Performance of Commercial Dermatoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
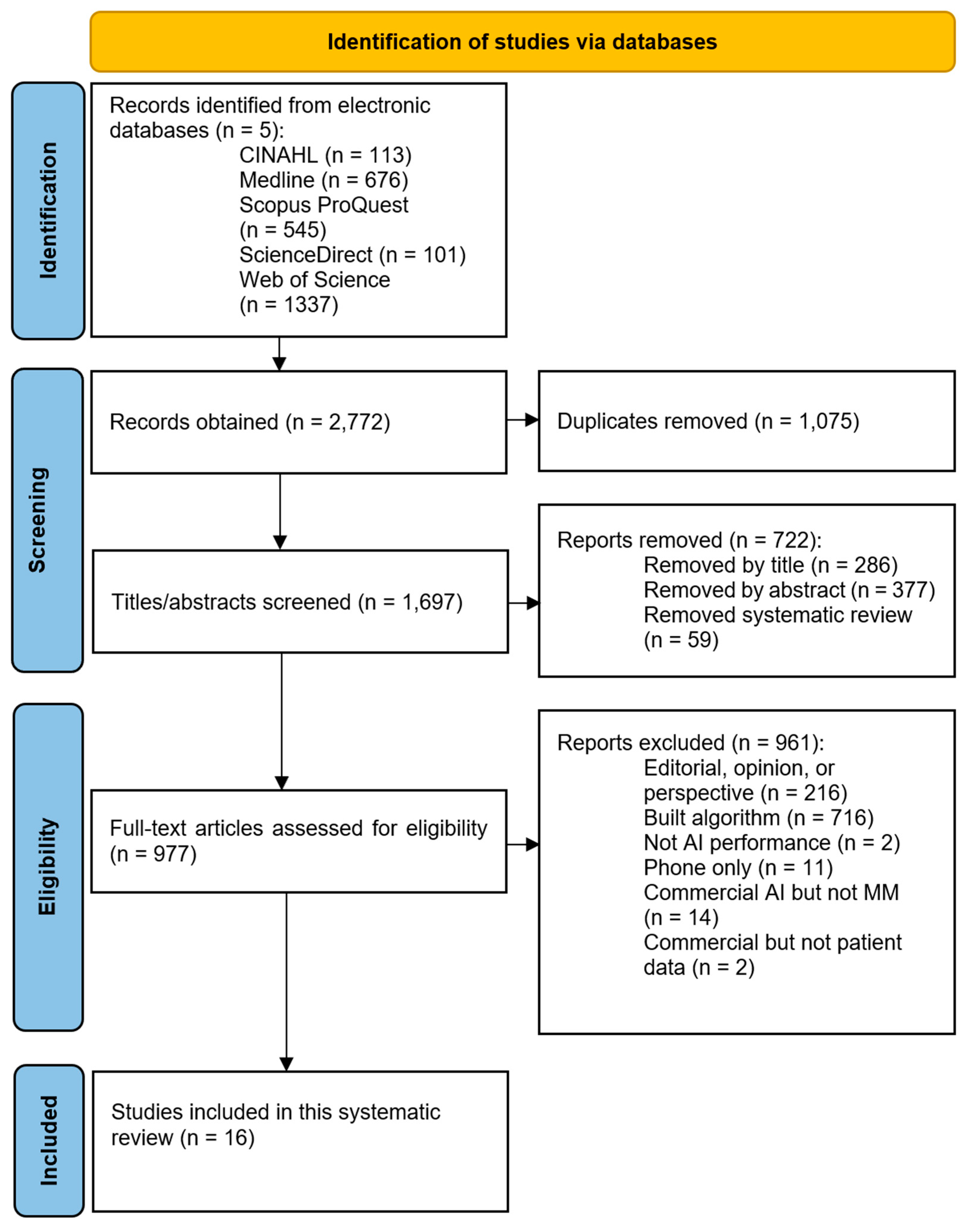
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Search Strategy
2.3. Reference Software and Study Selection
2.4. Data Extraction and Data Synthesis
2.5. Critical Appraisal
3. Results
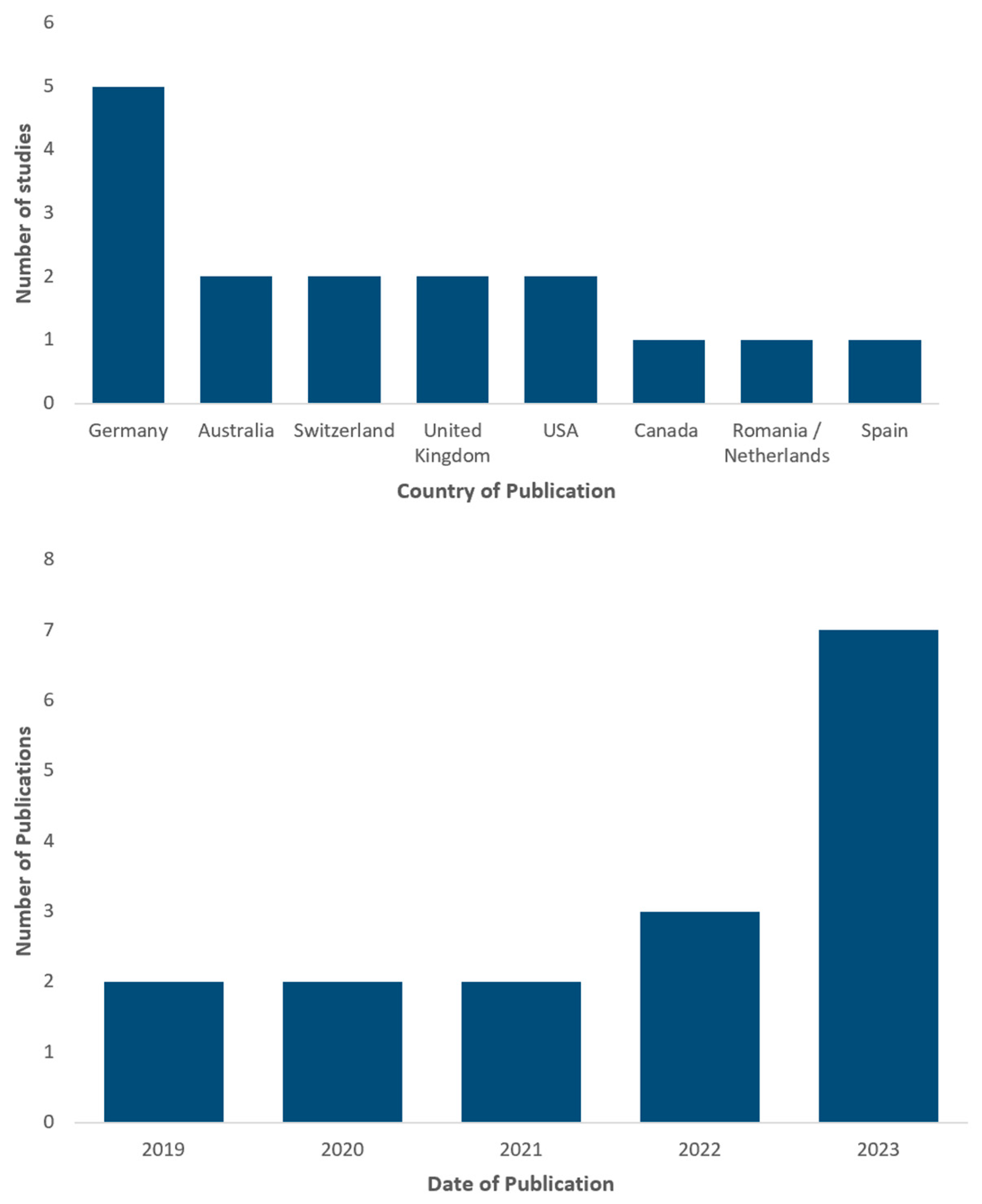
3.1. Study Characteristics
3.2. Quality of Studies
3.3. Assessment of Performance Metrics, Mobile Applications
3.4. Assessment of Performance Metrics, 3D TBP
3.5. Assessment of Performance Metrics, 2D TBP
3.6. Assessment of Performance Metrics, CNN with Clinician
3.7. Assessment of Performance Metrics, CNN
3.8. Assessment of Performance Metrics, CNN versus Clinician
3.9. Summary of Performance Metrics
4. Discussion
4.1. Sensitivity and Specificity: Which Is More Important?
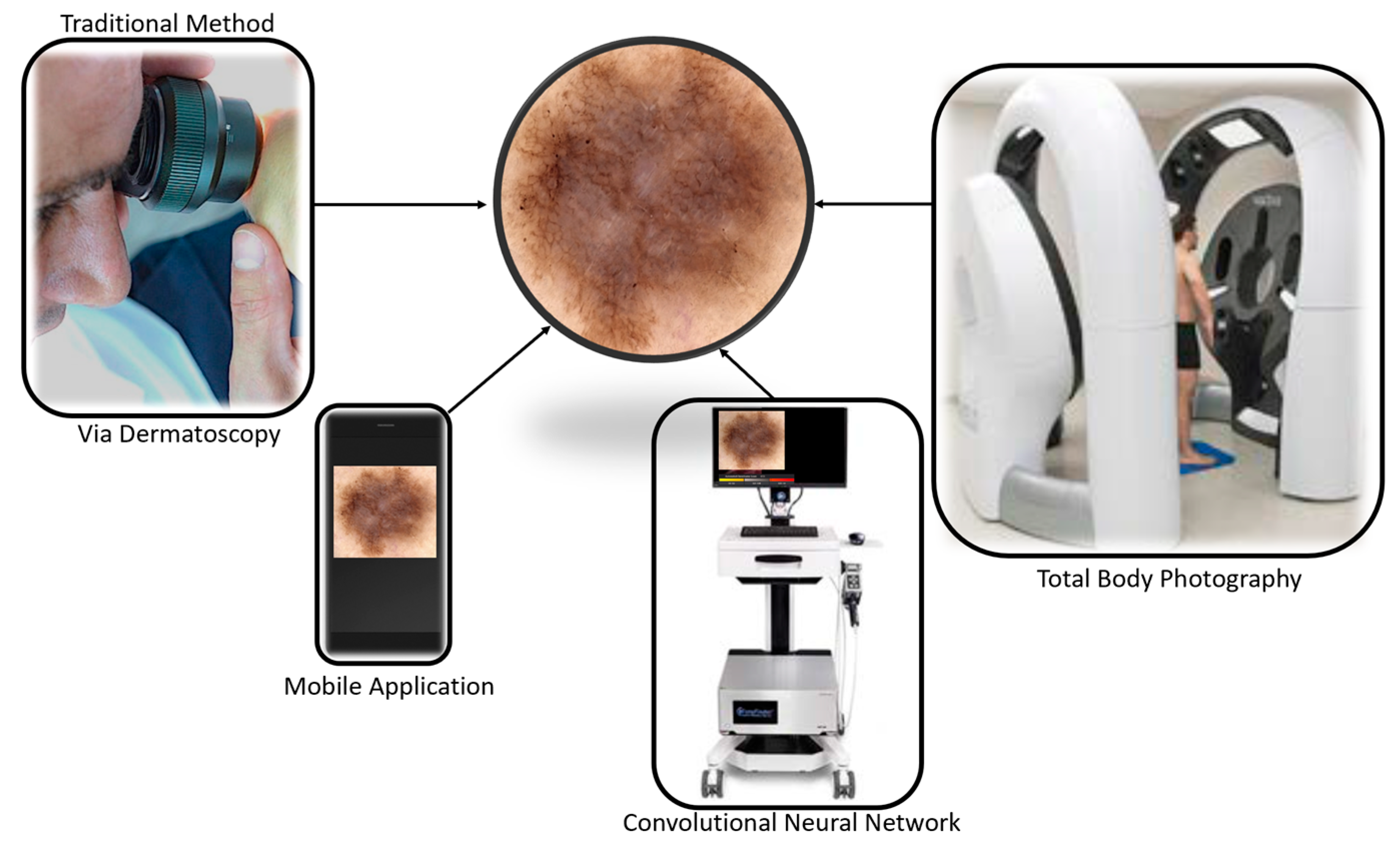
4.2. Mobile Applications and Their Performance
4.3. Total Body Photography
4.4. Clinicians and AI, Working in Unison
4.5. Performance of Bedside CNN versus Clinicians
4.6. Future Directions
4.7. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Olsen, C.M.; Green, A.C.; Pandeya, N.; Whiteman, D.C. Trends in Melanoma Incidence Rates in Eight Susceptible Populations through 2015. J. Investig. Dermatol. 2019, 139, 1392–1395. [Google Scholar] [CrossRef] [PubMed]
- Watts, C.G.; Dieng, M.; Morton, R.L.; Mann, G.J.; Menzies, S.W.; Cust, A.E. Clinical practice guidelines for identification, screening and follow-up of individuals at high risk of primary cutaneous melanoma: A systematic review. Br. J. Dermatol. 2015, 172, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, D.C.; Olsen, C.M.; MacGregor, S.; Law, M.H.; Thompson, B.; Dusingize, J.C.; Green, A.C.; Neale, R.E.; Pandeya, N.; Study, Q.S. The effect of screening on melanoma incidence and biopsy rates. Br. J. Dermatol. 2022, 187, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Henrikson, N.B.; Ivlev, I.; Blasi, P.R.; Nguyen, M.B.; Senger, C.A.; Perdue, L.A.; Lin, J.S. Skin Cancer Screening: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA 2023, 329, 1296–1307. [Google Scholar] [CrossRef]
- Kittler, H. How to Combat Over, diagnosis of Melanoma. Dermatol. Pract. Concept. 2023, 13, e2023248. [Google Scholar] [CrossRef] [PubMed]
