Immune Escape Strategies in Head and Neck Cancer: Evade, Resist, Inhibit, Recruit
Simple Summary
Abstract
1. Introduction
2. Evade
3. Resist
4. Inhibit
5. Recruit
5.1. Dendritic Cells (DCs)
5.2. Macrophages
5.3. Regulatory T Cells (Tregs)
5.4. Myeloid-Derived Suppressor Cells (MDSCs)
5.5. Neutrophils
5.6. Cancer-Associated Fibroblasts (CAFs)
6. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L.; Daniel, R.W.; Viglione, M.; Symer, D.E.; et al. Evidence for a Causal Association Between Human Papillomavirus and a Subset of Head and Neck Cancers. JNCI 2000, 92, 709–720. [Google Scholar] [CrossRef]
- Harari, P.M.; Huang, S. Radiation combined with EGFR signal inhibitors: Head and neck cancer focus. Semin. Radiat. Oncol. 2006, 16, 38–44. [Google Scholar] [CrossRef]
- Harari, P.M.; Wheeler, D.L.; Grandis, J.R. Molecular target approaches in head and neck cancer: Epidermal growth factor receptor and beyond. Semin. Radiat. Oncol. 2009, 19, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-M.; Bock, J.M.; Harari, P.M. Epidermal Growth Factor Receptor Blockade with C225 Modulates Proliferation, Apoptosis, and Radiosensitivity in Squamous Cell Carcinomas of the Head and Neck1. Cancer Res. 1999, 59, 1935–1940. [Google Scholar] [PubMed]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Azarnia, N.; Shin, D.M.; Cohen, R.B.; Jones, C.U.; Sur, R.; Raben, D.; Jassem, J.; et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2006, 354, 567–578. [Google Scholar] [CrossRef]
- Bonner, J.A.; Harari, P.M.; Giralt, J.; Cohen, R.B.; Jones, C.U.; Sur, R.K.; Raben, D.; Baselga, J.; Spencer, S.A.; Zhu, J.; et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010, 11, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Akagi, K.; Xiao, W.; Jiang, B.; Pickard, R.K.L.; Li, J.; Swanson, B.J.; Agrawal, A.D.; Zucker, M.; Stache-Crain, B.; et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 2019, 29, 1–17. [Google Scholar] [CrossRef]
- Mehanna, H.; Robinson, M.; Hartley, A.; Kong, A.; Foran, B.; Fulton-Lieuw, T.; Dalby, M.; Mistry, P.; Sen, M.; O’Toole, L.; et al. Radiotherapy plus cisplatin or cetuximab in low-risk human papillomavirus-positive oropharyngeal cancer (De-ESCALaTE HPV): An open-label randomised controlled phase 3 trial. Lancet 2019, 393, 51–60. [Google Scholar] [CrossRef]
- Hoang, T.; Huang, S.; Armstrong, E.; Eickhoff, J.C.; Harari, P.M. Enhancement of radiation response with bevacizumab. J. Exp. Clin. Cancer Res. 2012, 31, 37. [Google Scholar] [CrossRef]
- Lee, N.Y.; Harris, J.; Kim, J.; Garden, A.; Mechalakos, J.; Pfister, D.G.; Chan, A.T.C.; Hu, K.; Colevas, A.D.; Frank, S.; et al. Long-term Outcomes of Bevacizumab and Chemoradiation for Locoregionally Advanced Nasopharyngeal Carcinoma: A Nonrandomized Controlled Trial. JAMA Netw. Open 2023, 6, e2316094. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, S.; Armstrong, E.A.; Fowler, J.F.; Harari, P.M. Angiogenesis and radiation response modulation after vascular endothelial growth factor receptor-2 (VEGFR2) blockade. Int. J. Radiat. Oncol. Biol. Phys. 2005, 62, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulieres, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Baste, N.; Neupane, P.; Bratland, A.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.E.W.; Soulières, D.; Le Tourneau, C.; Dinis, J.; Licitra, L.; Ahn, M.J.; Soria, A.; Machiels, J.P.; Mach, N.; Mehra, R.; et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 2019, 393, 156–167. [Google Scholar] [CrossRef]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.Y.; Ferris, R.L.; Psyrri, A.; Haddad, R.I.; Tahara, M.; Bourhis, J.; Harrington, K.; Chang, P.M.; Lin, J.C.; Razaq, M.A.; et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: A randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol. 2021, 22, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.; Gillison, M.L. Nivolumab for Squamous-Cell Cancer of Head and Neck. N. Engl. J. Med. 2017, 376, 596. [Google Scholar] [CrossRef]
- Ran, X.; Yang, K. Inhibitors of the PD-1/PD-L1 axis for the treatment of head and neck cancer: Current status and future perspectives. Drug Des. Devel Ther. 2017, 11, 2007–2014. [Google Scholar] [CrossRef]
- Brunet, J.F.; Denizot, F.; Luciani, M.F.; Roux-Dosseto, M.; Suzan, M.; Mattei, M.G.; Golstein, P. A new member of the immunoglobulin superfamily—CTLA-4. Nature 1987, 328, 267–270. [Google Scholar] [CrossRef]
- Krummel, M.F.; Allison, J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995, 182, 459–465. [Google Scholar] [CrossRef]
- Walunas, T.L.; Lenschow, D.J.; Bakker, C.Y.; Linsley, P.S.; Freeman, G.J.; Green, J.M.; Thompson, C.B.; Bluestone, J.A. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994, 1, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. Embo J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhu, G.; Tamada, K.; Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5, 1365–1369. [Google Scholar] [CrossRef]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef]
- Traynor, K. Ipilimumab approved for metastatic melanoma. Am. J. Health Syst. Pharm. 2011, 68, 768. [Google Scholar] [CrossRef]
- Dobosz, P.; Dzieciątkowski, T. The Intriguing History of Cancer Immunotherapy. Front. Immunol. 2019, 10, 2965. [Google Scholar] [CrossRef]
- Ehrlich, P. Über den jetzigen Stand der Karzinomforschung. Ned. Tijdschr. Geneeskd. 1909, 5, 273–290. [Google Scholar]
- Burnet, F.M. Immunological Surveillance in Neoplasia. Immunol. Rev. 1971, 7, 3–25. [Google Scholar] [CrossRef]
- Burnet, M. Cancer—A Biological Approach. Br. Med. J. 1957, 1, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Burnet, M. Immunological factors in the process of carcinogenesis. Br. Med. Bull. 1964, 20, 154–158. [Google Scholar] [CrossRef]
- Verdonck, E.; Schaap, K.; Thomas, L.C. A discussion of the principles and applications of Modulated Temperature DSC (MTDSC). Int. J. Pharm. 1999, 192, 3–20. [Google Scholar] [CrossRef]
- Burnet, F. The concept of immunological surveillance. Immunol. Asp. Neoplasia 1970, 13, 1–27. [Google Scholar]
- Smyth, M.J.; Godfrey, D.I.; Trapani, J.A. A fresh look at tumor immunosurveillance and immunotherapy. Nat. Immunol. 2001, 2, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The three Es of cancer immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef]
- Smyth, M.J.; Thia, K.Y.; Street, S.E.; MacGregor, D.; Godfrey, D.I.; Trapani, J.A. Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. J. Exp. Med. 2000, 192, 755–760. [Google Scholar] [CrossRef]
- Wilczynski, J.R.; Nowak, M. Cancer immunoediting: Elimination, equilibrium, and immune escape in solid tumors. In Interaction of Immune and Cancer Cells; Springer: Vienna, Austria, 2014; pp. 143–205. [Google Scholar]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Whiteside, T.L. Immune responses to malignancies. J. Allergy Clin. Immunol. 2010, 125, S272–S283. [Google Scholar] [CrossRef]
- Naumov, G.N.; Akslen, L.A.; Folkman, J. Role of angiogenesis in human tumor dormancy: Animal models of the angiogenic switch. Cell Cycle 2006, 5, 1779–1787. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.W.; Swann, J.B.; Koebel, C.M.; Schreiber, R.D.; Smyth, M.J. Immune-mediated dormancy: An equilibrium with cancer. J. Leukoc. Biol. 2008, 84, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.T.; Judd, N.P.; Bui, J.D.; Uppaluri, R. The clinical implications of antitumor immunity in head and neck cancer. Laryngoscope 2012, 122, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Leethanakul, C.; Patel, V.; Gillespie, J.; Pallente, M.; Ensley, J.F.; Koontongkaew, S.; Liotta, L.A.; Emmert-Buck, M.; Gutkind, J.S. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene 2000, 19, 3220–3224. [Google Scholar] [CrossRef]
