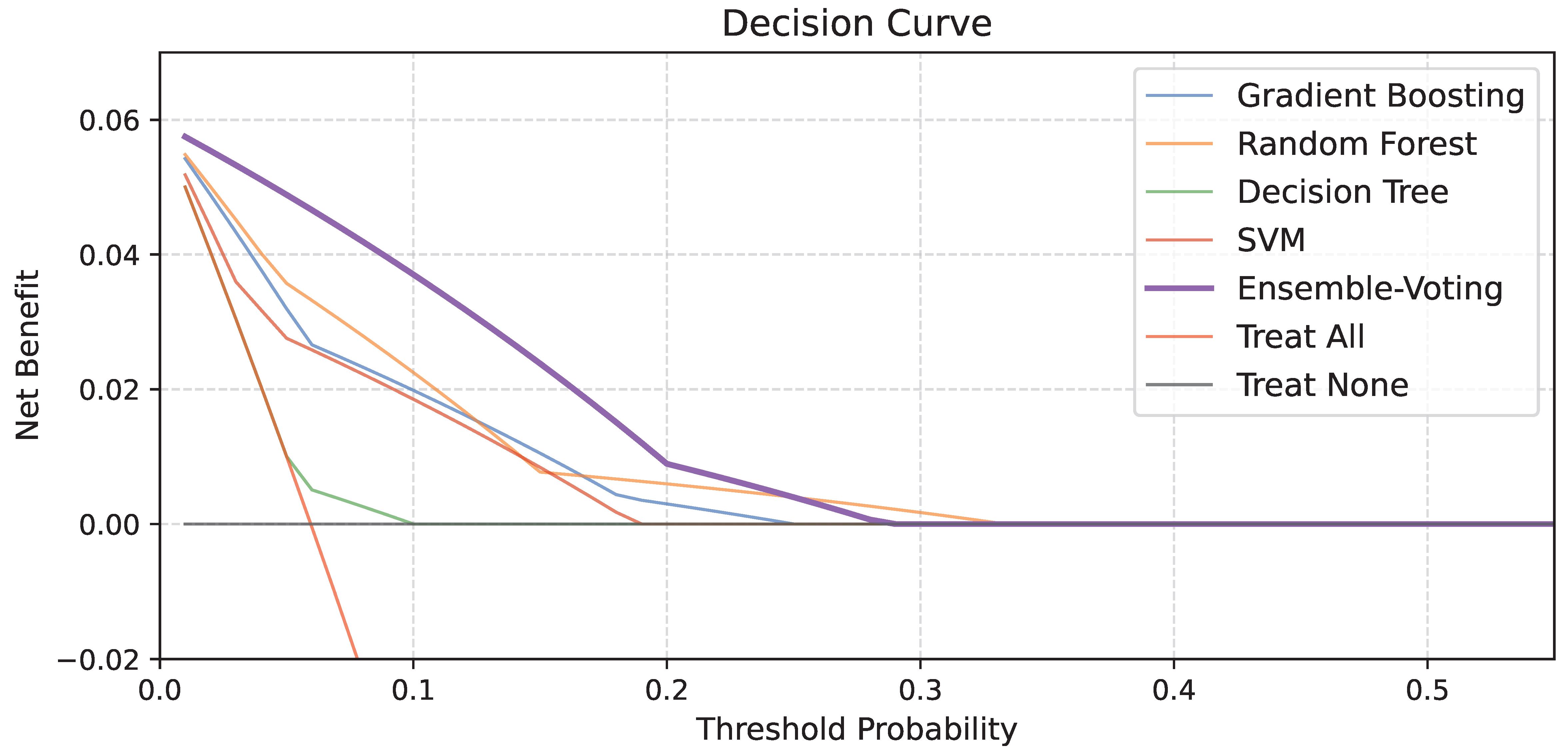
| AUC | Sensitivity | Specificity | NPV | PPV | |
|---|---|---|---|---|---|
| Soft-voting Ensemble Model | 0.873 (0.672−1.000) | 1.000 (1.000−1.000) | 0.785 (0.694−0.875) | 1.000 (1.000−1.000) | 0.227 (0.052−0.402) |
| Stacking Ensemble Model | 0.828 (0.602−1.000) | 1.000 (1.000−1.000) | 0.684 (0.581−0.786) | 1.000 (1.000−1.000) | 0.167 (0.033−0.300) |
| Gradient Boosting | 0.727 (0.468−0.985) | 0.600 (0.171−1.000) | 0.848 (0.769−0.927) | 0.971 (0.931−1.000) | 0.200 (0.000−0.402) |
| Random Forest | 0.794 (0.554−1.000) | 0.800 (0.449−1.000) | 0.759 (0.665−0.854) | 0.984 (0.952−1.000) | 0.174 (0.019−0.329) |
| Decision Tree | 0.543 (0.275−0.811) | 0.200 (0.000−0.551) | 0.886 (0.816−0.956) | 0.946 (0.894−0.997) | 0.100 (0.000−0.286) |
| Support Vector Machine | 0.678 (0.412−0.945) | 0.600 (0.171−1.000) | 0.835 (0.754−0.917) | 0.971 (0.930−1.000) | 0.188 (0.000−0.379) |
| Logistic Regression | 0.466 (0.209−0.722) | 0.400 (0.000−0.829) | 0.823 (0.739−0.907) | 0.956 (0.907−1.000) | 0.125 (0.000−0.287) |
| Multi-Layer Perceptron Classifier | 0.572 (0.302−0.842) | 0.600 (0.171−1.000) | 0.696 (0.595−0.798) | 0.965 (0.917−1.000) | 0.111 (0.000−0.230) |
| K-Nearest Neighbors | 0.627 (0.356−0.897) | 0.600 (0.171−1.000) | 0.620 (0.513−0.727) | 0.961 (0.908−1.000) | 0.091 (0.000−0.189) |
