Deep-Learning-Based Predictive Imaging Biomarker Model for EGFR Mutation Status in Non-Small Cell Lung Cancer from CT Imaging
Abstract
Simple Summary
Abstract
1. Introduction
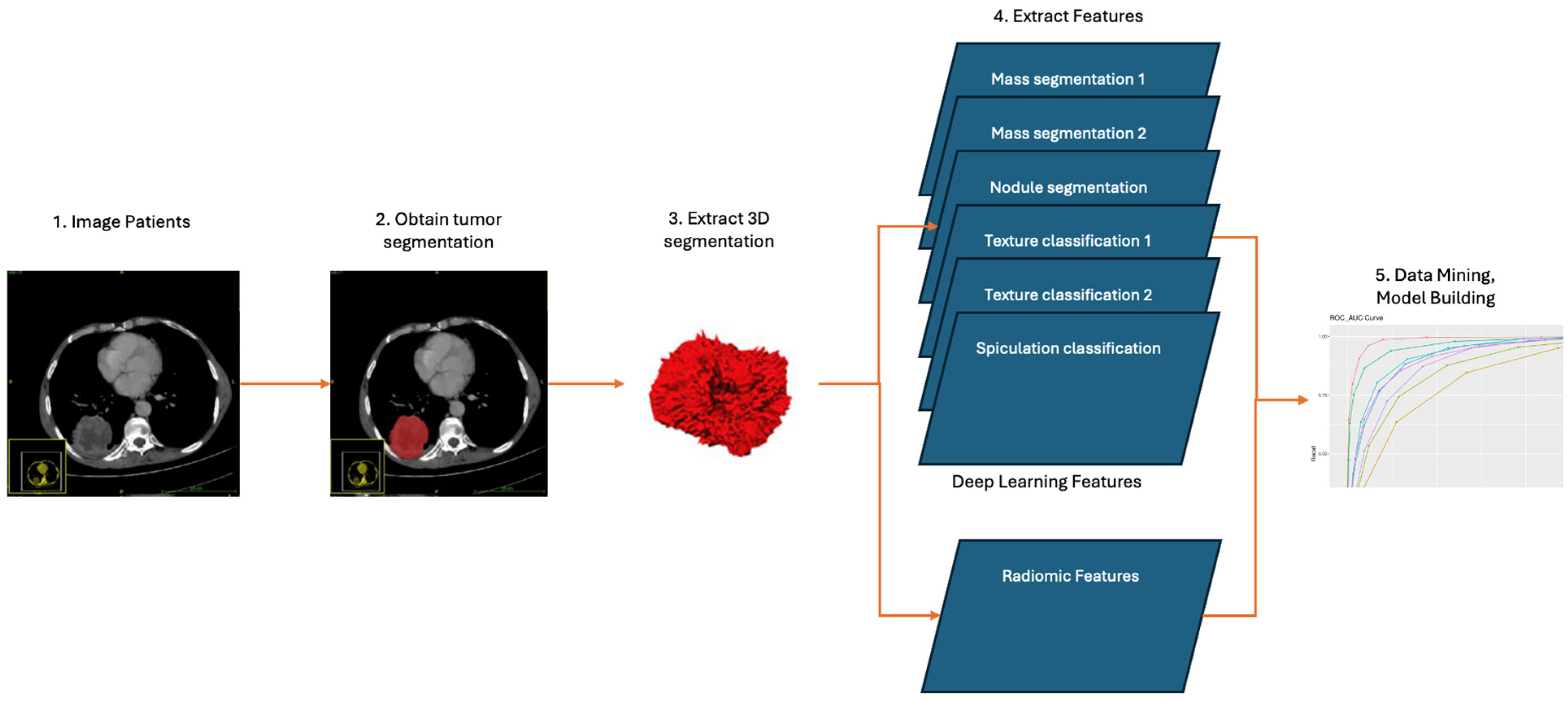
2. Materials and Methods
2.1. Radiology Review
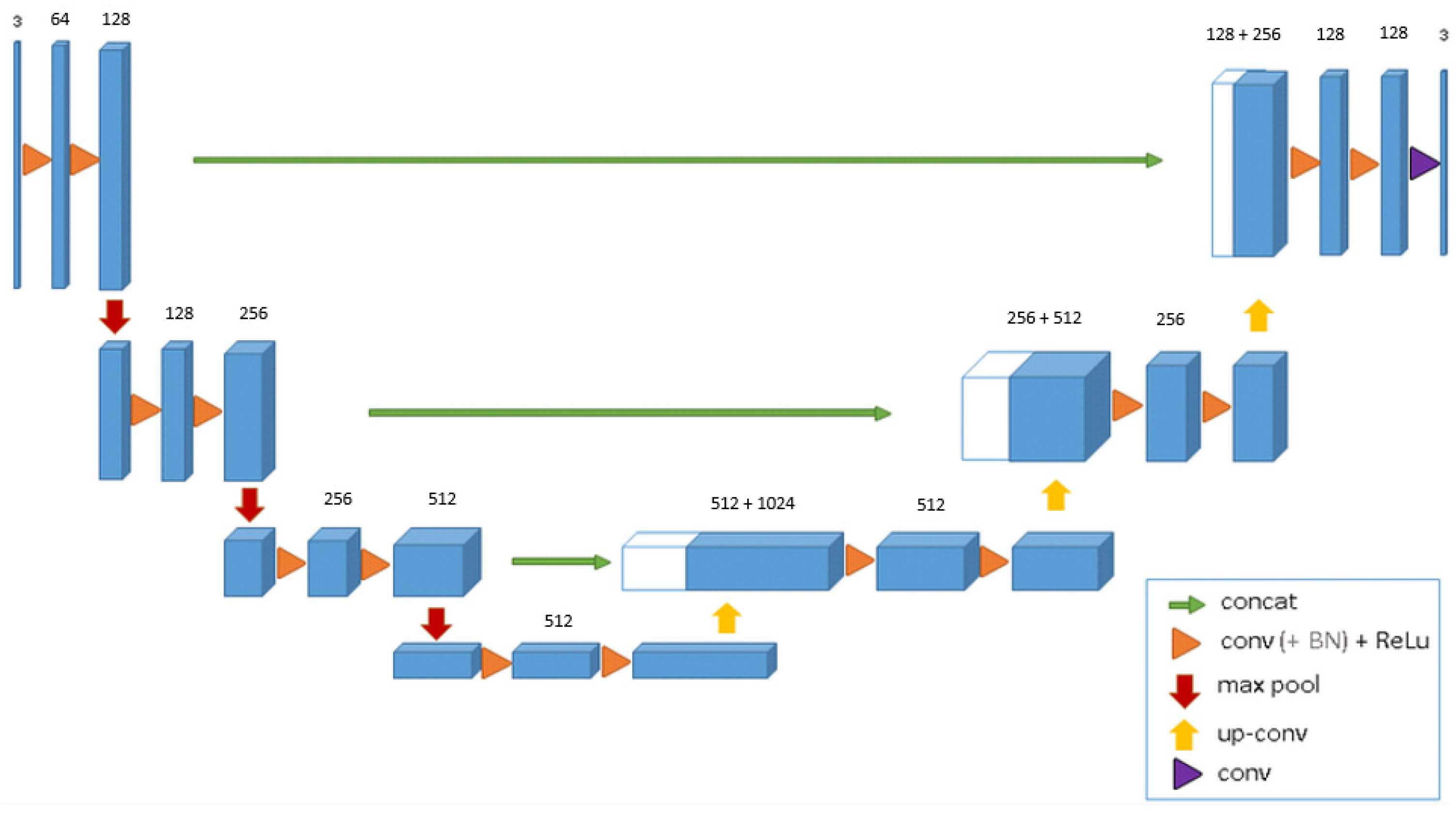
2.2. Development of the DL Model
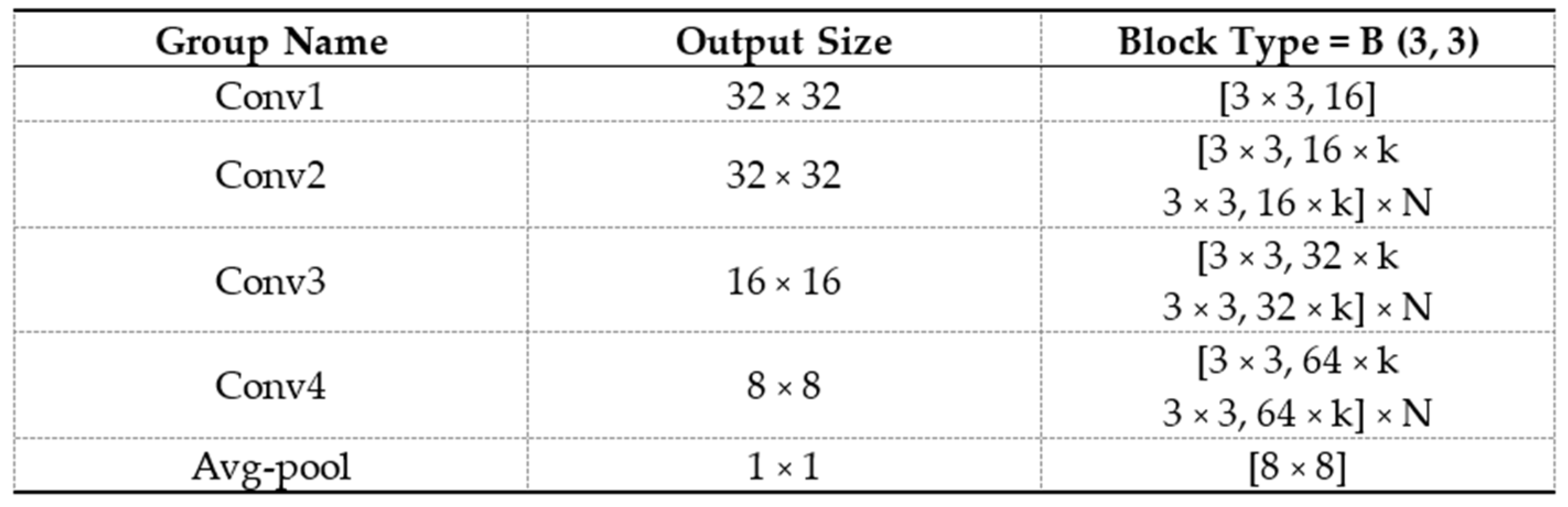
2.3. Development of Radiomics Model
2.4. Combining DL and Radiomic Features
2.5. Statistical Analysis
3. Results
3.1. Patient’s Characteristics
3.2. Correlation of EGFR Mutation Status with Clinical Features
3.3. Correlation of EGFR Mutation Status with Semantic Features
3.4. Radiomics Model Used in Predicting EGFR Mutation
3.5. DL Model in Predicting EGFR Mutation
3.6. Combining DL and Radiomic Features
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, H.-J.; Kim, Y.T.; Kang, C.H.; Zhao, B.; Tan, Y.; Schwartz, L.H.; Persigehl, T.; Jeon, Y.K.; Chung, D.H. Epidermal growth factor receptor mutation in lung adenocarcinomas: Relationship with CT characteristics and histologic subtypes. Radiology 2013, 268, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Oxnard, G.R.; Lo, P.C.; Nishino, M.; Dahlberg, S.E.; Lindeman, N.I.; Butaney, M.; Jackman, D.M.; Johnson, B.E.; Jänne, P.A. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J. Thorac. Oncol. 2013, 8, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, F.A.; Rodrigues Pereira, J.; Ciuleanu, T.