- Navarrete-Dechent, C.; Lallas, A. Overdiagnosis of Melanoma: Is It a Real Problem? Dermatol. Pract. Concept. 2023, 13, e2023246. [Google Scholar] [CrossRef]
- Janda, M.; Cust, A.E.; Neale, R.E.; Aitken, J.F.; Baade, P.D.; Green, A.C.; Khosrotehrani, K.; Mar, V.; Soyer, H.P.; Whiteman, D.C. Early detection of melanoma: A consensus report from the Australian Skin and Skin Cancer Research Centre Melanoma Screening Summit. Aust. N. Z. J. Public Health 2020, 44, 111–115. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Chuchu, N.; Ferrante di Ruffano, L.; Matin, R.N.; Thomson, D.R.; Wong, K.Y.; Aldridge, R.B.; Abbott, R.; Fawzy, M.; et al. Dermoscopy, with and without visual inspection, for diagnosing melanoma in adults. Cochrane Database Syst. Rev. 2018, 12, CD011902. [Google Scholar] [CrossRef]
- Kittler, H. Evolution of the Clinical, Dermoscopic and Pathologic Diagnosis of Melanoma. Dermatol. Pract. Concept. 2021, 11, e2021163S. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Cowen, E.A.; Kurtansky, N.R.; Weber, J.; Dauscher, M.; DeFazio, J.; Deng, L.; Dusza, S.W.; Haliasos, H.; Halpern, A.C.; et al. Prospective validation of dermoscopy-based open-source artificial intelligence for melanoma diagnosis (PROVE-AI study). NPJ Digit. Med. 2023, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Jutzi, T.B.; Krieghoff-Henning, E.I.; Holland-Letz, T.; Utikal, J.S.; Hauschild, A.; Schadendorf, D.; Sondermann, W.; Fröhling, S.; Hekler, A.; Schmitt, M.; et al. Artificial Intelligence in Skin Cancer Diagnostics: The Patients’ Perspective. Front. Med. 2020, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P. Artificial intelligence for melanoma diagnosis. Ital. J. Dermatol. Venereol. 2021, 156, 289–299. [Google Scholar] [CrossRef]
- Menzies, S.W.; Bischof, L.; Talbot, H.; Gutenev, A.; Avramidis, M.; Wong, L.; Lo, S.K.; Mackellar, G.; Skladnev, V.; McCarthy, W.; et al. The Performance of SolarScan: An Automated Dermoscopy Image Analysis Instrument for the Diagnosis of Primary Melanoma. Arch. Dermatol. 2005, 141, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Melarkode, N.; Srinivasan, K.; Qaisar, S.M.; Plawiak, P. AI-Powered Diagnosis of Skin Cancer: A Contemporary Review, Open Challenges and Future Research Directions. Cancers 2023, 15, 1183. