Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation
Abstract
Simple Summary
Abstract
Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.fossilhunters.xyz/natural-history-2/micromutations-or-macromutations.html (accessed on 14 January 2024).
- Heng, H.H. Genome Chaos: Rethinking Genetics, Evolution and Molecular Medicine; Academic Press: San Diego, CA, USA, 2019. [Google Scholar] [CrossRef]
- Pellestor, F.; Gatinois, V. Chromoanagenesis: A piece of the macroevolution scenario. Mol. Cytogenet. 2020, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.J.; Eldredge, N. Punctuated equilibrium comes of age. Nature 1993, 366, 223–227. [Google Scholar] [CrossRef]
- Gould, S.J. The Structure of Evolutionary Theory; Belknap Press of Harvard University Press: Cambridge MA, USA, 2002; 1464p. [Google Scholar] [CrossRef]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; Macdonald, T.Y.; Ghandi, M.; et al. Punctuated Evolution of Prostate Cancer Genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with re-duced response to immunotherapy. Science 2017, 355, eaaf83. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Wilson, G.A.; McGranahan, N.; Birkbak, N.J.; Watkins, T.B.K.; Veeriah, S.; Shafi, S.; Johnson, D.H.; Mitter, R.; Rosenthal, R.; et al. Tracking the Evolution of Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2109–2121. [Google Scholar] [CrossRef] [PubMed]
- Wikipedia “Cretaceous–Paleogene Extinction Event”. Available online: https://en.wikipedia.org/wiki/Cretaceous%E2%80%93Paleogene_extinction_event (accessed on 14 January 2024).
- McClintock, B. The stability of broken ends of chromosomes in Zea mays. Genetics 1941, 26, 234–282. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Hu, Q.; Mazagatti, A.; Valle-Inclan, J.E.; Maurais, E.G.; Dahiya, R.; Guyer, A.; Sanders, J.T.; Engel, J.L.; Nguyen, G.; et al. Mitotic clustering of pulverized chromosomes from micronuclei. Nature 2023, 618, 1041–1048. [Google Scholar] [CrossRef]
- Mazzagatti, A.; Engel, J.L.; Ly, P. Boveri and beyond: Chromothripsis and genomic instability from mitotic errors. Mol. Cell 2024, 84, 55–69. [Google Scholar] [CrossRef]
- Trivedi, P.; Steele, C.D.; Au, K.C.; Alexandrov, L.B.; Cleveland, D.W. Mitotic tethering enables inheritance of shattered micronuclear chromosomes. Nature 2023, 618, 1049–1056. [Google Scholar] [CrossRef]