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Campos, D.; Maoleekoonpiroj, S.; Smylie, M.; Martins, R.; et al. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 2005, 353, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Kris, M.G.; Natale, R.B.; Herbst, R.S.; Lynch, T.J.; Prager, D.; Belani, C.P.; Schiller, J.H.; Kelly, K.; Spiridonidis, H.; Sandler, A.; et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: A randomized trial. JAMA 2003, 290, 2149–2158. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Okami, J.; Kodama, K.; Higashiyama, M.; Kato, K. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008, 99, 929–935. [Google Scholar] [CrossRef]
- Vaidya, T.; Agrawal, A.; Mahajan, S.; Thakur, M.H.; Mahajan, A. The Continuing Evolution of Molecular Functional Imaging in Clinical Oncology: The Road to Precision Medicine and Radiogenomics (Part II). Mol. Diagn. Ther. 2019, 23, 27–51. [Google Scholar] [CrossRef]
- Vaidya, T.; Agrawal, A.; Mahajan, S.; Thakur, M.H.; Mahajan, A. The Continuing Evolution of Molecular Functional Imaging in Clinical Oncology: The Road to Precision Medicine and Radiogenomics (Part I). Mol. Diagn. Ther. 2019, 23, 1–26. [Google Scholar] [CrossRef]
- Chakrabarty, N.; Mahajan, A. Imaging Analytics using Artificial Intelligence in Oncology: A Comprehensive Review. Clin. Oncol. R. Coll. Radiol. 2023. [Google Scholar] [CrossRef]