- Wijetunga, N.A.; Yu, Y.; Morris, L.G.; Lee, N.; Riaz, N. The head and neck cancer genome in the era of immunotherapy. Oral Oncol. 2021, 112, 105040. [Google Scholar] [CrossRef]
- Luke, J.J.; Bao, R.; Sweis, R.F.; Spranger, S.; Gajewski, T.F. WNT/beta-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin. Cancer Res. 2019, 25, 3074–3083. [Google Scholar] [CrossRef] [PubMed]
- Koukourakis, I.M.; Gkegka, A.G.; Xanthopoulou, E.; Nanos, C.; Giatromanolaki, A.; Koukourakis, M.I. Prognostic and Predictive Relevance of Tumor-Infiltrating Lymphocytes in Squamous Cell Head-Neck Cancer Patients Treated with Radical Radiotherapy/Chemo-Radiotherapy. Curr. Oncol. 2022, 29, 4274–4284. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, J.; Chen, X.; Jiang, F.; Sun, Y.; Pan, Y.; Zhang, W.; Zhang, J. MiR-34a suppresses HNSCC growth through modulating cell cycle arrest and senescence. Neoplasma 2017, 64, 543–553. [Google Scholar] [CrossRef]
- Wu, X.; Cheng, Y.L.; Matthen, M.; Yoon, A.; Schwartz, G.K.; Bala, S.; Taylor, A.M.; Momen-Heravi, F. Down-regulation of the tumor suppressor miR-34a contributes to head and neck cancer by up-regulating the MET oncogene and modulating tumor immune evasion. J. Exp. Clin. Cancer Res. 2021, 40, 70. [Google Scholar] [CrossRef]
- Karakas, B.; Bachman, K.E.; Park, B.H. Mutation of the PIK3CA oncogene in human cancers. Br. J. Cancer 2006, 94, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Lui, V.W.; Hedberg, M.L.; Li, H.; Vangara, B.S.; Pendleton, K.; Zeng, Y.; Lu, Y.; Zhang, Q.; Du, Y.; Gilbert, B.R.; et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013, 3, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Chen, J.Q.; Liu, C.; Malu, S.; Creasy, C.; Tetzlaff, M.T.; Xu, C.; McKenzie, J.A.; Zhang, C.; Liang, X.; et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016, 6, 202–216. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.; Liu, Y.; Huang, S.; Ding, J.; Wang, B.; Dong, P.; Sun, Z.; Chen, L. CSMD1 suppresses cancer progression by inhibiting proliferation, epithelial-mesenchymal transition, chemotherapy-resistance and inducing immunosuppression in esophageal squamous cell carcinoma. Exp. Cell Res. 2022, 417, 113220. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.; Li, M.; Jiang, Z.; Liu, Z.; Wang, X. Correlate the TP53 Mutation and the HRAS Mutation with Immune Signatures in Head and Neck Squamous Cell Cancer. Comput. Struct. Biotechnol. J. 2019, 17, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Siemers, N.O.; Holloway, J.L.; Chang, H.; Chasalow, S.D.; Ross-MacDonald, P.B.; Voliva, C.F.; Szustakowski, J.D. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS ONE 2017, 12, e0179726. [Google Scholar] [CrossRef]
- Duray, A.; Demoulin, S.; Hubert, P.; Delvenne, P.; Saussez, S. Immune suppression in head and neck cancers: A review. Clin. Dev. Immunol. 2010, 2010, 701657. [Google Scholar] [CrossRef]
- Moy, J.D.; Moskovitz, J.M.; Ferris, R.L. Biological mechanisms of immune escape and implications for immunotherapy in head and neck squamous cell carcinoma. Eur. J. Cancer 2017, 76, 152–166. [Google Scholar] [CrossRef]
- Ferris, R.L. Immunology and Immunotherapy of Head and Neck Cancer. J. Clin. Oncol. 2015, 33, 3293–3304. [Google Scholar] [CrossRef]
- Campoli, M.; Ferrone, S. HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance. Oncogene 2008, 27, 5869–5885. [Google Scholar] [CrossRef] [PubMed]
- Grandis, J.R.; Falkner, D.M.; Melhem, M.F.; Gooding, W.E.; Drenning, S.D.; Morel, P.A. Human Leukocyte Antigen Class I Allelic and Haplotype Loss in Squamous Cell Carcinoma of the Head and Neck: Clinical and Immunogenetic Consequences. Clin. Cancer Res. 2000, 6, 2794–2802. [Google Scholar]
- Ferris, R.L.; Hunt, J.L.; Ferrone, S. Human Leukocyte Antigen (HLA) Class I Defects in Head and Neck Cancer. Immunol. Res. 2005, 33, 113–133. [Google Scholar] [CrossRef]
- Greene, S.; Patel, P.; Allen, C.T. How patients with an intact immune system develop head and neck cancer. Oral Oncol. 2019, 92, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Yang, G.-y.; Song, Y.; Zhao, X.; So, C.; Liao, J.; Wang, L.-D.; Yang, C.S. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis 2001, 22, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Pollack, B.P.; Sapkota, B.; Cartee, T.V. Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes. Clin. Cancer Res. 2011, 17, 4400–4413. [Google Scholar] [CrossRef] [PubMed]
- Bandoh, N.; Ogino, T.; Katayama, A.; Takahara, M.; Katada, A.; Hayashi, T.; Harabuchi, Y. HLA class I antigen and transporter associated with antigen processing downregulation in metastatic lesions of head and neck squamous cell carcinoma as a marker of poor prognosis. Oncol. Rep. 2010, 23, 933–939. [Google Scholar] [CrossRef]
- Chang, C.C.; Campoli, M.; Ferrone, S. Classical and nonclassical HLA class I antigen and NK Cell-activating ligand changes in malignant cells: Current challenges and future directions. Adv. Cancer Res. 2005, 93, 189–234. [Google Scholar] [CrossRef]
- Meissner, M.; Reichert, T.E.; Kunkel, M.; Gooding, W.; Whiteside, T.L.; Ferrone, S.; Seliger, B. Defects in the Human Leukocyte Antigen Class I Antigen-Processing Machinery in Head and Neck Squamous Cell Carcinoma: Association with Clinical Outcome. Clin. Cancer Res. 2005, 11, 2552–2560. [Google Scholar] [CrossRef]
- Ogino, T.; Shigyo, H.; Ishii, H.; Katayama, A.; Miyokawa, N.; Harabuchi, Y.; Ferrone, S. HLA class I antigen down-regulation in primary laryngeal squamous cell carcinoma lesions as a poor prognostic marker. Cancer Res. 2006, 66, 9281–9289. [Google Scholar] [CrossRef]
- Horton, J.D.; Knochelmann, H.M.; Day, T.A.; Paulos, C.M.; Neskey, D.M. Immune Evasion by Head and Neck Cancer: Foundations for Combination Therapy. Trends Cancer 2019, 5, 208–232. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Albaitero, A.; Nayak, J.V.; Ogino, T.; Machandia, A.; Gooding, W.; DeLeo, A.B.; Ferrone, S.; Ferris, R.L. Role of antigen-processing machinery in the in vitro resistance of squamous cell carcinoma of the head and neck cells to recognition by CTL. J. Immunol. 2006, 176, 3402–3409. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Allen, C.T. Mechanisms of resistance to T cell-based immunotherapy in head and neck cancer. Head. Neck 2020, 42, 2722–2733. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Whiteside, T.L.; Ferrone, S. Immune escape associated with functional defects in antigen-processing machinery in head and neck cancer. Clin. Cancer Res. 2006, 12, 3890–3895. [Google Scholar] [CrossRef]
- Seliger, B. Molecular mechanisms of MHC class I abnormalities and APM components in human tumors. Cancer Immunol. Immunother. 2008, 57, 1719–1726. [Google Scholar] [CrossRef]
- Setiadi, A.F.; David, M.D.; Seipp, R.P.; Hartikainen, J.A.; Gopaul, R.; Jefferies, W.A. Epigenetic control of the immune escape mechanisms in malignant carcinomas. Mol. Cell Biol. 2007, 27, 7886–7894. [Google Scholar] [CrossRef]
- Tomasi, T.B.; Magner, W.J.; Khan, A.N. Epigenetic regulation of immune escape genes in cancer. Cancer Immunol. Immunother. 2006, 55, 1159–1184. [Google Scholar] [CrossRef]
- Campbell, J.D.; Yau, C.; Bowlby, R.; Liu, Y.; Brennan, K.; Fan, H.; Taylor, A.M.; Wang, C.; Walter, V.; Akbani, R.; et al. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell Rep. 2018, 23, 194–212.e196. [Google Scholar] [CrossRef]
- Zhou, L.; Mudianto, T.; Ma, X.; Riley, R.; Uppaluri, R. Targeting EZH2 Enhances Antigen Presentation, Antitumor Immunity, and Circumvents Anti-PD-1 Resistance in Head and Neck Cancer. Clin. Cancer Res. 2020, 26, 290–300. [Google Scholar] [CrossRef]
- Leibowitz, M.S.; Andrade Filho, P.A.; Ferrone, S.; Ferris, R.L. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol. Immunother. 2011, 60, 525–535. [Google Scholar] [CrossRef]