| Gaussian Naive Bayes | 0.537 (0.270−0.804) | 0.400 (0.000−0.829) | 0.810 (0.724−0.897) | 0.955 (0.906−1.000) | 0.118 (0.000−0.271) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Helis, C.; Cramer, C.; Munley, M.; Choi, A.R.; Tan, J.; Xing, F.; Lyu, Q.; Whitlow, C.; Willey, J.; et al. MRI-Based Radiomics Ensemble Model for Predicting Radiation Necrosis in Brain Metastasis Patients Treated with Stereotactic Radiosurgery and Immunotherapy. Cancers 2025, 17, 1974. https://doi.org/10.3390/cancers17121974
Chen Y, Helis C, Cramer C, Munley M, Choi AR, Tan J, Xing F, Lyu Q, Whitlow C, Willey J, et al. MRI-Based Radiomics Ensemble Model for Predicting Radiation Necrosis in Brain Metastasis Patients Treated with Stereotactic Radiosurgery and Immunotherapy. Cancers. 2025; 17(12):1974. https://doi.org/10.3390/cancers17121974
Chicago/Turabian StyleChen, Yijun, Corbin Helis, Christina Cramer, Michael Munley, Ariel Raimundo Choi, Josh Tan, Fei Xing, Qing Lyu, Christopher Whitlow, Jeffrey Willey, and et al. 2025. "MRI-Based Radiomics Ensemble Model for Predicting Radiation Necrosis in Brain Metastasis Patients Treated with Stereotactic Radiosurgery and Immunotherapy" Cancers 17, no. 12: 1974. https://doi.org/10.3390/cancers17121974
APA StyleChen, Y., Helis, C., Cramer, C., Munley, M., Choi, A. R., Tan, J., Xing, F., Lyu, Q., Whitlow, C., Willey, J., Chan, M., & Jiang, Y. (2025). MRI-Based Radiomics Ensemble Model for Predicting Radiation Necrosis in Brain Metastasis Patients Treated with Stereotactic Radiosurgery and Immunotherapy. Cancers, 17(12), 1974. https://doi.org/10.3390/cancers17121974