- Mahajan, A.; Gurukrishna, B.; Wadhwa, S.; Agarwal, U.; Baid, U.; Talbar, S.; Janu, A.K.; Patil, V.; Noronha, V.; Mummudi, N.; et al. Deep learning based automated epidermal growth factor receptor and anaplastic lymphoma kinase status prediction of brain metastasis in non-small cell lung cancer. Explor. Target. Anti-Tumor Ther. 2023, 4, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, A.; Chakrabarty, N.; Majithia, J.; Ahuja, A.; Agarwal, U.; Suryavanshi, S.; Biradar, M.; Sharma, P.; Raghavan, B.; Arafath, R.; et al. Multisystem Imaging Recommendations/Guidelines: In the Pursuit of Precision Oncology. Indian J. Med. Paediatr. Oncol. 2023, 44, 2–25. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Andersen, J.N.; Futreal, P.A. Cancer genomics: From discovery science to personalized medicine. Nat. Med. 2011, 17, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Garraway, L.A.; Verweij, J.; Ballman, K.V. Precision oncology: An overview. J. Clin. Oncol. 2013, 31, 1803–1805. [Google Scholar] [CrossRef] [PubMed]
- Hazlett, H.C.; Gu, H.; Munsell, B.C.; Kim, S.H.; Styner, M.; Wolff, J.J.; Elison, J.T.; Swanson, M.R.; Zhu, H.; Botteron, K.N.; et al. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017, 542, 348–351. [Google Scholar] [CrossRef]
- Lakhani, P.; Sundaram, B. Deep Learning at Chest Radiography: Automated Classification of Pulmonary Tuberculosis by Using Convolutional Neural Networks. Radiology 2017, 284, 574–582. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.W.; Cheung, C.Y.-L.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; San Yeo, I.Y.; Lee, S.Y.; et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA 2017, 318, 2211–2223. [Google Scholar] [CrossRef]