- Leibowitz, M.S.; Srivastava, R.M.; Andrade Filho, P.A.; Egloff, A.M.; Wang, L.; Seethala, R.R.; Ferrone, S.; Ferris, R.L. SHP2 is overexpressed and inhibits pSTAT1-mediated APM component expression, T-cell attracting chemokine secretion, and CTL recognition in head and neck cancer cells. Clin. Cancer Res. 2013, 19, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.G.T.; Chandramohan, R.; West, L.; Zehir, A.; Chakravarty, D.; Pfister, D.G.; Wong, R.J.; Lee, N.Y.; Sherman, E.J.; Baxi, S.S.; et al. The Molecular Landscape of Recurrent and Metastatic Head and Neck Cancers: Insights From a Precision Oncology Sequencing Platform. JAMA Oncol. 2017, 3, 244–255. [Google Scholar] [CrossRef]
- Badoual, C.; Sandoval, F.; Pere, H.; Hans, S.; Gey, A.; Merillon, N.; Van Ryswick, C.; Quintin-Colonna, F.; Bruneval, P.; Brasnu, D.; et al. Better understanding tumor-host interaction in head and neck cancer to improve the design and development of immunotherapeutic strategies. Head. Neck 2010, 32, 946–958. [Google Scholar] [CrossRef]
- Freiser, M.E.; Serafini, P.; Weed, D.T. The immune system and head and neck squamous cell carcinoma: From carcinogenesis to new therapeutic opportunities. Immunol. Res. 2013, 57, 52–69. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Whiteside, T.L.; Lebeau, A.; Zeidler, R.; Mack, B.; Wollenberg, B. Impairment of T-Cell Activation in Head and Neck Cancer In Situ and In Vitro. Arch. Otolaryngol. Head. Neck Surg. 1999, 125, 82–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wollenberg, B.; Zeidler, R.; Lebeau, A.; Mack, B.; Lang, S. Lack of B7.1 and B7.2 on head and neck cancer cells and possible significance for gene therapy. Int. J. Mol. Med. 1998, 2, 167–338. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.F.; Yu, J.M.; Zuo, W.S.; Wang, S.Z. Expression of CD80, CD86, TGF-beta1 and IL-10 mRNA in the esophageal carcinoma. Zhonghua Zhong Liu Za Zhi 2006, 28, 762–765. [Google Scholar]
- Thomas, G.R.; Chen, Z.; Leukinova, E.; Van Waes, C.; Wen, J. Cytokines IL-1 alpha, IL-6, and GM-CSF constitutively secreted by oral squamous carcinoma induce down-regulation of CD80 costimulatory molecule expression: Restoration by interferon gamma. Cancer Immunol. Immunother. 2004, 53, 33–40. [Google Scholar] [CrossRef]
- Chen, Z.; Malhotra, P.S.; Thomas, G.R.; Ondrey, F.G.; Duffey, D.C.; Smith, C.W.; Enamorado, I.; Yeh, N.T.; Kroog, G.S.; Rudy, S.; et al. Expression of Proinflammatory and Proangiogenic Cytokines in Patients with Head and Neck Cancer1. Clin. Cancer Res. 1999, 5, 1369–1379. [Google Scholar]
- Lathers, D.M.; Young, M.R. Increased aberrance of cytokine expression in plasma of patients with more advanced squamous cell carcinoma of the head and neck. Cytokine 2004, 25, 220–228. [Google Scholar] [CrossRef]
- Loro, L.L.; Vintermyr, O.K.; Johannessen, A.C.; Liavaag, P.G.; Jonsson, R. Suppression of Fas receptor and negative correlation of Fas ligand with differentiation and apoptosis in oral squamous cell carcinoma. J. Oral Pathol. Med. 1999, 28, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Bie, T.; Zhang, X. Higher Expression of SPP1 Predicts Poorer Survival Outcomes in Head and Neck Cancer. J. Immunol. Res. 2021, 2021, 8569575. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.X.; Li, Z.J.; Jiang, X.O.; Lum, Y.L.; Khin, E.; Lee, N.P.; Wu, G.H.; Luk, J.M. Osteopontin as potential biomarker and therapeutic target in gastric and liver cancers. World J. Gastroenterol. 2012, 18, 3923–3930. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; Czystowska, M.; Szajnik, M.; Harasymczuk, M.; Boyiadzis, M.; Kruk-Zagajewska, A.; Szyfter, W.; Zeromski, J.; Whiteside, T.L. Triggering of Toll-like receptor 4 expressed on human head and neck squamous cell carcinoma promotes tumor development and protects the tumor from immune attack. Cancer Res. 2009, 69, 3105–3113. [Google Scholar] [CrossRef] [PubMed]
- Medema, J.P.; De Jong, J.; Peltenburg, L.T.C.; Verdegaal, E.M.E.; Gorter, A.; Bres, S.A.; Franken, K.L.M.C.; Hahne, M.; Albar, J.P.; Melief, C.J.M.; et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proc. Natl. Acad. Sci. USA 2001, 98, 11515–11520. [Google Scholar] [CrossRef]
- van Kempen, P.M.; Noorlag, R.; Swartz, J.E.; Bovenschen, N.; Braunius, W.W.; Vermeulen, J.F.; Van Cann, E.M.; Grolman, W.; Willems, S.M. Oropharyngeal squamous cell carcinomas differentially express granzyme inhibitors. Cancer Immunol. Immunother. 2016, 65, 575–585. [Google Scholar] [CrossRef]
- Wieckowski, E.; Atarashi, Y.; Stanson, J.; Sato, T.A.; Whiteside, T.L. FAP-1-mediated activation of NF-kappaB induces resistance of head and neck cancer to Fas-induced apoptosis. J. Cell Biochem. 2007, 100, 16–28. [Google Scholar] [CrossRef]
- Li, X.; Pan, X.; Zhang, H.; Lei, D.; Liu, D.; Xu, F.; Luan, X. Overexpression of cFLIP in head and neck squamous cell carcinoma and its clinicopathologic correlations. J. Cancer Res. Clin. Oncol. 2008, 134, 609–615. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Nguyen, D.; Chea, C.; Miyauchi, M.; Fujii, M.; Takata, T. Interaction between N-cadherin and decoy receptor-2 regulates apoptosis in head and neck cancer. Oncotarget 2018, 9, 31516–31530. [Google Scholar] [CrossRef][Green Version]
- Yoldas, B.; Ozer, C.; Ozen, O.; Canpolat, T.; Dogan, I.; Griffith, T.S.; Sanlioglu, S.; Ozluoglu, L.N. Clinical significance of TRAIL and TRAIL receptors in patients with head and neck cancer. Head. Neck 2011, 33, 1278–1284. [Google Scholar] [CrossRef]
- Zhao, L.Y.; Xu, W.L.; Xu, Z.Q.; Qi, C.; Li, Y.; Cheng, J.; Liu, L.K.; Wu, Y.N.; Gao, J.; Ye, J.H. The overexpressed functional transient receptor potential channel TRPM2 in oral squamous cell carcinoma. Sci. Rep. 2016, 6, 38471. [Google Scholar] [CrossRef] [PubMed]
- Reichert, T.E.; Kashii, Y.; Stanson, J.; Zeevi, A.; Whiteside, T.L. The role of endogenous interleukin-2 in proliferation of human carcinoma cell lines. Br. J. Cancer 1999, 81, 822–831. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alam, M.; Kashyap, T.; Pramanik, K.K.; Singh, A.K.; Nagini, S.; Mishra, R. The elevated activation of NFκB and AP-1 is correlated with differential regulation of Bcl-2 and associated with oral squamous cell carcinoma progression and resistance. Clin. Oral Investig. 2017, 21, 2721–2731. [Google Scholar] [CrossRef]
- Niklander, S.E.; Murdoch, C.; Hunter, K.D. IL-1/IL-1R Signaling in Head and Neck Cancer. Front. Oral Health 2021, 2, 722676. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Egloff, A.M.; Sen, M.; Grandis, J.R.; Johnson, D.E. Caspase-8 mutations in head and neck cancer confer resistance to death receptor-mediated apoptosis and enhance migration, invasion, and tumor growth. Mol. Oncol. 2014, 8, 1220–1230. [Google Scholar] [CrossRef]
- Teng, M.S.; Brandwein-Gensler, M.S.; Teixeira, M.S.; Martignetti, J.A.; Duffey, D.C. A Study of TRAIL Receptors in Squamous Cell Carcinoma of the Head and Neck. Arch. Otolaryngol. Head. Neck Surg. 2005, 131, 407–412. [Google Scholar] [CrossRef][Green Version]
- Vitale, I.; Galluzzi, L.; Castedo, M.; Kroemer, G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011, 12, 385–392. [Google Scholar] [CrossRef]
- Chen, G.; Shi, L.; Litchfield, D.W.; Greenberg, A.H. Rescue from Granzyme B-induced Apoptosis by Wee1 Kinase. J. Exp. Med. 1995, 181, 2295–2300. [Google Scholar] [CrossRef]
- Friedman, J.; Morisada, M.; Sun, L.; Moore, E.C.; Padget, M.; Hodge, J.W.; Schlom, J.; Gameiro, S.R.; Allen, C.T. Inhibition of WEE1 kinase and cell cycle checkpoint activation sensitizes head and neck cancers to natural killer cell therapies. J. Immunother. Cancer 2018, 6, 59. [Google Scholar] [CrossRef]