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset.; a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 2021, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.S.; Rozeira, J. PH2—A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Marchetti, M.A.; Liopyris, K.; Dusza, S.W.; Codella, N.C.F.; Gutman, D.A.; Helba, B.; Kalloo, A.; Halpern, A.C.; Soyer, H.P.; Curiel-Lewandrowski, C.; et al. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J. Am. Acad. Dermatol. 2020, 82, 622–627. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef]
- Kourounis, G.; Elmahmudi, A.A.; Thomson, B.; Hunter, J.; Ugail, H.; Wilson, C. Computer image analysis with artificial intelligence: A practical introduction to convolutional neural networks for medical professionals. Postgrad. Med. J. 2023, 99, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 1. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional neural networks in medical image understanding: A survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Dick, V.; Sinz, C.; Mittlböck, M.; Kittler, H.; Tschandl, P. Accuracy of Computer-Aided Diagnosis of Melanoma: A Meta-analysis. JAMA Dermatol. 2019, 155, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Chuchu, N.; Takwoingi, Y.; Dinnes, J.; Matin, R.N.; Bassett, O.; Moreau, J.F.; Bayliss, S.E.; Davenport, C.; Godfrey, K.; O’Connell, S.; et al. Smartphone applications for triaging adults with skin lesions that are suspicious for melanoma. Cochrane Database Syst. Rev. 2018, 2018, 12. [Google Scholar] [CrossRef]
- Jones, O.T.; Matin, R.N.; van der Schaar, M.; Prathivadi Bhayankaram, K.; Ranmuthu, C.K.I.; Islam, M.S.; Behiyat, D.; Boscott, R.; Calanzani, N.; Emery, J.; et al. Artificial intelligence and machine learning algorithms for early detection of skin cancer in community and primary care settings: A systematic review. Lancet Digit. 2022, 4, e466–e476. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesthesiol. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Downes, M.J.; Brennan, M.L.; Williams, H.C.; Dean, R.S. Development of a critical appraisal tool to assess the quality of cross-sectional studies (AXIS). BMJ Open 2016, 6, e011458. [Google Scholar] [CrossRef]
- National Health and Medical Research Council. NHMRC Evidence Hierarchy: Designations of ‘Levels of Evidence’ According to Type of Research Questions. 2009. Available online: https://www.nhmrc.gov.au/ (accessed on 29 January 2024).