- Schubert, I. Boon and bane of DNA double-strand breaks. Int. J. Mol. Sci. 2021, 22, 5171. [Google Scholar] [CrossRef]
- Thomas, C.A. The genetic organization of chromosomes. Annu. Rev. Genet. 1971, 5, 237–256. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Vu, G.T.H. Genome stability and evolution: Attempting a holistic view. Trends Plant Sci. 2016, 21, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Orel, N.; Puchta, H. Differences in processing DNA ends in Arabidopsis thaliana and tobacco: Possible implications for genome evolution. Plant Mol. Biol. 2003, 51, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Vu, G.T.H.; Cao, H.X.; Watanabe, K.; Hensel, G.; Blattner, F.R.; Kumlehn, J.; Schubert, I. Repair of site-specific DNA double-strand breaks in barley occurs via diverse pathways, primarily involving the sister chromatid. Plant Cell 2014, 26, 2156–2167. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Sun, x.; Cormack, B.P.; Boeke, J.D. Karyotype engineering by chromosome fusion leads to reproductive isolation in yeast. Nature 2018, 560, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Heng, H.H. Elimination of altered karyotypes by sexual reproduction preserves species identity. Genome 2007, 50, 517–524. [Google Scholar] [CrossRef] [PubMed]
- King, M. Species Evolution: The Rule of Chromosome Change; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Heng, J.; Heng, H.H. Karyotype coding: The creation and maintenance of system information for complexity and biodiversity. BioSystems 2021, 208, 104476. [Google Scholar] [CrossRef]
- Heng, J.; Heng, H.H. Karyotype as code of codes; An inheritance platform to shape the pattern and scale of evolution. BioSystems 2023, 233, 1–12. [Google Scholar] [CrossRef]
- Hoang, P.N.T.; Schubert, I. Reconstruction of chromosome rearrangements between the two most ancestral duckweed species Spirodela polyrhiza and S. intermedia. Chromosoma 2017, 126, 729–739. [Google Scholar] [CrossRef]
- Hoang, P.N.T.; Fuchs, J.; Schubert, V.; Tran, T.B.N.; Schubert, I. Chromosome numbers and genome sizes of all 36 duckweed species (Lemnaceae). Plants 2022, 11, 2674. [Google Scholar] [CrossRef]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 2007–2016. [Google Scholar] [CrossRef]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Endo, T.R. The gametocidal chromosome as a tool for chromosome manipulation in wheat. Chromosome Res. 2007, 15, 67–75. [Google Scholar] [CrossRef]