- Sun, L.; Moore, E.; Berman, R.; Clavijo, P.E.; Saleh, A.; Chen, Z.; Van Waes, C.; Davies, J.; Friedman, J.; Allen, C.T. WEE1 kinase inhibition reverses G2/M cell cycle checkpoint activation to sensitize cancer cells to immunotherapy. Oncoimmunology 2018, 7, e1488359. [Google Scholar] [CrossRef]
- Ludwig, S.; Floros, T.; Theodoraki, M.N.; Hong, C.S.; Jackson, E.K.; Lang, S.; Whiteside, T.L. Suppression of Lymphocyte Functions by Plasma Exosomes Correlates with Disease Activity in Patients with Head and Neck Cancer. Clin. Cancer Res. 2017, 23, 4843–4854. [Google Scholar] [CrossRef] [PubMed]
- Theodoraki, M.N.; Hoffmann, T.K.; Jackson, E.K.; Whiteside, T.L. Exosomes in HNSCC plasma as surrogate markers of tumour progression and immune competence. Clin. Exp. Immunol. 2018, 194, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Gastman, B.R.; Atarashi, Y.; Reichert, T.E.; Saito, T.; Balkir, L.; Rabinowich, H.; Whiteside, T.L. Fas Ligand Is Expressed on Human Squamous Cell Carcinomas of the Head and Neck, and It Promotes Apoptosis of T Lymphocytes1. Cancer Res. 1999, 59, 5356–5364. [Google Scholar] [PubMed]
- Gastman, B.R.; Johnson, D.E.; Whiteside, T.L.; Rabinowich, H. Tumor-induced apoptosis of T lymphocytes: Elucidation of intracellular apoptotic events. Blood 2000, 95, 2015–2023. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.K.; Dworacki, G.; Tsukihiro, T.; Meidenbauer, N.; Gooding, W.; Johnson, J.T.; Whiteside, T.L. Spontaneous apoptosis of circulating T lymphocytes in patients with head and neck cancer and its clinical importance. Clin. Cancer Res. 2002, 8, 2553–2562. [Google Scholar] [PubMed]
- Whiteside, T.L. Tumor-induced death of immune cells: Its mechanisms and consequences. Semin. Cancer Biol. 2002, 12, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kassouf, N.; Thornhill, M.H. Oral cancer cell lines can use multiple ligands, including Fas-L, TRAIL and TNF-alpha, to induce apoptosis in Jurkat T cells: Possible mechanisms for immune escape by head and neck cancers. Oral Oncol. 2008, 44, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Saussez, S.; Camby, I.; Toubeau, G.; Kiss, R. Galectins as modulators of tumor progression in head and neck squamous cell carcinomas. Head. Neck 2007, 29, 874–884. [Google Scholar] [CrossRef]
- Strome, S.E.; Dong, H.; Tamura, H.; Voss, S.G.; Flies, D.B.; Tamada, K.; Salomao, D.; Cheville, J.; Hirano, F.; Lin, W.; et al. B7-H1 Blockade Augments Adoptive T-Cell Immunotherapy for Squamous Cell Carcinoma1. Cancer Res. 2003, 63, 6501–6505. [Google Scholar]
- Nambiar, D.K.; Aguilera, T.; Cao, H.; Kwok, S.; Kong, C.; Bloomstein, J.; Wang, Z.; Rangan, V.S.; Jiang, D.; von Eyben, R.; et al. Galectin-1-driven T cell exclusion in the tumor endothelium promotes immunotherapy resistance. J. Clin. Investig. 2019, 129, 5553–5567. [Google Scholar] [CrossRef]
- Concha-Benavente, F.; Srivastava, R.M.; Trivedi, S.; Lei, Y.; Chandran, U.; Seethala, R.R.; Freeman, G.J.; Ferris, R.L. Identification of the Cell-Intrinsic and -Extrinsic Pathways Downstream of EGFR and IFNγ That Induce PD-L1 Expression in Head and Neck Cancer. Cancer Res. 2016, 76, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Seiwert, T.Y.; Burtness, B.; Mehra, R.; Weiss, J.; Berger, R.; Eder, J.P.; Heath, K.; McClanahan, T.; Lunceford, J.; Gause, C.; et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): An open-label, multicentre, phase 1b trial. Lancet Oncol. 2016, 17, 956–965. [Google Scholar] [CrossRef] [PubMed]
- Zandberg, D.P.; Strome, S.E. The role of the PD-L1:PD-1 pathway in squamous cell carcinoma of the head and neck. Oral Oncol. 2014, 50, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Chen, L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008, 8, 467–477. [Google Scholar] [CrossRef]
- Bellmunt, À.M.; López-Puerto, L.; Lorente, J.; Closa, D. Involvement of extracellular vesicles in the macrophage-tumor cell communication in head and neck squamous cell carcinoma. PLoS ONE 2019, 14, e0224710. [Google Scholar] [CrossRef]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Sharpe, A.H.; Freeman, G.J. Programmed Death-1 Ligand 1 Interacts Specifically with the B7-1 Costimulatory Molecule to Inhibit T Cell Responses. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef]
- Eppihimer, M.J.; Gunn, J.; Freeman, G.J.; Greenfield, E.A.; Chernova, T.; Erickson, J.; Leonard, J.P. Expression and regulation of the PD-L1 immunoinhibitory molecule on micro vascular endothelial cells. Microcirculation 2002, 9, 133–145. [Google Scholar] [CrossRef]
- Chen, S.; Crabill, G.A.; Pritchard, T.S.; McMiller, T.L.; Wei, P.; Pardoll, D.M.; Pan, F.; Topalian, S.L. Mechanisms regulating PD-L1 expression on tumor and immune cells. J. Immunother. Cancer 2019, 7, 305. [Google Scholar] [CrossRef]
- Cui, B.; Chen, J.; Luo, M.; Wang, L.; Chen, H.; Kang, Y.; Wang, J.; Zhou, X.; Feng, Y.; Zhang, P. Protein kinase D3 regulates the expression of the immunosuppressive protein, PD-L1, through STAT1/STAT3 signaling. Int. J. Oncol. 2020, 56, 909–920. [Google Scholar] [CrossRef]
- Kondoh, N.; Mizuno-Kamiya, M. The Role of Immune Modulatory Cytokines in the Tumor Microenvironments of Head and Neck Squamous Cell Carcinomas. Cancers 2022, 14, 2884. [Google Scholar] [CrossRef]
- Kuo, C.-S.; Yang, C.-Y.; Lin, C.-K.; Lin, G.-J.; Sytwu, H.-K.; Chen, Y.-W. Triptolide suppresses oral cancer cell PD-L1 expression in the interferon-γ-modulated microenvironment in vitro, in vivo, and in clinical patients. Biomed. Pharmacother. 2021, 133, 111057. [Google Scholar] [CrossRef] [PubMed]
- Parsa, A.T.; Waldron, J.S.; Panner, A.; Crane, C.A.; Parney, I.F.; Barry, J.J.; Cachola, K.E.; Murray, J.C.; Tihan, T.; Jensen, M.C.; et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007, 13, 84–88. [Google Scholar] [CrossRef]
- Skinner, H.D.; Giri, U.; Yang, L.P.; Kumar, M.; Liu, Y.; Story, M.D.; Pickering, C.R.; Byers, L.A.; Williams, M.D.; Wang, J.; et al. Integrative Analysis Identifies a Novel AXL-PI3 Kinase-PD-L1 Signaling Axis Associated with Radiation Resistance in Head and Neck Cancer. Clin. Cancer Res. 2017, 23, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Zhao, J.; Song, C.; Yuan, Q.; Liu, X. TRPM8 facilitates proliferation and immune evasion of esophageal cancer cells. Biosci. Rep. 2019, 39, BSR20191878. [Google Scholar] [CrossRef] [PubMed]
- Mullins, R.D.Z.; Pal, A.; Barrett, T.F.; Heft Neal, M.E.; Puram, S.V. Epithelial-Mesenchymal Plasticity in Tumor Immune Evasion. Cancer Res. 2022, 82, 2329–2343. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef]
- Noman, M.Z.; Janji, B.; Abdou, A.; Hasmim, M.; Terry, S.; Tan, T.Z.; Mami-Chouaib, F.; Thiery, J.P.; Chouaib, S. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology 2017, 6, e1263412. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Nayak, S.P.; Hari, K.; Purkait, P.; Mandal, S.; Kishore, A.; Levine, H.; Jolly, M.K. Immunosuppressive Traits of the Hybrid Epithelial/Mesenchymal Phenotype. Front. Immunol. 2021, 12, 797261. [Google Scholar] [CrossRef]
- Dhupar, R.; Van Der Kraak, L.; Pennathur, A.; Schuchert, M.J.; Nason, K.S.; Luketich, J.D.; Lotze, M.T. Targeting Immune Checkpoints in Esophageal Cancer: A High Mutational Load Tumor. Ann. Thorac. Surg. 2017, 103, 1340–1349. [Google Scholar] [CrossRef]
- Yearley, J.H.; Gibson, C.; Yu, N.; Moon, C.; Murphy, E.; Juco, J.; Lunceford, J.; Cheng, J.; Chow, L.Q.M.; Seiwert, T.Y.; et al. PD-L2 Expression in Human Tumors: Relevance to Anti-PD-1 Therapy in Cancer. Clin. Cancer Res. 2017, 23, 3158–3167. [Google Scholar] [CrossRef]
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Gao, Y.; Wei, W. Post-translational regulations of PD-L1 and PD-1: Mechanisms and opportunities for combined immunotherapy. Semin. Cancer Biol. 2022, 85, 246–252. [Google Scholar] [CrossRef]