- Wang, K.; Lu, X.; Zhou, H.; Gao, Y.; Zheng, J.; Tong, M.; Wu, C.; Liu, C.; Huang, L.; Jiang, T.; et al. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: A prospective multicentre study. Gut 2019, 68, 729–741. [Google Scholar] [CrossRef]
- Parmar, C.; Leijenaar, R.T.H.; Grossmann, P.; Rios Velazquez, E.; Bussink, J.; Rietveld, D.; Rietbergen, M.M.; Haibe-Kains, B.; Lambin, P.; Aerts, H.J.W.L. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci. Rep. 2015, 5, 11044. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Noronha, V.; Patil, V.M.; Joshi, A.; Menon, N.; Chougule, A.; Mahajan, A.; Janu, A.; Purandare, N.; Kumar, R.; More, S.; et al. Gefitinib Versus Gefitinib Plus Pemetrexed and Carboplatin Chemotherapy in EGFR-Mutated Lung Cancer. J. Clin. Oncol. 2020, 38, 124–136. [Google Scholar] [CrossRef]
- Patil, V.M.; Noronha, V.; Joshi, A.; Choughule, A.B.; Bhattacharjee, A.; Kumar, R.; Goud, S.; More, S.; Ramaswamy, A.; Karpe, A.; et al. Phase III study of gefitinib or pemetrexed with carboplatin in EGFR-mutated advanced lung adenocarcinoma. ESMO Open 2017, 2, e000168. [Google Scholar] [CrossRef]
- AIM-Harvard/Pyradiomics. Artificial Intelligence in Medicine (AIM) Program. 2024. Available online: https://github.com/AIM-Harvard/pyradiomics (accessed on 16 February 2024).