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Tejani, I.; Jarmain, T.; Kellett, L.; Moy, R.L. Artificial Intelligence vs Medical Providers in the Dermoscopic Diagnosis of Melanoma. Cutis 2023, 111, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Jahn, A.S.; Navarini, A.A.; Cerminara, S.E.; Kostner, L.; Huber, S.M.; Kunz, M.; Maul, J.T.; Dummer, R.; Sommer, S.; Neuner, A.D.; et al. Over-Detection of Melanoma-Suspect Lesions by a CE-Certified Smartphone App: Performance in Comparison to Dermatologists, 2D and 3D Convolutional Neural Networks in a Prospective Data Set of 1204 Pigmented Skin Lesions Involving Patients’ Perception. Cancers 2022, 14, 3829. [Google Scholar] [CrossRef] [PubMed]
- Udrea, A.; Mitra, G.D.; Costea, D.; Noels, E.C.; Wakkee, M.; Siegel, D.M.; de Carvalho, T.M.; Nijsten, T.E.C. Accuracy of a smartphone application for triage of skin lesions based on machine learning algorithms. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Cerminara, S.E.; Cheng, P.; Kostner, L.; Huber, S.; Kunz, M.; Maul, J.T.; Böhm, J.S.; Dettwiler, C.F.; Geser, A.; Jakopović, C.; et al. Diagnostic performance of augmented intelligence with 2D and 3D total body photography and convolutional neural networks in a high-risk population for melanoma under real-world conditions: A new era of skin cancer screening? Eur. J. Cancer 2023, 190, 112954. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.A.; Nazir, Z.H.; Nanda, J.K.; Dusza, S.W.; D’Alessandro, B.M.; DeFazio, J.; Halpern, A.C.; Rotemberg, V.M.; Marghoob, A.A. 3D Whole-body skin imaging for automated melanoma detection. J. Eur. Acad. Dermatol. Venereol. 2023, 37, 945–950. [Google Scholar] [CrossRef]
- Winkler, J.K.; Blum, A.; Kommoss, K.; Enk, A.; Toberer, F.; Rosenberger, A.; Haenssle, H.A. Assessment of Diagnostic Performance of Dermatologists Cooperating with a Convolutional Neural Network in a Prospective Clinical Study: Human with Machine. JAMA Dermatol. 2023, 159, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; Stolz, W.; et al. Association Between Surgical Skin Markings in Dermoscopic Images and Diagnostic Performance of a Deep Learning Convolutional Neural Network for Melanoma Recognition. JAMA Dermatol. 2019, 155, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Abassi, M.S.; Fuchs, T.; Haenssle, H.A. Association between different scale bars in dermoscopic images and diagnostic performance of a market-approved deep learning convolutional neural network for melanoma recognition. Eur. J. Cancer 2021, 145, 146–154. [Google Scholar] [CrossRef]
- Winkler, J.K.; Tschandl, P.; Toberer, F.; Sies, K.; Fink, C.; Enk, A.; Kittler, H.; Haenssle, H.A. Monitoring patients at risk for melanoma: May convolutional neural networks replace the strategy of sequential digital dermoscopy? Eur. J. Cancer 2022, 160, 180–188. [Google Scholar] [CrossRef]
- Fink, C.; Blum, A.; Buhl, T.; Mitteldorf, C.; Hofmann-Wellenhof, R.; Deinlein, T.; Stolz, W.; Trennheuser, L.; Cussigh, C.; Deltgen, D.; et al. Diagnostic performance of a deep learning convolutional neural network in the differentiation of combined naevi and melanomas. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- MacLellan, A.N.; Price, E.L.; Publicover-Brouwer, P.; Matheson, K.; Ly, T.Y.; Pasternak, S.; Walsh, N.M.; Gallant, C.J.; Oakley, A.; Hull, P.R.; et al. The use of noninvasive imaging techniques in the diagnosis of melanoma: A prospective diagnostic accuracy study. J. Am. Acad. Dermatol. 2021, 85, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gonzalez, M.; Azcarraga, C.; Martin-Gil, A.; Carpena-Torres, C.; Jaen, P. Efficacy of a Deep Learning Convolutional Neural Network System for Melanoma Diagnosis in a Hospital Population. Int. J. Environ. Res. Public Health 2022, 19, 3892. [Google Scholar] [CrossRef] [PubMed]
- Menzies, S.W.; Sinz, C.; Menzies, M.; Lo, S.N.; Yolland, W.; Lingohr, J.; Razmara, M.; Tschandl, P.; Guitera, P.; Scolyer, R.A.; et al. Comparison of humans versus mobile phone-powered artificial intelligence for the diagnosis and management of pigmented skin cancer in secondary care: A multicentre, prospective, diagnostic, clinical trial. Lancet Digit. 2023, 5, e679–e691. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.J.; Stapelberg, M.; Rosic, N.; Hudson, J.; Coxon, P.; Furness, J.; Walsh, J.; Climstein, M. Implementation of artificial intelligence for the detection of cutaneous melanoma within a primary care setting: Prevalence and types of skin cancer in outdoor enthusiasts. PeerJ 2023, 11, e15737. [Google Scholar] [CrossRef]
- Phillips, M.; Marsden, H.; Jaffe, W.; Matin, R.N.; Wali, G.N.; Greenhalgh, J.; McGrath, E.; James, R.; Ladoyanni, E.; Bewley, A.; et al. Assessment of Accuracy of an Artificial Intelligence Algorithm to Detect Melanoma in Images of Skin Lesions. JAMA Netw. Open 2019, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.; Hyde, C.; Mullarkey, D.; Greenhalgh, J.; Kalsi, D.; Ko, J. Real-world post-deployment performance of a novel machine learning-based digital health technology for skin lesion assessment and suggestions for post-market surveillance. Front. Med. 2023, 10, 1264846. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, S.; Donnelly, D.; Call, M.; Johannet, P.; Moran, U.; Polsky, D.; Shapiro, R.; Berman, R.; Pavlick, A.; Weber, J.; et al. Melanoma Prognosis: Accuracy of the American Joint Committee on Cancer Staging Manual Eighth Edition. J. Natl. Cancer Inst. 2020, 112, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Sangers, T.E.; Kittler, H.; Blum, A.; Braun, R.P.; Barata, C.; Cartocci, A.; Combalia, M.; Esdaile, B.; Guitera, P.; Haenssle, H.A.; et al. Position statement of the EADV Artificial Intelligence (AI) Task Force on AI-assisted smartphone apps and web-based services for skin disease. J. Eur. Acad. Dermatol. Venereol. 2023, 38, 22–30. [Google Scholar] [CrossRef]
- Melanoma Research Alliance. Melanoma: Confirming the Diagnosis. 2024. Available online: https://www.curemelanoma.org/patient-eng/diagnosing-melanoma/confirming-the-diagnosis (accessed on 28 March 2024).