- Hu, X.; Lin, Z.; Wang, Z.; Zhou, Q. Emerging role of PD-L1 modification in cancer immunotherapy. Am. J. Cancer Res. 2021, 11, 3832–3840. [Google Scholar]
- Hu, X.; Wang, J.; Chu, M.; Liu, Y.; Wang, Z.-w.; Zhu, X. Emerging Role of Ubiquitination in the Regulation of PD-1/PD-L1 in Cancer Immunotherapy. Mol. Ther. 2021, 29, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gao, Z.; Hu, R.; Wang, Y.; Wang, Y.; Su, Z.; Zhang, X.; Yang, J.; Mei, M.; Ren, Y.; et al. PD-L2 glycosylation promotes immune evasion and predicts anti-EGFR efficacy. J. Immunother. Cancer 2021, 9, e002699. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.M.; Luo, Y.F.; Zeng, F.F.; Su, C.; Liu, X.; Li, X.P.; Lu, J. TGF-beta1-Mediated PD-L1 Glycosylation Contributes to Immune Escape via c-Jun/STT3A Pathway in Nasopharyngeal Carcinoma. Front. Oncol. 2022, 12, 815437. [Google Scholar] [CrossRef]
- Blum, A.E.; Venkitachalam, S.; Ravillah, D.; Chelluboyina, A.K.; Kieber-Emmons, A.M.; Ravi, L.; Kresak, A.; Chandar, A.K.; Markowitz, S.D.; Canto, M.I.; et al. Systems Biology Analyses Show Hyperactivation of Transforming Growth Factor-β and JNK Signaling Pathways in Esophageal Cancer. Gastroenterology 2019, 156, 1761–1774. [Google Scholar] [CrossRef] [PubMed]
- Gholamin, M.; Moaven, O.; Memar, B.; Farshchian, M.; Naseh, H.; Malekzadeh, R.; Sotoudeh, M.; Rajabi-Mashhadi, M.T.; Forghani, M.N.; Farrokhi, F.; et al. Overexpression and interactions of interleukin-10, transforming growth factor beta, and vascular endothelial growth factor in esophageal squamous cell carcinoma. World J. Surg. 2009, 33, 1439–1445. [Google Scholar] [CrossRef]
- Hargadon, K.M. Tumor-altered dendritic cell function: Implications for anti-tumor immunity. Front. Immunol. 2013, 4, 192. [Google Scholar] [CrossRef]
- Chen, Y.; Di, C.; Zhang, X.; Wang, J.; Wang, F.; Yan, J.-f.; Xu, C.; Zhang, J.; Zhang, Q.; Li, H.; et al. Transforming growth factor β signaling pathway: A promising therapeutic target for cancer. J. Cell. Physiol. 2020, 235, 1903–1914. [Google Scholar] [CrossRef]
- Moutsopoulos, N.M.; Wen, J.; Wahl, S.M. TGF-beta and tumors—An ill-fated alliance. Curr. Opin. Immunol. 2008, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Suzuki, S.; Takahara, T.; Ono, S.; Goto, M.; Miyabe, S.; Sugita, Y.; Ogawa, T.; Ito, H.; Satou, A.; et al. Improving function of cytotoxic T-lymphocytes by transforming growth factor-β inhibitor in oral squamous cell carcinoma. Cancer Sci. 2021, 112, 4037–4049. [Google Scholar] [CrossRef] [PubMed]
- Duffey, D.C.; Chen, Z.; Dong, G.; Ondrey, F.G.; Wolf, J.S.; Brown, K.; Siebenlist, U.; Van Waes, C. Expression of a dominant-negative mutant inhibitor-kappaBalpha of nuclear factor-kappaB in human head and neck squamous cell carcinoma inhibits survival, proinflammatory cytokine expression, and tumor growth in vivo. Cancer Res. 1999, 59, 3468–3474. [Google Scholar] [PubMed]
- Varilla, V.; Atienza, J.; Dasanu, C.A. Immune alterations and immunotherapy prospects in head and neck cancer. Expert Opin. Biol. Ther. 2013, 13, 1241–1256. [Google Scholar] [CrossRef]
- Riedel, F.; Zaiss, I.; Herzog, D.; Götte, K.; Naim, R.; Hörmann, K. Serum levels of interleukin-6 in patients with primary head and neck squamous cell carcinoma. Anticancer Res. 2005, 25, 2761–2765. [Google Scholar]
- Vinocha, A.; Grover, R.K.; Deepak, R. Clinical significance of interleukin-6 in diagnosis of lung, oral, esophageal, and gall bladder carcinomas. J. Cancer Res. Ther. 2018, 14, S758–S760. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gao, F.X.; Wang, C.; Qin, M.; Han, F.; Xu, T.; Hu, Z.; Long, Y.; He, X.M.; Deng, X.; et al. IL-6 and IL-8 secreted by tumour cells impair the function of NK cells via the STAT3 pathway in oesophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 321. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, H.W.; Cuenca, A.; Huang, M.; Ghansah, T.; Brayer, J.; Kerr, W.G.; Takeda, K.; Akira, S.; Schoenberger, S.P.; et al. A critical role for Stat3 signaling in immune tolerance. Immunity 2003, 19, 425–436. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Sun, Y.; Chin, Y.E.; Weisiger, E.; Malter, C.; Tawara, I.; Toubai, T.; Gatza, E.; Mascagni, P.; Dinarello, C.A.; Reddy, P. Cutting edge: Negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J. Immunol. 2009, 182, 5899–5903. [Google Scholar] [CrossRef]
- Barton, B.E. STAT3: A potential therapeutic target in dendritic cells for the induction of transplant tolerance. Expert. Opin. Ther. Targets 2006, 10, 459–470. [Google Scholar] [CrossRef]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Nakagawa, T.; Kitamura, H.; Atsumi, T.; Kamon, H.; Sawa, S.; Kamimura, D.; Ueda, N.; Iwakura, Y.; Ishihara, K.; et al. IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J. Immunol. 2004, 173, 3844–3854. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Cohen, E.E. Targeting angiogenesis in head and neck cancer. Semin. Oncol. 2008, 35, 274–285. [Google Scholar] [CrossRef]
- Vassilakopoulou, M.; Psyrri, A.; Argiris, A. Targeting angiogenesis in head and neck cancer. Oral Oncol. 2015, 51, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.; Ishida, T.; Oyama, T.; Ran, S.; Kravtsov, V.; Nadaf, S.; Carbone, D.P. Vascular endothelial growth factor inhibits the development of dendritic cells and dramatically affects the differentiation of multiple hematopoietic lineages in vivo. Blood 1998, 92, 4150–4166. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Chen, H.L.; Girgis, K.R.; Cunningham, H.T.; Meny, G.M.; Nadaf, S.; Kavanaugh, D.; Carbone, D.P. Production of vascular endothelial growth factor by human tumors inhibits the functional maturation of dendritic cells. Nat. Med. 1996, 2, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.F.; Clay, T.M.; Hobeika, A.C.; Lyerly, H.K.; Morse, M.A. Vascular endothelial growth factor and immunosuppression in cancer: Current knowledge and potential for new therapy. Expert. Opin. Biol. Ther. 2007, 7, 449–460. [Google Scholar] [CrossRef]
- Strauss, L.; Volland, D.; Kunkel, M.; Reichert, T.E. Dual role of VEGF family members in the pathogenesis of head and neck cancer (HNSCC): Possible link between angiogenesis and immune tolerance. Med. Sci. Monit. 2005, 11, Br280–Br292. [Google Scholar]
- Ohm, J.E.; Shurin, M.R.; Esche, C.; Lotze, M.T.; Carbone, D.P.; Gabrilovich, D.I. Effect of vascular endothelial growth factor and FLT3 ligand on dendritic cell generation in vivo. J. Immunol. 1999, 163, 3260–3268. [Google Scholar] [CrossRef]
- Brossart, P.; Zobywalski, A.; Grünebach, F.; Behnke, L.; Stuhler, G.; Reichardt, V.L.; Kanz, L.; Brugger, W. Tumor necrosis factor alpha and CD40 ligand antagonize the inhibitory effects of interleukin 10 on T-cell stimulatory capacity of dendritic cells. Cancer Res. 2000, 60, 4485–4492. [Google Scholar] [PubMed]
- Hadjigol, S.; Shah, B.A.; O’Brien-Simpson, N.M. The ‘Danse Macabre’-Neutrophils the Interactive Partner Affecting Oral Cancer Outcomes. Front. Immunol. 2022, 13, 894021. [Google Scholar] [CrossRef] [PubMed]
- Morse, M.A.; Mosca, P.J.; Clay, T.M.; Lyerly, H.K. Dendritic cell maturation in active immunotherapy strategies. Expert. Opin. Biol. Ther. 2002, 2, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.S.; Lattime, E.C. Tumor-induced interleukin 10 suppresses the ability of splenic dendritic cells to stimulate CD4 and CD8 T-cell responses. Cancer Res. 2003, 63, 2150–2157. [Google Scholar] [PubMed]
- Steinbrink, K.; Wölfl, M.; Jonuleit, H.; Knop, J.; Enk, A.H. Induction of tolerance by IL-10-treated dendritic cells. J. Immunol. 1997, 159, 4772–4780. [Google Scholar] [CrossRef]