- Yoon, H.J.; Choi, J.; Kim, E.; Um, S.-W.; Kang, N.; Kim, W.; Kim, G.; Park, H.; Lee, H.Y. Deep learning analysis to predict EGFR mutation status in lung adenocarcinoma manifesting as pure ground-glass opacity nodules on CT. Front. Oncol. 2022, 12, 951575. [Google Scholar] [CrossRef]
- Kim, S.; Lim, J.H.; Kim, C.-H.; Roh, J.; You, S.; Choi, J.-S.; Lim, J.H.; Kim, L.; Chang, J.W.; Park, D.; et al. Deep learning–radiomics integrated noninvasive detection of epidermal growth factor receptor mutations in non-small cell lung cancer patients. Sci. Rep. 2024, 14, 922. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.S.; Ho, D.K.N.; Nguyen, N.N.; Tran, H.M.; Tam, K.-W.; Le, N.Q.K. Predicting EGFR Mutation Status in Non–Small Cell Lung Cancer Using Artificial Intelligence: A Systematic Review and Meta-Analysis. Acad. Radiol. 2024, 31, 660–683. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Cai, H.; Wang, Y.; Cui, R.; Huo, L.; Lee, E.Y.-P.; Liang, Y.; Li, X.; Hu, Z.; Chen, L.; et al. Deep learning for predicting epidermal growth factor receptor mutations of non-small cell lung cancer on PET/CT images. Quant. Imaging Med. Surg. 2023, 13, 1286–1299. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Hou, L.; Yang, W.; Han, J.; Wang, J.; Qiang, Y.; Zhao, J.; Hou, J.; Song, K.; Ma, Y.; et al. Multi-channel multi-task deep learning for predicting EGFR and KRAS mutations of non-small cell lung cancer on CT images. Quant. Imaging Med. Surg. 2021, 11, 2354–2375. [Google Scholar] [CrossRef] [PubMed]





| Feature Vector | Model | Patch Size | Processing | Feature Vector Size |
| 1 | Mass segmentation 1 | 132 × 132 × 132 | Original image is at 1× zoom | 512 |
| 2 | Mass segmentation 1 | 132 × 132 × 132 | Original image is at 2× zoom | 512 |
| 3 | Mass segmentation 1 | 132 × 132 × 132 | Original image is at 0.5× zoom | 512 |
| 4 | Mass segmentation 2 | 132 × 132 × 132 | Original image at 0.5× zoom | 512 |
| 5 | Nodule segmentation | 132 × 132 × 132 | Original image is 0.5× zoom | 512 |
| 6 | Texture classification 1 | 64 × 64 × 64 | Original image is at 0.5× zoom | 320 |
| 7 | Texture classification 1 | 32 × 32 × 32 | Original image is at 0.5× zoom | 320 |
| 8 | Texture classification 2 | 64 × 64 × 64 | Original image is at 0.5× zoom | 320 |
| 9 | Spiculation classification | 64 × 64 × 64 | Original image is at 0.5× zoom | 512 |
| Radiomics Features | Importance | Normalized Importance (%) |
|---|---|---|
| Entropy | 0.046 | 71.3 |
| Variance | 0.03 | 56.4 |
| Enhance Count | 0.036 | 57.4 |
| Core Count | 0.032 | 49.70 |
| Cluster Shade | 0.033 | 46.2 |
| Core Count | 0.031 | 44.4 |
| Two-step Cluster Number Based on Age | 0.032 | 42.10 |
| Edema Count | 0.030 | 42.6 |
| Dissimilarity | 0.027 | 46.3 |
| Core Count | 0.03 | 42.8 |
| Difference in Entropy | 0.028 | 41.9 |
| Enhance Count | 0.025 | 42.9 |
| Variance | 0.026 | 38.0 |
| Maximum Probability | 0.029 | 36.9 |