- Brinker, T.J.; Hekler, A.; Hauschild, A.; Berking, C.; Schilling, B.; Enk, A.H.; Haferkamp, S.; Karoglan, A.; von Kalle, C.; Weichenthal, M.; et al. Comparing artificial intelligence algorithms to 157 German dermatologists: The melanoma classification benchmark. Eur. J. Cancer 2019, 111, 30–37. [Google Scholar] [CrossRef]
- Koh, U.; Horsham, C.; Soyer, H.P.; Loescher, L.J.; Gillespie, N.; Vagenas, D.; Janda, M. Consumer Acceptance and Expectations of a Mobile Health Application to Photograph Skin Lesions for Early Detection of Melanoma. Dermatology 2019, 235, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Hornung, A.; Steeb, T.; Wessely, A.; Brinker, T.J.; Breakell, T.; Erdmann, M.; Berking, C.; Heppt, M.V. The value of total body photography for the early detection of melanoma: A systematic review. Int. J. Environ. Res. Public Health 2021, 18, 1726. [Google Scholar] [CrossRef] [PubMed]
- Betz-Stablein, B.; Soyer, H.P. Overdiagnosis in Melanoma Screening: Is It a Real Problem? Dermatol. Pract. Concept. 2023, 13, e2023247. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Way, D.; Gupta, V.; Gao, Y.; De Oliveira Marinho, G.; Hartford, J.; Sayres, R.; Kanada, K.; Eng, C.; Nagpal, K.; et al. Development and Assessment of an Artificial Intelligence-Based Tool for Skin Condition Diagnosis by Primary Care Physicians and Nurse Practitioners in Teledermatology Practices. JAMA Netw. Open 2021, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human-computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Pandeya, N.; Olsen, C.M.; Shalit, M.M.; Dusingize, J.C.; Neale, R.E.; Whiteman, D.C. The diagnosis and initial management of melanoma in Australia: Findings from the prospective, population-based QSkin Study. Med. J. Aust. 2023, 218, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Goodman, G.J.; Armour, K.S.; Kolodziejczyk, J.K.; Santangelo, S.; Gallagher, C.J. Comparison of self-reported signs of facial ageing among Caucasian women in Australia versus those in the USA, the UK and Canada. Australas. J. Dermatol. 2018, 59, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Petty, A.J.; Ackerson, B.; Garza, R.; Peterson, M.; Liu, B.; Green, C.; Pavlis, M. Meta-analysis of number needed to treat for diagnosis of melanoma by clinical setting. J. Am. Acad. Dermatol. 2020, 82, 1158–1165. [Google Scholar] [CrossRef] [PubMed]
- Rosendahl, C.; Cameron, A.; McColl, I.; Wilkinson, D. Dermatoscopy in routine practice—‘chaos and clues’. Aust. Fam. Physician 2012, 41, 482–487. [Google Scholar]
- Polap, D. Analysis of Skin Marks Through the Use of Intelligent Things. IEEE Access 2019, 7, 149355–149363. [Google Scholar] [CrossRef]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar]
- Crawford, M.E.; Kamali, K.; Dorey, R.A.; MacIntyre, O.C.; Cleminson, K.; MacGillivary, M.L.; Green, P.J.; Langley, R.G.; Purdy, K.S.; DeCoste, R.C.; et al. Using Artificial Intelligence as a Melanoma Screening Tool in Self-Referred Patients. J. Cutan. Med. Surg. 2023, 28, 12034754231216967. [Google Scholar] [CrossRef] [PubMed]
- Goessinger, E.V.; Cerminara, S.E.; Mueller, A.M.; Gottfrois, P.; Huber, S.; Amaral, M.; Wenz, F.; Kostner, L.; Weiss, L.; Kunz, M.; et al. Consistency of convolutional neural networks in dermoscopic melanoma recognition: A prospective real-world study about the pitfalls of augmented intelligence. J. Eur. Acad. Dermatol. Venereol. 1977, 19777. [Google Scholar]
- Haenssle, H.A.; Fink, C.; Toberer, F.; Winkler, J.; Stolz, W.; Deinlein, T.; Hofmann-Wellenhof, R.; Lallas, A.; Emmert, S.; Buhl, T.; et al. Man against machine reloaded: Performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions. Ann. Oncol. 2020, 31, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.A.; Winkler, J.K.; Fink, C.; Toberer, F.; Enk, A.; Stolz, W.; Deinlein, T.; Hofmann-Wellenhof, R.; Kittler, H.; Tschandl, P.; et al. Skin lesions of face and scalp—Classification by a market-approved convolutional neural network in comparison with 64 dermatologists. Eur. J. Cancer 2021, 144, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Kommoss, K.S.; Winkler, J.K.; Mueller-Christmann, C.; Bardehle, F.; Toberer, F.; Stolz, W.; Kraenke, T.; Hofmann-Wellenhof, R.; Blum, A.; Enk, A.; et al. Observational study investigating the level of support from a convolutional neural network in face and scalp lesions deemed diagnostically ‘unclear’ by dermatologists. Eur. J. Cancer 2023, 185, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Fei, W.M.; Shen, C.B.; Wang, Z.Y.; Jing, Y.; Meng, R.S.; Cui, Y. Diagnostic capacity of skin tumor artificial intelligence-assisted decision-making software in real-world clinical settings. China Med. J. 2020, 133, 2020–2026. [Google Scholar] [CrossRef]