- Zhou, L.; Nazarian, A.A.; Smale, S.T. Interleukin-10 inhibits interleukin-12 p40 gene transcription by targeting a late event in the activation pathway. Mol. Cell Biol. 2004, 24, 2385–2396. [Google Scholar] [CrossRef] [PubMed]
- Loaiza, N.; Demaria, M. Cellular senescence and tumor promotion: Is aging the key? Biochim. Biophys. Acta (BBA) Rev. Cancer 2016, 1865, 155–167. [Google Scholar] [CrossRef]
- Wolf, J.S.; Chen, Z.; Dong, G.; Sunwoo, J.B.; Bancroft, C.C.; Capo, D.E.; Yeh, N.T.; Mukaida, N.; Van Waes, C. IL (interleukin)-1alpha promotes nuclear factor-kappaB and AP-1-induced IL-8 expression, cell survival, and proliferation in head and neck squamous cell carcinomas. Clin. Cancer Res. 2001, 7, 1812–1820. [Google Scholar]
- Woods, K.V.; Adler-Storthz, K.; Clayman, G.L.; Francis, G.M.; Grimm, E.A. Interleukin-1 regulates interleukin-6 secretion in human oral squamous cell carcinoma in vitro: Possible influence of p53 but not human papillomavirus E6/E7. Cancer Res. 1998, 58, 3142–3149. [Google Scholar]
- Pries, R.; Wollenberg, B. Cytokines in head and neck cancer. Cytokine Growth Factor. Rev. 2006, 17, 141–146. [Google Scholar] [CrossRef]
- Zeidler, R.; Csanady, M.; Gires, O.; Lang, S.; Schmitt, B.; Wollenberg, B. Tumor cell-derived prostaglandin E2 inhibits monocyte function by interfering with CCR5 and Mac-1. FASEB J. 2000, 14, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Tourkova, I.L.; Shurin, G.V.; Chatta, G.S.; Perez, L.; Finke, J.; Whiteside, T.L.; Ferrone, S.; Shurin, M.R. Restoration by IL-15 of MHC class I antigen-processing machinery in human dendritic cells inhibited by tumor-derived gangliosides. J. Immunol. 2005, 175, 3045–3052. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, S.; Heitger, A.; Shen, W.; Liu, Y.; Taylor, B.; Ladisch, S. Mechanisms of ganglioside inhibition of APC function. J. Immunol. 2003, 171, 1676–1683. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Ladisch, S. Ganglioside GD1a impedes lipopolysaccharide-induced maturation of human dendritic cells. Cell Immunol. 2002, 220, 125–133. [Google Scholar] [CrossRef]
- Shurin, G.V.; Shurin, M.R.; Bykovskaia, S.; Shogan, J.; Lotze, M.T.; Barksdale, E.M., Jr. Neuroblastoma-derived gangliosides inhibit dendritic cell generation and function. Cancer Res. 2001, 61, 363–369. [Google Scholar]
- Whiteside, T.L.; Stanson, J.; Shurin, M.R.; Ferrone, S. Antigen-processing machinery in human dendritic cells: Up-regulation by maturation and down-regulation by tumor cells. J. Immunol. 2004, 173, 1526–1534. [Google Scholar] [CrossRef]
- Wölfl, M.; Batten, W.Y.; Posovszky, C.; Bernhard, H.; Berthold, F. Gangliosides inhibit the development from monocytes to dendritic cells. Clin. Exp. Immunol. 2002, 130, 441–448. [Google Scholar] [CrossRef]
- Li, B.; Song, T.-N.; Wang, F.-R.; Yin, C.; Li, Z.; Lin, J.-P.; Meng, Y.-Q.; Feng, H.-M.; Jing, T. Tumor-derived exosomal HMGB1 promotes esophageal squamous cell carcinoma progression through inducing PD1+ TAM expansion. Oncogenesis 2019, 8, 17. [Google Scholar] [CrossRef]
- Qian, W.; Huang, P.; Liang, X.; Chen, Y.; Guan, B. High expression of carcinoembryonic antigen-associated cell adhesion molecule 1 is associated with microangiogenesis in esophageal squamous cell carcinoma. Transl. Cancer Res. 2020, 9, 4762. [Google Scholar] [CrossRef]
- Vermeer, D.W.; Spanos, W.C.; Vermeer, P.D.; Bruns, A.M.; Lee, K.M.; Lee, J.H. Radiation-induced loss of cell surface CD47 enhances immune-mediated clearance of human papillomavirus-positive cancer. Int. J. Cancer 2013, 133, 120–129. [Google Scholar] [CrossRef]
- Yu, G.T.; Bu, L.L.; Zhao, Y.Y.; Mao, L.; Deng, W.W.; Wu, T.F.; Zhang, W.F.; Sun, Z.J. CTLA4 blockade reduces immature myeloid cells in head and neck squamous cell carcinoma. Oncoimmunology 2016, 5, e1151594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Pan, K.; Weng, D.S.; Chen, C.L.; Wang, Q.J.; Zhao, J.J.; Pan, Q.Z.; Liu, Q.; Jiang, S.S.; Li, Y.Q.; et al. Cytotoxic T lymphocyte antigen-4 expression in esophageal carcinoma: Implications for prognosis. Oncotarget 2016, 7, 26670–26679. [Google Scholar] [CrossRef]
- Elmusrati, A.; Wang, J.; Wang, C.Y. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma. Int. J. Oral Sci. 2021, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Y.; Jia, L.; Kim, J.k.; Li, J.; Deng, P.; Zhang, W.; Krebsbach, P.H.; Wang, C.-Y. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell 2021, 28, 1597–1613.e7. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Xiong, F.; Wang, L.; Chen, K.; Wu, P.; Hua, L.; Zhang, Z. Down-regulation of MLLT1 super elongation complex subunit impairs the anti-tumor activity of natural killer cells in esophageal cancer. Immunobiology 2022, 227, 152238. [Google Scholar] [CrossRef] [PubMed]
- Barclay, A.N.; Berg, T.K.v.d. The Interaction Between Signal Regulatory Protein Alpha (SIRPα) and CD47: Structure, Function, and Therapeutic Target. Annu. Rev. Immunol. 2014, 32, 25–50. [Google Scholar] [CrossRef] [PubMed]
- Blazar, B.R.; Lindberg, F.P.; Ingulli, E.; Panoskaltsis-Mortari, A.; Oldenborg, P.-A.; Iizuka, K.; Yokoyama, W.M.; Taylor, P.A. Cd47 (Integrin-Associated Protein) Engagement of Dendritic Cell and Macrophage Counterreceptors Is Required to Prevent the Clearance of Donor Lymphohematopoietic Cells. J. Exp. Med. 2001, 194, 541–550. [Google Scholar] [CrossRef]
- Engelhardt, J.J.; Sullivan, T.J.; Allison, J.P. CTLA-4 Overexpression Inhibits T Cell Responses through a CD28-B7-Dependent Mechanism1. J. Immunol. 2006, 177, 1052–1061. [Google Scholar] [CrossRef]
- Collins, A.V.; Brodie, D.W.; Gilbert, R.J.; Iaboni, A.; Manso-Sancho, R.; Walse, B.; Stuart, D.I.; van der Merwe, P.A.; Davis, S.J. The interaction properties of costimulatory molecules revisited. Immunity 2002, 17, 201–210. [Google Scholar] [CrossRef]
- Kontos, F.; Michelakos, T.; Kurokawa, T.; Sadagopan, A.; Schwab, J.H.; Ferrone, C.R.; Ferrone, S. B7-H3: An Attractive Target for Antibody-based Immunotherapy. Clin. Cancer Res. 2021, 27, 1227–1235. [Google Scholar] [CrossRef]
- Ackerman, D.; Simon, M.C. Hypoxia, lipids, and cancer: Surviving the harsh tumor microenvironment. Trends Cell Biol. 2014, 24, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Pouysségur, J.; Dayan, F.; Mazure, N.M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 2006, 441, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Ariffin, A.B.; Forde, P.F.; Jahangeer, S.; Soden, D.M.; Hinchion, J. Releasing pressure in tumors: What do we know so far and where do we go from here? A review. Cancer Res. 2014, 74, 2655–2662. [Google Scholar] [CrossRef] [PubMed]
- Le, Q.T.; Shi, G.; Cao, H.; Nelson, D.W.; Wang, Y.; Chen, E.Y.; Zhao, S.; Kong, C.; Richardson, D.; O’Byrne, K.J.; et al. Galectin-1: A link between tumor hypoxia and tumor immune privilege. J. Clin. Oncol. 2005, 23, 8932–8941. [Google Scholar] [CrossRef]
- Sitkovsky, M.; Lukashev, D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat. Rev. Immunol. 2005, 5, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Sitkovsky, M.V.; Kjaergaard, J.; Lukashev, D.; Ohta, A. Hypoxia-adenosinergic immunosuppression: Tumor protection by T regulatory cells and cancerous tissue hypoxia. Clin. Cancer Res. 2008, 14, 5947–5952. [Google Scholar] [CrossRef]
- Brand, A.; Singer, K.; Koehl, G.E.; Kolitzus, M.; Schoenhammer, G.; Thiel, A.; Matos, C.; Bruss, C.; Klobuch, S.; Peter, K.; et al. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab. 2016, 24, 657–671. [Google Scholar] [CrossRef]
- Pilon-Thomas, S.; Kodumudi, K.N.; El-Kenawi, A.E.; Russell, S.; Weber, A.M.; Luddy, K.; Damaghi, M.; Wojtkowiak, J.W.; Mulé, J.J.; Ibrahim-Hashim, A.; et al. Neutralization of Tumor Acidity Improves Antitumor Responses to Immunotherapy. Cancer Res. 2016, 76, 1381–1390. [Google Scholar] [CrossRef]