| Sum of Variance | 0.027 | 36.7 |
| Homogeneity | 0.026 | 36.5 |
| Minimum Probability | 0.026 | 35.4 |
| Correlation | 0.022 | 32.6 |
| Inverse Difference | 0.024 | 32.3 |
| Contrast | 0.023 | 29.5 |
| Cluster Shade | 0.018 | 26.6 |
| Correlation | 0.017 | 24.5 |
| Variance | 0.017 | 22.7 |
| Maximum Probability | 0.015 | 19.5 |
| Cluster Prominence | 0.016 | 18.5 |
| Dissimilarity | 0.013 | 18.9 |
| Auto-Correlation | 0.015 | 18.5 |
| Inverse Difference | 0.013 | 18.4 |
| Sum of Squares Variance | 0.011 | 18.2 |
| Difference in Entropy | 0.013 | 17.5 |
| Average | 0.015 | 17.2 |
| Maximum Probability | 0.013 | 16.3 |
| Homogeneity | 0.01 | 15.4 |
| Difference in Entropy | 0.008 | 13.9 |
| Mean | 0.008 | 13.4 |
| Cluster Prominence | 0.008 | 11.2 |
| Sum Average | 0.008 | 11.1 |
| Inverse Difference | 0.007 | 9 |
| Minimum | 0.006 | 7.8 |
| Contrast | 0.005 | 6.4 |
| Sum of Intensities | 0.003 | 6.3 |
| Contrast | 0.004 | 4.7 |
| Homogeneity | 0.002 | 2.5 |
| Contrast | 0.002 | 1.7 |
| Dissimilarity | 0.001 | 1.5 |
| Sr. No. | Variables | N | EGFR Wild Type | EGFR Mutant Type | Mean Age | p Value ^ | Univariate OR {CI} | |
|---|---|---|---|---|---|---|---|---|
| Total Patients | 223 | 102 | 121 | |||||
| 1 | Median Age (years) | 57 (48–62.8) | 54 (46–59) | 0.095 | 0.981 {0.95, 1.007} | |||
| 2 | Gender [%] | Male | 143 | 79 [77.55%] | 68 [56.25%] | 54.7 (28–80) | ||
| Female | 76 | 23 [22.5%] | 53 [43.8%] | 52.7 (31–75) | 0.001 | 2.6 {1.48, 4.81} | ||
| 3 | Smoking status [%] | Yes | 44 [45.1%] | 22 [18.1%] | 0.001 | 3.5 {1.94, 6.50} | ||
| No | 57 [55.9%] | 99 [81.8%] | ||||||
| 4 | Tumour stage [%] | III | 12 [11.8%] | 1 [0.8%] | ||||
| IV | 90 [88.2%] | 120 [99.2%] | 0.008 | 16 {2.04, 125.31} |
| Variables | EGFR Wild Type | EGFR Mutant Type | p Value ^ | OR (Odds Ratio) | ||
|---|---|---|---|---|---|---|
| 1 | Tumour size | ≤5 CM (Ref.) | 60 (58.8) | 65 (53.7) | Reference | |
| >5 CM | 42 (41.2) | 56 (46.3) | 0.44 | 1.231 {0.72, 2.09} | ||
| 2 | Tumour lobe location | Right upper lobe (Ref.) | 25 (24.5) | 36 (29.8) | Reference | |
| Right middle lobe | 7 (6.9) | 9 (7.4) | 0.84 | 0.893 {0.29, 2.71} | ||
| Right lower lobe | 17 (16.7) | 21 (17.4) | 0.71 | 0.858 {0.37, 1.94} | ||
| Left upper lobe | 34 (33.3) | 30 (24.8) | 0.17 | 0.613 {0.30, 1.24} | ||
| Left lower lobe | 19 (18.6) | 25 (20.7) | 0.82 | 0.914 {0.41, 2.003} | ||
| 3 | Tumour distribution | Central | 9 (8.8) | 14 (11.6) | 0.87 | 0.929 {0.36, 2.34} |
| Peripheral | 53 (52.0) | 40 (33.1) | 0.01 | 0.451 {0.25, 0.79} | ||
| Both (Ref.) | 40 (39.2) | 67 (55.4) | Reference | |||
| 4 | Contour (%) | Round/oval (Ref.) | 0 (0.0) | 1 (0.8) | Reference | |
| Irregular | 99 (97.1) | 117 (96.7) | 0.87 | 0.886 {0.19, 4.05} | ||
| 5 | Margins | Well defined (Ref.) | 25 (24.5) | 24 (19.8) | Reference | |
| Poorly defined | 77 (75.5) | 97 (80.2) | 0.40 | 1.312 {0.69, 2.47} | ||