- Simakov, O.; Bredeson, J.; Berkoff, K.; Marletaz, F.; Mitros, T.; Schultz, D.T.; O’Connell, B.L.; Dear, P.; Martinez, D.E.; Steele, R.E.; et al. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci. Adv. 2022, 8, eabi5884. [Google Scholar] [CrossRef]
- Sacerdot, C.; Louis, A.; Bon, C.; Berthelot, C.; Roest Crollius, H. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 2018, 19, 166. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I. Chromosome evolution. Curr. Opin. Plant Biol. 2007, 10, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Mojica, E.A.; Kültz, D. Physiological mechanisms of stress-induced evolution. J. Exp. Biol. 2022, 225 (Suppl. S1), jeb243264. [Google Scholar] [CrossRef] [PubMed]
- Heng, J.; Heng, H.H. Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 2022, 81, 160–175. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.C.; Horne, S.; Zhang, J.Z.; Jackson, L.; Heng, H.H. Therapy Induced Genome Chaos: A Novel Mechanism of Rapid Cancer Drug Resistance. Front. Cell Dev. Biol. 2021, 9, 676344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mercado-Uribe, I.; Xing, Z.; Sun, B.; Kuang, J.; Liu, J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 2014, 33, 116–128. [Google Scholar] [CrossRef]
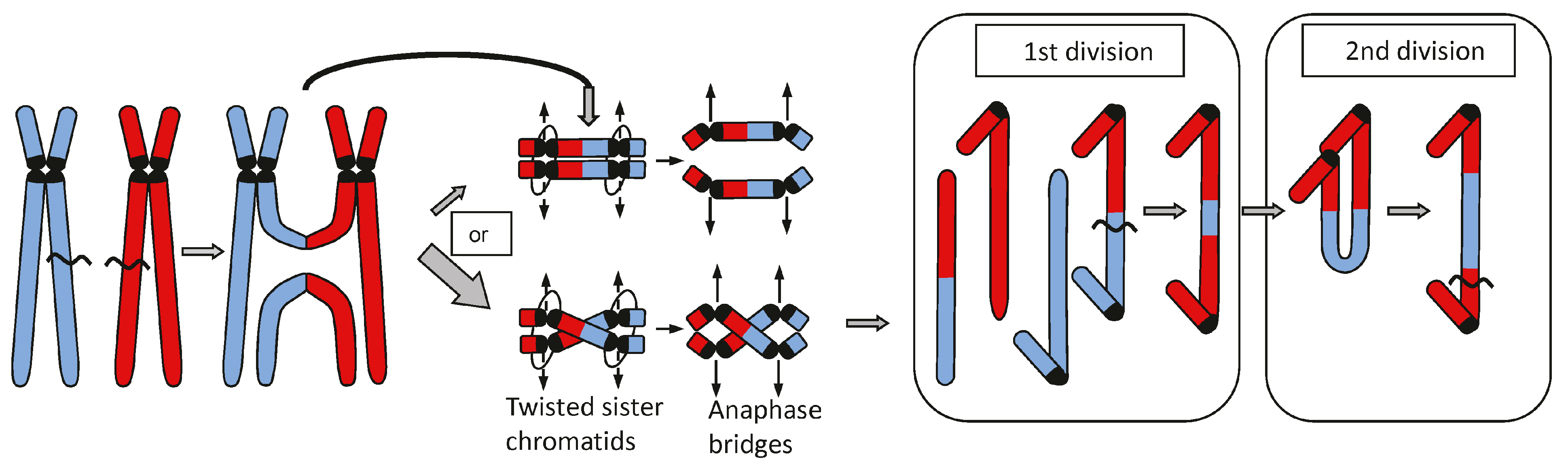
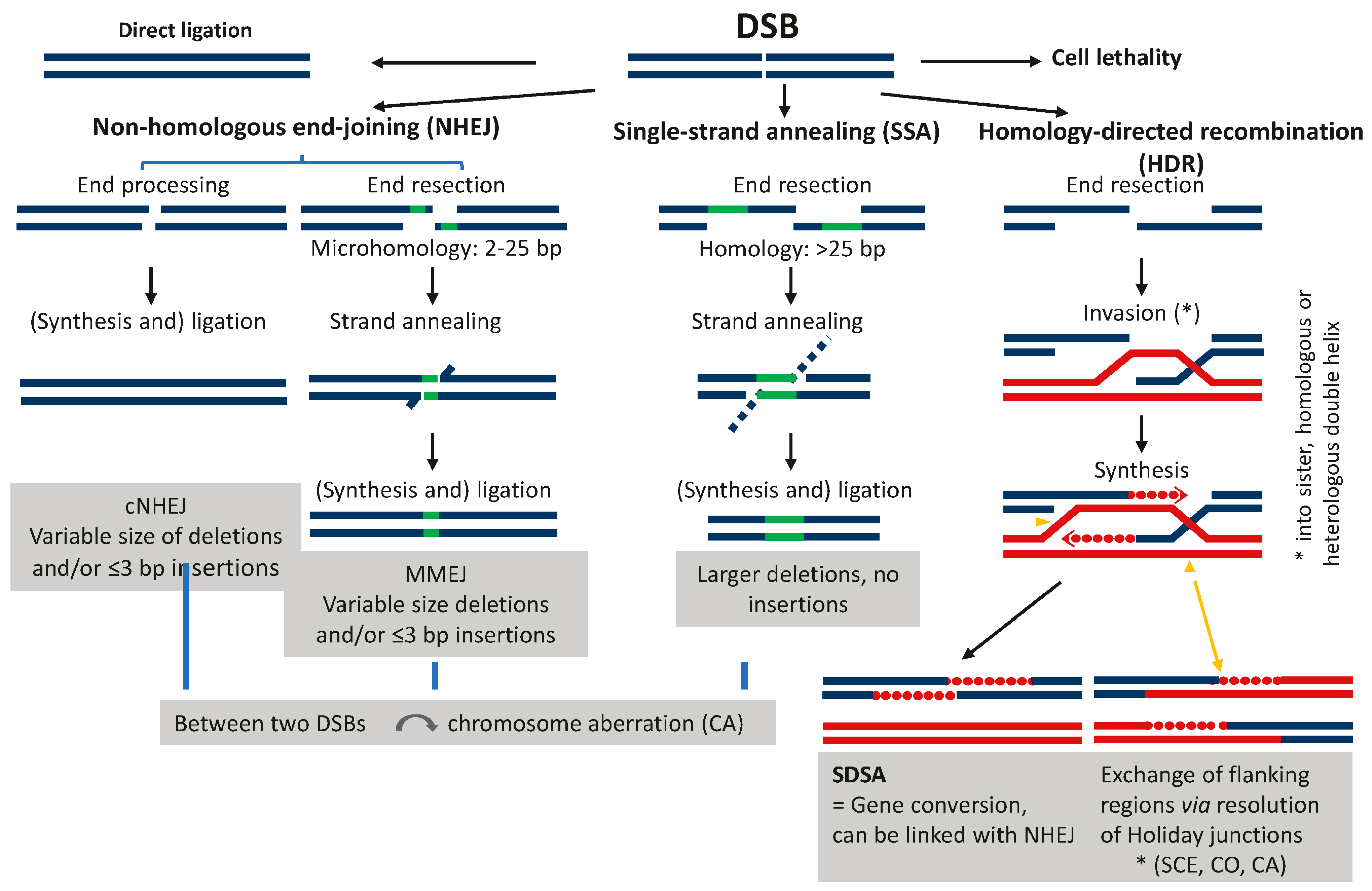



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schubert, I. Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation. Cancers 2024, 16, 554. https://doi.org/10.3390/cancers16030554
Schubert I. Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation. Cancers. 2024; 16(3):554. https://doi.org/10.3390/cancers16030554
Chicago/Turabian StyleSchubert, Ingo. 2024. "Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation" Cancers 16, no. 3: 554. https://doi.org/10.3390/cancers16030554
APA StyleSchubert, I. (2024). Macromutations Yielding Karyotype Alterations (and the Process(es) behind Them) Are the Favored Route of Carcinogenesis and Speciation. Cancers, 16(3), 554. https://doi.org/10.3390/cancers16030554