- Xie, H.; Hanai, J.; Ren, J.G.; Kats, L.; Burgess, K.; Bhargava, P.; Signoretti, S.; Billiard, J.; Duffy, K.J.; Grant, A.; et al. Targeting lactate dehydrogenase—A inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab. 2014, 19, 795–809. [Google Scholar] [CrossRef]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef]
- Ho, P.C.; Bihuniak, J.D.; Macintyre, A.N.; Staron, M.; Liu, X.; Amezquita, R.; Tsui, Y.C.; Cui, G.; Micevic, G.; Perales, J.C.; et al. Phosphoenolpyruvate Is a Metabolic Checkpoint of Anti-tumor T Cell Responses. Cell 2015, 162, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Ottensmeier, C.H.; Perry, K.L.; Harden, E.L.; Stasakova, J.; Jenei, V.; Fleming, J.; Wood, O.; Woo, J.; Woelk, C.H.; Thomas, G.J.; et al. Upregulated Glucose Metabolism Correlates Inversely with CD8+ T-cell Infiltration and Survival in Squamous Cell Carcinoma. Cancer Res. 2016, 76, 4136–4148. [Google Scholar] [CrossRef] [PubMed]
- Kiyozumi, Y.; Baba, Y.; Okadome, K.; Yagi, T.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Watanabe, M.; Komohara, Y.; et al. IDO1 Expression Is Associated With Immune Tolerance and Poor Prognosis in Patients With Surgically Resected Esophageal Cancer. Ann. Surg. 2019, 269, 1101–1108. [Google Scholar] [CrossRef]
- Lin, D.J.; Ng, J.C.K.; Huang, L.; Robinson, M.; O’Hara, J.; Wilson, J.A.; Mellor, A.L. The immunotherapeutic role of indoleamine 2,3-dioxygenase in head and neck squamous cell carcinoma: A systematic review. Clin. Otolaryngol. 2021, 46, 919–934. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Shafizadeh, E.; Attwood, J.T.; Bondarev, I.; Pashine, A.; Mellor, A.L. Inhibition of T Cell Proliferation by Macrophage Tryptophan Catabolism. J. Exp. Med. 1999, 189, 1363–1372. [Google Scholar] [CrossRef]
- Uyttenhove, C.; Pilotte, L.; Théate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Deaglio, S.; Robson, S.C. Ectonucleotidases as regulators of purinergic signaling in thrombosis, inflammation, and immunity. Adv. Pharmacol. 2011, 61, 301–332. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Yuan, Y.X.; Ji, T.; Zhang, C.P. CD73 as a novel marker for poor prognosis of oral squamous cell carcinoma. Oncol. Lett. 2016, 12, 556–562. [Google Scholar] [CrossRef]
- Xue, F.; Wang, T.; Shi, H.; Feng, H.; Feng, G.; Wang, R.; Yao, Y.; Yuan, H. CD73 facilitates invadopodia formation and boosts malignancy of head and neck squamous cell carcinoma via the MAPK signaling pathway. Cancer Sci. 2022, 113, 2704–2715. [Google Scholar] [CrossRef]
- Marafon, F.; Bonadiman, B.; de Rocco Donassolo, S.; Marins, K.; Zanchi, M.M.; Kosvosky, G.C.; Basso, H.F.; Zamoner, A.; Bagatini, M.D. Deregulation of purinergic ectoenzyme activity in head and neck cancer promotes immunosuppression. Mol. Biol. Rep. 2022, 49, 7687–7695. [Google Scholar] [CrossRef]
- Allard, B.; Beavis, P.A.; Darcy, P.K.; Stagg, J. Immunosuppressive activities of adenosine in cancer. Curr. Opin. Pharmacol. 2016, 29, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Cekic, C.; Day, Y.J.; Sag, D.; Linden, J. Myeloid expression of adenosine A2A receptor suppresses T and NK cell responses in the solid tumor microenvironment. Cancer Res. 2014, 74, 7250–7259. [Google Scholar] [CrossRef] [PubMed]
- Hoskin, D.W.; Mader, J.S.; Furlong, S.J.; Conrad, D.M.; Blay, J. Inhibition of T cell and natural killer cell function by adenosine and its contribution to immune evasion by tumor cells (Review). Int. J. Oncol. 2008, 32, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, J.P.; Reis e Sousa, C. The Role of Type 1 Conventional Dendritic Cells in Cancer Immunity. Trends Cancer 2018, 4, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Schuon, R.; Brieger, J.; Franke, R.L.; Jakob, R.; Mann, W.J. Increased PGE2 levels in nonmalignant mucosa adjacent to squamous cell carcinoma of the head and neck. ORL J. Otorhinolaryngol. Relat. Spec. 2005, 67, 96–100. [Google Scholar] [CrossRef]
- Young, M.R. Trials and tribulations of immunotherapy as a treatment option for patients with squamous cell carcinoma of the head and neck. Cancer Immunol. Immunother. 2004, 53, 375–382. [Google Scholar] [CrossRef]
- Kaliński, P.; Smits, H.H.; Schuitemaker, J.H.; Vieira, P.L.; van Eijk, M.; de Jong, E.C.; Wierenga, E.A.; Kapsenberg, M.L. IL-4 is a mediator of IL-12p70 induction by human Th2 cells: Reversal of polarized Th2 phenotype by dendritic cells. J. Immunol. 2000, 165, 1877–1881. [Google Scholar] [CrossRef]
- Brocks, C.P.; Pries, R.; Frenzel, H.; Ernst, M.; Schlenke, P.; Wollenberg, B. Functional alteration of myeloid dendritic cells through head and neck cancer. Anticancer Res. 2007, 27, 817–824. [Google Scholar]
- Pahne-Zeppenfeld, J.; Schröer, N.; Walch-Rückheim, B.; Oldak, M.; Gorter, A.; Hegde, S.; Smola, S. Cervical cancer cell-derived interleukin-6 impairs CCR7-dependent migration of MMP-9-expressing dendritic cells. Int. J. Cancer 2014, 134, 2061–2073. [Google Scholar] [CrossRef]
- Ratta, M.; Fagnoni, F.; Curti, A.; Vescovini, R.; Sansoni, P.; Oliviero, B.; Fogli, M.; Ferri, E.; Della Cuna, G.R.; Tura, S.; et al. Dendritic cells are functionally defective in multiple myeloma: The role of interleukin-6. Blood 2002, 100, 230–237. [Google Scholar] [CrossRef]
- Chaux, P.; Favre, N.; Martin, M.; Martin, F. Tumor-infiltrating dendritic cells are defective in their antigen-presenting function and inducible B7 expression in rats. Int. J. Cancer 1997, 72, 619–624. [Google Scholar] [CrossRef]
- French, J.D. Revisiting immune-based therapies for aggressive follicular cell-derived thyroid cancers. Thyroid. 2013, 23, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D. Mechanisms and functional significance of tumour-induced dendritic-cell defects. Nat. Rev. Immunol. 2004, 4, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Hawiger, D.; Inaba, K.; Dorsett, Y.; Guo, M.; Mahnke, K.; Rivera, M.; Ravetch, J.V.; Steinman, R.M.; Nussenzweig, M.C. Dendritic cells induce peripheral T cell unresponsiveness under steady state conditions in vivo. J. Exp. Med. 2001, 194, 769–779. [Google Scholar] [CrossRef]
- Jonuleit, H.; Schmitt, E.; Schuler, G.; Knop, J.; Enk, A.H. Induction of interleukin 10-producing, nonproliferating CD4(+) T cells with regulatory properties by repetitive stimulation with allogeneic immature human dendritic cells. J. Exp. Med. 2000, 192, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Kel, J.M.; Girard-Madoux, M.J.; Reizis, B.; Clausen, B.E. TGF-beta is required to maintain the pool of immature Langerhans cells in the epidermis. J. Immunol. 2010, 185, 3248–3255. [Google Scholar] [CrossRef] [PubMed]
- Steinbrink, K.; Jonuleit, H.; Müller, G.; Schuler, G.; Knop, J.; Enk, A.H. Interleukin-10-treated human dendritic cells induce a melanoma-antigen-specific anergy in CD8(+) T cells resulting in a failure to lyse tumor cells. Blood 1999, 93, 1634–1642. [Google Scholar] [CrossRef]
- Yamazaki, S.; Inaba, K.; Tarbell, K.V.; Steinman, R.M. Dendritic cells expand antigen-specific Foxp3+ CD25+ CD4+ regulatory T cells including suppressors of alloreactivity. Immunol. Rev. 2006, 212, 314–329. [Google Scholar] [CrossRef]
- Oliveira-Neto, H.H.; Silva, E.T.; Leles, C.R.; Mendonça, E.F.; Alencar Rde, C.; Silva, T.A.; Batista, A.C. Involvement of CXCL12 and CXCR4 in lymph node metastases and development of oral squamous cell carcinomas. Tumour Biol. 2008, 29, 262–271. [Google Scholar] [CrossRef]
- Parikh, A.; Shin, J.; Faquin, W.; Lin, D.T.; Tirosh, I.; Sunwoo, J.B.; Puram, S.V. Malignant cell-specific CXCL14 promotes tumor lymphocyte infiltration in oral cavity squamous cell carcinoma. J. Immunother. Cancer 2020, 8, e001048. [Google Scholar] [CrossRef]