| 6 | Spiculations (%) | Absent (Ref.) | 28 (27.5) | 23 (19.0) | Reference | |
| Fine spiculations | 38 (37.3) | 45 (37.2) | 0.30 | 1.442 {0.71, 2.90} | ||
| Coarse spiculations | 36 (35.3) | 53 (43.8) | 0.10 | 1.792 {0.89, 3.59} | ||
| 7 | Enhancement pattern | Homogeneous (Ref.) | 14 (13.7) | 13 (10.7) | Reference | |
| Mild/moderate heterogeneous | 43 (42.2) | 41 (33.9) | 0.95 | 1.027 {0.43, 2.44} | ||
| Marked heterogeneous | 45 (44.1) | 67 (55.4) | 0.27 | 1.603 {0.68, 3.73} | ||
| 8 | Enhancement heterogeneity | Maximum Enhancement | 60.5 [50.25–75.5] | 71 [59–87] | 0.001 | 1.024 {1.01, 1.03} |
| Minimum Enhancement | 35 [28–48] | 44 [35–55] | 0.002 | 1.026 {1.009, 1.04} | ||
| Average Enhancement A. Average Enhancement < 54 HU B. Average Enhancement > 54 HU | 48 [41–60] 64 (62.7) 38(37.3) | 57 [48–66] 47 (38.8) 74 (61.2) | 0.004 Reference <0.001 | 2.652 {1.54, 4.55} | ||
| Relative Enhancement to reference artery | 0.32 [0.24–0.4] | 0.35 [0.28–0.41] | 0.116 | 9.733 | ||
| 9 | Texture | Predominant solid with associated GGO component | 21 (20.6) | 11 (9.1) | Reference | |
| Pure Solid (no associated ground glass component) | 81 (79.4) | 109 (90.1) | 0.028 | 2.355 {1.09, 5.06} | ||
| 10 | Air bronchogram | Absent (Ref.) | 65 (63.7) | 63 (52.1) | ||
| Present | 37 (36.3) | 58 (47.9) | 0.08 | 1.617 {0.94, 2.77} | ||
| 11 | Bubble like lucency | Absent (Ref.) | 94 (92.2) | 110 (90.9) | Reference | |
| Present | 8 (7.8) | 11 (9.1) | 0.73 | 1.175 {0.45, 3.04} | ||
| 12 | Cavitation | Absent (Ref.) | 99 (97.1) | 114 (94.2) | Reference | |
| Present | 3 (2.9) | 7 (5.8) | 0.31 | 2.026 {0.51, 8.04} | ||
| 13 | Peripheral emphysema | Absent (Ref.) | 85 (83.3) | 114 (94.2) | Reference | |
| Mild/moderate | 14 (13.7) | 6 (5.0) | 0.024 | 0.320 {0.11, 0.86} | ||
| Marked | 3 (2.9) | 1 (0.8) | 0.23 | 0.249 {0.025, 2.43} | ||
| 14 | Peripheral fibrosis | Absent (Ref.) | 67 (65.7) | 73 (60.3) | Reference | |
| Mild/Moderate | 27 (26.5) | 38 (31.4) | 0.39 | 1.292 {0.71, 2.34} | ||
| Marked | 8 (7.8) | 10 (8.3) | 0.78 | 1.147 {0.42, 3.07} | ||
| 15 | Fissure attachment | Absent (Ref.) | 43 (42.2) | 25 (20.7) | Reference | |
| Present | 59 (57.8) | 96 (79.3) | 0.001 | 2.799 {1.55, 5.04} | ||
| 16 | Pleural attachment | Absent (Ref.) | 12 (11.8) | 13 (10.7) | Reference | |
| Present | 90 (88.2) | 108 (89.3) | 0.80 | 1.108 {0.48, 2.54} | ||
| 17 | Pleural retraction | Absent (Ref.) | 38 (37.3) | 24 (19.8) | Reference | |
| Present | 64 (62.7) | 97 (80.2) | 0.004 | 2.400 {1.31, 4.37} | ||
| 18 | Vascular convergence | Absent (Ref.) | 101 (99.0) | 119 (98.3) | Reference | |
| Present | 1 (1.0) | 2 (1.7) | 0.66 | 1.697 {0.15, 18.99} | ||
| 19 | Thickened Broncho vascular bundle | Absent (Ref.) | 50 (49.0) | 50 (41.3) | Reference | |
| Present | 52 (51.0) | 71 (58.7) | 0.25 | 1.365 {0.80, 2.32} | ||
| 20 | Calcifications | Absent (Ref.) | 100 (98.0) | 115 (95.0) | 2.609 | |