- Maguire, W.F.; Haley, P.H.; Dietz, C.M.; Hoffelder, M.; Brandt, C.S.; Joyce, R.; Fitzgerald, G.; Minnier, C.; Sander, C.; Ferris, L.K.; et al. Development and Narrow Validation of Computer Vision Approach to Facilitate Assessment of Change in Pigmented Cutaneous Lesions. JID Innov. 2023, 3, 100181. [Google Scholar] [CrossRef] [PubMed]
- Marsden, H.; Morgan, C.; Austin, S.; DeGiovanni, C.; Venzi, M.; Kemos, P.; Greenhalgh, J.; Mullarkey, D.; Palamaras, I. Effectiveness of an image analyzing AI-based Digital Health Technology to identify Non-Melanoma Skin Cancer and other skin lesions: Results of the DERM-003 study. Front. Med. 2023, 10, 1288521. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-López, C.; Ramírez-Cornejo, C.; Marchetti, M.A.; Han, S.S.; Del Barrio-Díaz, P.; Jaque, A.; Uribe, P.; Majerson, D.; Curi, M.; Del Puerto, C.; et al. Performance of a deep neural network in teledermatology: A single-centre prospective diagnostic study. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 546–553. [Google Scholar] [CrossRef]
- Sies, K.; Winkler, J.K.; Fink, C.; Bardehle, F.; Toberer, F.; Buhl, T.; Enk, A.; Blum, A.; Rosenberger, A.; Haenssle, H.A. Past and present of computer-assisted dermoscopic diagnosis: Performance of a conventional image analyser versus a convolutional neural network in a prospective data set of 1,981 skin lesions. Eur. J. Cancer 2020, 135, 39–46. [Google Scholar] [CrossRef]
- Sies, K.; Winkler, J.K.; Fink, C.; Bardehle, F.; Toberer, F.; Buhl, T.; Enk, A.; Blum, A.; Stolz, W.; Rosenberger, A.; et al. Does sex matter? Analysis of sex-related differences in the diagnostic performance of a market-approved convolutional neural network for skin cancer detection. Eur. J. Cancer 2022, 164, 88–94. [Google Scholar] [CrossRef]
- Sies, K.; Winkler, J.K.; Fink, C.; Bardehle, F.; Toberer, F.; Kommoss, F.K.F.; Buhl, T.; Enk, A.; Rosenberger, A.; Haenssle, H.A. Dark corner artefact and diagnostic performance of a market-approved neural network for skin cancer classification. J. Dtsch. Dermatol. Ges. 2021, 19, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Q.; Zhang, X.Y.; Liu, J.; Tao, C.; Zhu, C.Y.; Shu, C.; Xu, T.; Jin, H.Z. Deep learning-based.; computer-aided classifier developed with dermoscopic images shows comparable performance to 164 dermatologists in cutaneous disease diagnosis in the Chinese population. China Med. J. 2020, 133, 2027–2036. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Abassi, M.S.; Fuchs, T.; Blum, A.; Stolz, W.; Coras-Stepanek, B.; et al. Collective human intelligence outperforms artificial intelligence in a skin lesion classification task. J. Dtsch. Dermatol. Ges. 2021, 19, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Deinlein, T.; Hofmann-Wellenhof, R.; Thomas, L.; Lallas, A.; Blum, A.; et al. Melanoma recognition by a deep learning convolutional neural network-Performance in different melanoma subtypes and localisations. Eur. J. Cancer 2020, 127, 21–29. [Google Scholar] [CrossRef] [PubMed]

| Author, Year, Country | Patients | Technology | True Positive | False Negative | False Positive | True Negative | Sensitivity [95% CI] * | Specificity [95% CI] * | Accuracy [95% CI] * | AUROC [95% CI] * |
|---|---|---|---|---|---|---|---|---|---|---|
| Anderson et al., 2023, USA [34] | MM (n = 20), Other (n = 80 | CNN (triage; mobile application) | 16 | 4 | 4 | 76 | 80.0 | 95.0 | 92.0 | |
| Jahn et al., 2022, Switzerland [35] | MM (n = 6), Other (n = 55) | CNN (SkinVision; mobile application) | 5 | 1 | 22 | 33 | 83.3 | 60.0 | 62.3 | 0.717 |
| Udrea et al., 2020, Romania/Netherlands [36] | MM (n = 138), Other (n = 6000) | CNN SkinVision; mobile application) | 128 | 10 | 1302 | 4698 | 92.8 [87.8–96.5] | 78.3 [77.2–79.3] | 78.6 |
| Author, Year, Country | Patients | Technology | True Positive | False Negative | False Positive | True Negative | Sensitivity [95% CI] * | Specificity [95% CI] * | Accuracy [95% CI] * | AUROC [95% CI] * |
|---|---|---|---|---|---|---|---|---|---|---|
| Cerminara et al., 2023, Switzerland [37] | MM (n = 10), Other (n = 65) | CNN (Canfield; 3D Vectra WB360) | 9 | 1 | 23 | 42 | 90.0 | 64.6 | 68.0 | 0.92 [0.85–1.00] |
| Jahn et al., 2022, Switzerland [35] | MM (n = 6), Other (n = 55) | CNN (Canfield; 3D Vectra WB360) | 5 | 1 | 20 | 35 | 83.3 | 63.6 | 65.6 | |