| Present | 2 (2.0) | 6 (5.0) | 0.24 | {0.51, 13.2} | ||
| 21 | Lymphadenopathy | Absent (Ref.) | 28 (27.5) | 29 (24.0) | Reference | |
| Present | 74 (72.5) | 92 (76.0) | 0.55 | 1.200 {0.65, 2.19} | ||
| 22 | Vascular involvement | Absent (Ref.) | 56 (54.9) | 39 (32.2) | Reference | |
| Present | 46 (45.1) | 82 (67.8) | 0.001 | 2.560 {1.48, 4.41} | ||
| 23 | Pleural effusion | Absent (Ref.) | 71 (69.6) | 67 (55.4) | Reference | |
| Present ipsilateral | 30 (29.4) | 51 (42.1) | 0.04 | 1.801 {1.02, 3.15} | ||
| Present contralateral | 1 (1.0) | 3 (2.5) | 0.32 | 3.179 {0.32, 31.32} | ||
| 24 | Lymphangitic spread | Absent (Ref.) | 76 (74.5) | 82 (67.8) | Reference | |
| Present | 26 (25.5) | 39 (32.2) | 0.27 | 1.390 {0.77, 2.49} | ||
| 25 | Pleural nodularity | Absent (Ref.) | 66 (64.7) | 71 (58.7) | Reference | |
| Present | 34 (33.3) | 50 (41.3) | 0.35 | 1.291 {0.74, 2.22} | ||
| 26 | Lobulations | Absent (Ref.) | 80 (78.4) | 98 (81.0) | Reference | |
| Present < 3 | 1 (1.0) | 4 (3.3) | 0.29 | 3.265 {0.35, 29.79} | ||
| Present > 3 | 21 (20.6) | 19 (15.7) | 0.38 | 0.739 {0.37, 1.46} | ||
| 27 | Tumour lobe metastatic nodule | Absent (Ref.) | 39 (38.2) | 30 (24.8) | Reference | |
| Present | 63 (61.8) | 91 (75.2) | 0.032 | 1.878 {1.05, 3.33} | ||
| 28 | Non-tumour lobe metastatic nodule | Absent (Ref.) | 42 (41.2) | 25 (20.7) | Reference | |
| Present | 60 (58.8) | 96 (79.3) | 0.001 | 2.688 {1.48, 4.85} |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahajan, A.; Kania, V.; Agarwal, U.; Ashtekar, R.; Shukla, S.; Patil, V.M.; Noronha, V.; Joshi, A.; Menon, N.; Kaushal, R.K.; et al. Deep-Learning-Based Predictive Imaging Biomarker Model for EGFR Mutation Status in Non-Small Cell Lung Cancer from CT Imaging. Cancers 2024, 16, 1130. https://doi.org/10.3390/cancers16061130
Mahajan A, Kania V, Agarwal U, Ashtekar R, Shukla S, Patil VM, Noronha V, Joshi A, Menon N, Kaushal RK, et al. Deep-Learning-Based Predictive Imaging Biomarker Model for EGFR Mutation Status in Non-Small Cell Lung Cancer from CT Imaging. Cancers. 2024; 16(6):1130. https://doi.org/10.3390/cancers16061130
Chicago/Turabian StyleMahajan, Abhishek, Vatsal Kania, Ujjwal Agarwal, Renuka Ashtekar, Shreya Shukla, Vijay Maruti Patil, Vanita Noronha, Amit Joshi, Nandini Menon, Rajiv Kumar Kaushal, and et al. 2024. "Deep-Learning-Based Predictive Imaging Biomarker Model for EGFR Mutation Status in Non-Small Cell Lung Cancer from CT Imaging" Cancers 16, no. 6: 1130. https://doi.org/10.3390/cancers16061130
APA StyleMahajan, A., Kania, V., Agarwal, U., Ashtekar, R., Shukla, S., Patil, V. M., Noronha, V., Joshi, A., Menon, N., Kaushal, R. K., Rane, S., Chougule, A., Vaidya, S., Kaluva, K., & Prabhash, K. (2024). Deep-Learning-Based Predictive Imaging Biomarker Model for EGFR Mutation Status in Non-Small Cell Lung Cancer from CT Imaging. Cancers, 16(6), 1130. https://doi.org/10.3390/cancers16061130