| Marchetti et al., 2023, USA [38] | MM (n = 43), Other (n = 22,489) | CNN (Canfield; 3D Vectra WB360) | 0.9399 [0.92–0.96] |
| Author, Year, Country | Patients | Technology | True Positive | False Negative | False Positive | True Negative | Sensitivity [95% CI] * | Specificity [95% CI] * | Accuracy [95% CI] * | AUROC [95% CI] * |
|---|---|---|---|---|---|---|---|---|---|---|
| Cerminara et al., 2023, Switzerland [37] | MM (n = 10), Other (n = 65) | CNN (FotoFinder; 2D TBP) | 7 | 3 | 39 | 26 | 70.0 | 40.0 | 44.0 | 0.68 [0.46–0.90] |
| Jahn et al., 2022, Switzerland [35] | MM (n = 6), Other (n = 55) | CNN (FotoFinder; 2D TBP) | 5 | 1 | 33 | 22 | 83.3 | 40.0 | 44.3 |
| Author, Year, Country | Patients | Clinician or Clinician with AI | True Positive | False Negative | False Positive | True Negative | Sensitivity [95% CI] * | Specificity [95% CI] * | Accuracy [95% CI] * | AUROC [95% CI] * |
|---|---|---|---|---|---|---|---|---|---|---|
| Cerminara et al., 2023, Switzerland [37] | MM (n = 10), Other (n = 65) | Dermatologist plus AI | 9 | 1 | 9 | 56 | 90.0 | 86.2 | 86.7 | 0.88 [0.80–1.00] |
| Jahn et al., 2022, Switzerland [35] | MM (n = 6), Other (n = 55) | Dermatologist (average) plus AI | 5 | 1 | 7 | 48 | 83.3 | 87.3 | 86.9 | |
| Beginner (<2 years) # | 4 | 1 | 6 | 34 | 80.0 | 85.0 | 84.4 | |||
| Skilled (2–5 years) # | 0 | 0 | 1 | 4 | - | 80.0 | 80.0 | |||
| Expert (>5 years) # | 1 | 0 | 0 | 10 | 100.0 | 100.0 | 100.0 | |||
| Winkler et al., 2023, Germany [39] | MM (n = 38), Other (n = 190) | Dermatologist plus AI | 38 | 0 | 31 | 159 | 100.0 [90.8–100] | 83.7 [77.8–88.3] | 86.4 [81.3–90.3] | 0.968 |
| Author, Year, Country | Patients | Technology | True Positive | False Negative | False Positive | True Negative | Sensitivity [95% CI] * | Specificity [95% CI] * | Accuracy [95% CI] * | AUROC [95% CI] * |
|---|---|---|---|---|---|---|---|---|---|---|
| Phillips et al., 2019, United Kingdom [48] | MM (n = 79), Other (n = 310) | CNN (SkinAnalytics; DERM with iPhone) | 62 | 17 | 78.5 | 0.879 | ||||
| MM (n = 76), Other (n = 300) | CNN (SkinAnalytics; DERM with Galaxy5) | 54 | 22 | 71.1 | 0.823 | |||||
| MM (n = 51), Other (n = 220) | CNN (SkinAnalytics; DERM with DSLR) | 38 | 13 | 74.5 | 0.850 | |||||
| Thomas et al., 2023, United Kingdom [49] | MM (n = 140), Other (n = 4495) | CNN (SkinAnalytics; DERMvA-UHB) | 133 | 7 | 1852 | 2643 | 95.0 [90.0–97.6] | 58.8 [57.4–60.2] | 59.9 | |
| MM (n = 33), Other (n = 676) | CNN (SkinAnalytics; DERMvA-WSFT) | 32 | 33 | 249 | 427 | 97.0 [84.7–99.5] | 63.2 [59.5–66.7] | 64.7 | ||
| MM (n = 58), Other (n = 2527) | CNN (SkinAnalytics; DERMvB-UHB) | 58 | 0 | 482 | 2045 | 100.0 [93.8–100] | 80.9 [79.3–82.4] | 81.4 | ||
| MM (n = 18), Other (n = 624) | CNN (SkinAnalytics; DERMvB-WSFT) | 18 | 0 | 122 | 502 | 100.0 [82.4–100] | 80.4 [77.2–83.4] | 81.0 | ||
| Fink et al., 2020, Germany [43] | MM (n = 36), Other (n = 36) | CNN (Fotofinder; MoleAnalyzer Pro) | 35 | 1 | 8 | 28 | 97.1 [82.7–99.6] | 78.8 [62.8–89.1] | 87.5 | |
| MacLellan et al., 2021, Canada [44] | MM (n = 59), Other (n = 150) | CNN (FotoFinder; MoleAnalyzer Pro) | 52 | 7 | 32 | 118 | 88.1 [79.4–96.9] | 78.8 [71.5–86.2] | 81.3 | |
| MM (n = 59), Other (n = 150) | CNN2 (FotoFinder; MoleAnalyzer Tuebinger) | 49 | 10 | 37 | 113 | 83.1 [72.6–93.6] | 75.2 [67.3–83.1] | 77.5 | ||
| Miller et al., 2023, Australia [47] | MM (n = 15), Other (n = 33) | CNN (FotoFinder; MoleAnalyzer Pro) | 8 | 7 | 15 | 33 | 53.3 | 54.4 | 54.2 | 0.540 |
| Winkler et al., 2023, Germany [39] | MM (n = 38), Other (n = 190) | CNN (FotoFinder; MoleAnalyzer Pro) | 31 | 7 | 21 | 169 | 81.6 [66.6–90.8] | 88.9 [77.8–88.3] | 87.7 [82.8–91.4] | 0.904 |
| Winkler et al., 2019, Germany [40] | MM (n = 23), Other (n = 107) | CNN (FotoFinder; MoleAnalyzer Pro) | 22 | 1 | 17 | 90 | 95.7 [79.0–99.2] | 84.1 [76.0–89.8] | 86.2 | 0.969 |
| Winkler et al., 2021, Germany [41] | MM (n = 23), Other (n = 107) | CNN (FotoFinder; MoleAnalyzer Pro) | 20 | 3 | 13 | 94 | 87.0 [67.9–95.5] | 87.9 [80.3–92.8] | 87.7 | 0.953 [0.914–0.992] |
| Winkler et al., 2022, Germany [42] | MM (n = 59), Other (n = 236) | CNN (FotoFinder; MoleAnalyzer Pro) | ||||||||
| Menzies et al., 2023, Australia [46] | MM (n = 55), Other (n = 117) | CNN (MetaOptima; 7-class) | 28 | 27 | 7 | 110 | 50.9 | 94.0 | 80.2 | |
| CNN (MetaOptima; ISIC) | 9 | 46 | 2 | 115 | 16.4 | 98.3 | 72.1 | |||
| Martin-Gonzalez et al., 2022, Spain [45] | MM (n = 55), Other (n = 177) | CNN (quantusSKIN) | 38 | 17 | 34 | 142 | 69.1 | 80.2 | 77.6 | 0.802 |
| Author, year, country | Patients | Clinician or clinician with AI | True positive | False negative | False positive | True negative | Sensitivity [95% CI] * | Specificity [95% CI] * | Accuracy [95% CI] * | AUROC [95% CI] * |
|---|---|---|---|---|---|---|---|---|---|---|
| Anderson et al., 2023, USA [34] | MM (n = 20), Other (n = 80) | Primary care providers | 67.0 | 48.0 | 52.0 | |||||
| Family physicians # | 78.0 | 41.0 | 48.0 | |||||||
| Mid-level provider # | 61.0 | 53.0 | 55.0 | |||||||
| Dermatologist | 77.0 | 57.0 | 61.0 | |||||||
| Cerminara et al., 2023, Switzerland [37] | MM (n = 10), Other (n = 65) | Dermatologist | 9 | 1 | 5 | 60 | 90.0 | 92.3 | 92.0 | 0.91 [0.80–1.00] |
| Fink et al., 2020, Germany [43] | MM (n = 36), Other (n = 36) | Dermatologist | 90.6 [84.1–94.7] | 71.0 [62.6–78.1] | ||||||
| Beginner (<2 years) # | 90.9 [82.4–95.5] | 55.1 [45.7–64.2] | ||||||||
| Skilled (2–5 years) # | 93.3 [86.3–96.9] | 74.2 [64.4–82.0] | ||||||||
| Expert (>5 years) # | 86.7 [77.7–92.4] | 80.6 [70.2–88.0] | ||||||||
| Jahn et al., 2022, Switzerland [35] | MM (n = 6), Other (n = 55) | Dermatologist (average) | 5 | 1 | 4 | 51 | 83.3 | 92.7 | 91.8 | |
| Beginner (<2 years) # | 4 | 1 | 3 | 37 | 80.0 | 92.5 | 91.1 | |||
| Skilled (2–5 years) # | 0 | 0 | 1 | 4 | - | 80.0 | 80.0 | |||
| Expert (>5 years) # | 1 | 0 | 0 | 10 | 100.0 | 100.0 | 100 | |||
| Maclellan et al., 2021, Canada [44] | MM (n = 59), Other (n = 150) | Dermatologist | 57 | 2 | 44 | 106 | 96.6 [91.9–100.0] | 32.2 [18.4–46.0] | 78.0 | |
| Menzies et al., 2023, Australia [46] | MM (n = 55), Other (n = 117) | Specialist | 34 | 21 | 17 | 100 | 61.8 | 85.5 | 77.9 | |
| Novice | 23 | 32 | 32 | 85 | 41.8 | 72.6 | 62.8 | |||
| Phillips et al., 2019, United Kingdom [48] | MM (n = 125), Other (n = 426) | Clinician | 84 | 41 | 67.2 | 0.778 | ||||
| Winkler et al., 2023, Germany [39] | MM (n = 38), Other (n = 190) | Dermatologist | 32 | 6 | 53 | 137 | 84.2 [69.9–92.6] | 72.1 [65.3–78.0] | 74.1 [68.1–79.4] | 0.895 |
| Type of Technology (n = Number of Studies) | Performance Metric | Lower Limit % | Upper Limit % |
|---|---|---|---|
| Mobile Applications (n = 3) [34,35,36] | Sensitivity | 80.0 [34] | 92.8 [36] |
| Specificity | 60.0 [35] | 95.0 [34] | |
| Accuracy | 62.3 [35] | 92.0 [34] | |
| AUROC | 0.717 [35] | 0.717 [35] | |
| 3D TBP (n = 3) [35,37,38] | Sensitivity | 83.3 [35] | 90.0 [37] |
| Specificity | 63.6 [35] | 64.6 [37] | |
| Accuracy | 65.6 [35] | 68.0 [37] | |
| AUROC | 0.92 [37] | 0.94 [38] | |
| 2D TBP (n = 2) [35,37] | Sensitivity | 70.0 [37] | 83.3 [35] |
| Specificity | 40.0 [35,37] | 40.0 [35,37] | |
| Accuracy | 44.0 [37] | 44.3 [35] | |
| AUROC | 0.68 [37] | 0.68 [37] | |
| Clinicians in unison with AI (n = 3) [35,37,39] | Sensitivity | 83.3 [35] | 100.0 [39] |
| Specificity | 83.7 [39] | 87.3 [35] | |
| Accuracy | 86.4 [39] | 86.9 [35] | |
| AUROC | 0.88 [37] | 0.968 [39] | |
| CNN (n = 11) [39,40,41,42,43,44,45,46,47,48,49] | Sensitivity | 16.4 [46] | 100.0 [49] |
| Specificity | 54.4 [47] | 98.3 [46] | |
| Accuracy | 54.2 [47] | 87.7 [39,41] | |
| AUROC | 0.540 [47] | 0.969 [40] | |
| Clinician, No AI (n = 8) [34,35,37,39,43,44,46,48] | Sensitivity | 41.8 [46] | 96.6 [44] |
| Specificity | 32.2 [44] | 92.7 [35] | |
| Accuracy | 52.0 [34] | 92.0 [37] | |
| AUROC | 0.778 [48] | 0.91 [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, I.; Rosic, N.; Stapelberg, M.; Hudson, J.; Coxon, P.; Furness, J.; Walsh, J.; Climstein, M. Performance of Commercial Dermatoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review. Cancers 2024, 16, 1443. https://doi.org/10.3390/cancers16071443
Miller I, Rosic N, Stapelberg M, Hudson J, Coxon P, Furness J, Walsh J, Climstein M. Performance of Commercial Dermatoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review. Cancers. 2024; 16(7):1443. https://doi.org/10.3390/cancers16071443
Chicago/Turabian StyleMiller, Ian, Nedeljka Rosic, Michael Stapelberg, Jeremy Hudson, Paul Coxon, James Furness, Joe Walsh, and Mike Climstein. 2024. "Performance of Commercial Dermatoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review" Cancers 16, no. 7: 1443. https://doi.org/10.3390/cancers16071443
APA StyleMiller, I., Rosic, N., Stapelberg, M., Hudson, J., Coxon, P., Furness, J., Walsh, J., & Climstein, M. (2024). Performance of Commercial Dermatoscopic Systems That Incorporate Artificial Intelligence for the Identification of Melanoma in General Practice: A Systematic Review. Cancers, 16(7), 1443. https://doi.org/10.3390/cancers16071443