- Thiel, A.; Pries, R.; Jeske, S.; Trenkle, T.; Wollenberg, B. Effect of head and neck cancer supernatant and CpG-oligonucleotides on migration and IFN-alpha production of plasmacytoid dendritic cells. Anticancer Res. 2009, 29, 3019–3025. [Google Scholar] [PubMed]
- Zhou, B.; Lawrence, T.; Liang, Y. The Role of Plasmacytoid Dendritic Cells in Cancers. Front. Immunol. 2021, 12, 749190. [Google Scholar] [CrossRef] [PubMed]
- Poropatich, K.; Dominguez, D.; Chan, W.C.; Andrade, J.; Zha, Y.; Wray, B.; Miska, J.; Qin, L.; Cole, L.; Coates, S.; et al. OX40+ plasmacytoid dendritic cells in the tumor microenvironment promote antitumor immunity. J. Clin. Investig. 2020, 130, 3528–3542. [Google Scholar] [CrossRef] [PubMed]
- Coffelt, S.B.; Hughes, R.; Lewis, C.E. Tumor-associated macrophages: Effectors of angiogenesis and tumor progression. Biochim. Biophys. Acta 2009, 1796, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Lamagna, C.; Aurrand-Lions, M.; Imhof, B.A. Dual role of macrophages in tumor growth and angiogenesis. J. Leukoc. Biol. 2006, 80, 705–713. [Google Scholar] [CrossRef]
- Lewis, C.E.; Pollard, J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006, 66, 605–612. [Google Scholar] [CrossRef]
- Sawa-Wejksza, K.; Kandefer-Szerszeń, M. Tumor-Associated Macrophages as Target for Antitumor Therapy. Arch. Immunol. Ther. Exp. 2018, 66, 97–111. [Google Scholar] [CrossRef]
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The cellular and molecular origin of tumor-associated macrophages. Science 2014, 344, 921–925. [Google Scholar] [CrossRef]
- Liss, C.; Fekete, M.J.; Hasina, R.; Lam, C.D.; Lingen, M.W. Paracrine angiogenic loop between head-and-neck squamous-cell carcinomas and macrophages. Int. J. Cancer 2001, 93, 781–785. [Google Scholar] [CrossRef]
- Liss, C.; Fekete, M.J.; Hasina, R.; Lingen, M.W. Retinoic acid modulates the ability of macrophages to participate in the induction of the angiogenic phenotype in head and neck squamous cell carcinoma. Int. J. Cancer 2002, 100, 283–289. [Google Scholar] [CrossRef]
- Murray, P.J.; Wynn, T.A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 2011, 11, 723–737. [Google Scholar] [CrossRef]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Ni, Y.H.; Ding, L.; Huang, X.F.; Dong, Y.C.; Hu, Q.G.; Hou, Y.Y. Microlocalization of CD68+ tumor-associated macrophages in tumor stroma correlated with poor clinical outcomes in oral squamous cell carcinoma patients. Tumour Biol. 2015, 36, 5291–5298. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef]
- Weber, M.; Büttner-Herold, M.; Hyckel, P.; Moebius, P.; Distel, L.; Ries, J.; Amann, K.; Neukam, F.W.; Wehrhan, F. Small oral squamous cell carcinomas with nodal lymphogenic metastasis show increased infiltration of M2 polarized macrophages—An immunohistochemical analysis. J. Craniomaxillofac Surg. 2014, 42, 1087–1094. [Google Scholar] [CrossRef]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Costa, N.L.; Valadares, M.C.; Souza, P.P.; Mendonça, E.F.; Oliveira, J.C.; Silva, T.A.; Batista, A.C. Tumor-associated macrophages and the profile of inflammatory cytokines in oral squamous cell carcinoma. Oral Oncol. 2013, 49, 216–223. [Google Scholar] [CrossRef]
- Marcus, B.; Arenberg, D.; Lee, J.; Kleer, C.; Chepeha, D.B.; Schmalbach, C.E.; Islam, M.; Paul, S.; Pan, Q.; Hanash, S.; et al. Prognostic factors in oral cavity and oropharyngeal squamous cell carcinoma. Cancer 2004, 101, 2779–2787. [Google Scholar] [CrossRef] [PubMed]
- Wolf, G.T.; Chepeha, D.B.; Bellile, E.; Nguyen, A.; Thomas, D.; McHugh, J. Tumor infiltrating lymphocytes (TIL) and prognosis in oral cavity squamous carcinoma: A preliminary study. Oral Oncol. 2015, 51, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Okamoto, M.; Goda, H.; Tano, T.; Nakashiro, K.; Sugita, A.; Fujita, T.; Koido, S.; Homma, S.; Kawakami, Y.; et al. Prognostic significance of interleukin-8 and CD163-positive cell-infiltration in tumor tissues in patients with oral squamous cell carcinoma. PLoS ONE 2014, 9, e110378. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Laoui, D.; Movahedi, K.; Van Overmeire, E.; Van den Bossche, J.; Schouppe, E.; Mommer, C.; Nikolaou, A.; Morias, Y.; De Baetselier, P.; Van Ginderachter, J.A. Tumor-associated macrophages in breast cancer: Distinct subsets, distinct functions. Int. J. Dev. Biol. 2011, 55, 861–867. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Petruzzi, M.N.; Cherubini, K.; Salum, F.G.; de Figueiredo, M.A. Role of tumour-associated macrophages in oral squamous cells carcinoma progression: An update on current knowledge. Diagn. Pathol. 2017, 12, 32. [Google Scholar] [CrossRef]
- Tymoszuk, P.; Evens, H.; Marzola, V.; Wachowicz, K.; Wasmer, M.H.; Datta, S.; Müller-Holzner, E.; Fiegl, H.; Böck, G.; van Rooijen, N.; et al. In situ proliferation contributes to accumulation of tumor-associated macrophages in spontaneous mammary tumors. Eur. J. Immunol. 2014, 44, 2247–2262. [Google Scholar] [CrossRef]
- Van Overmeire, E.; Stijlemans, B.; Heymann, F.; Keirsse, J.; Morias, Y.; Elkrim, Y.; Brys, L.; Abels, C.; Lahmar, Q.; Ergen, C.; et al. M-CSF and GM-CSF Receptor Signaling Differentially Regulate Monocyte Maturation and Macrophage Polarization in the Tumor Microenvironment. Cancer Res. 2016, 76, 35–42. [Google Scholar] [CrossRef]
- Barsoum, I.B.; Koti, M.; Siemens, D.R.; Graham, C.H. Mechanisms of hypoxia-mediated immune escape in cancer. Cancer Res. 2014, 74, 7185–7190. [Google Scholar] [CrossRef]
- Barsoum, I.B.; Smallwood, C.A.; Siemens, D.R.; Graham, C.H. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res. 2014, 74, 665–674. [Google Scholar] [CrossRef]
- Kai, K.; Moriyama, M.; Haque, A.; Hattori, T.; Chinju, A.; Hu, C.; Kubota, K.; Miyahara, Y.; Kakizoe-Ishiguro, N.; Kawano, S.; et al. Oral Squamous Cell Carcinoma Contributes to Differentiation of Monocyte-Derived Tumor-Associated Macrophages via PAI-1 and IL-8 Production. Int. J. Mol. Sci. 2021, 22, 9475. [Google Scholar] [CrossRef] [PubMed]
- Pries, R.; Thiel, A.; Brocks, C.; Wollenberg, B. Secretion of tumor-promoting and immune suppressive cytokines by cell lines of head and neck squamous cell carcinoma. In Vivo 2006, 20, 45–48. [Google Scholar] [PubMed]
- Squarize, C.H.; Castilho, R.M.; Sriuranpong, V.; Pinto, D.S., Jr.; Gutkind, J.S. Molecular cross-talk between the NFkappaB and STAT3 signaling pathways in head and neck squamous cell carcinoma. Neoplasia 2006, 8, 733–746. [Google Scholar] [CrossRef] [PubMed]
- Logullo, A.F.; Nonogaki, S.; Miguel, R.E.; Kowalski, L.P.; Nishimoto, I.N.; Pasini, F.S.; Federico, M.H.; Brentani, R.R.; Brentani, M.M. Transforming growth factor beta1 (TGFbeta1) expression in head and neck squamous cell carcinoma patients as related to prognosis. J. Oral Pathol. Med. 2003, 32, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, E.; McCrory, A.; Talbert, M.; Young, G.; Murphy-Ullrich, J.; Gladson, C. Elevated expression of TGF-beta1 in head and neck cancer-associated fibroblasts. Mol. Carcinog. 2004, 40, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.; Murdoch, C. Macrophage responses to hypoxia: Implications for tumor progression and anti-cancer therapies. Am. J. Pathol. 2005, 167, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Wennerberg, E.; Lhuillier, C.; Vanpouille-Box, C.; Pilones, K.A.; García-Martínez, E.; Rudqvist, N.P.; Formenti, S.C.; Demaria, S. Barriers to Radiation-Induced In Situ Tumor Vaccination. Front. Immunol. 2017, 8, 229. [Google Scholar] [CrossRef]
- Chen, P.; Zuo, H.; Xiong, H.; Kolar, M.J.; Chu, Q.; Saghatelian, A.; Siegwart, D.J.; Wan, Y. Gpr132 sensing of lactate mediates tumor-macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. USA 2017, 114, 580–585. [Google Scholar] [CrossRef]
- Gallina, G.; Dolcetti, L.; Serafini, P.; De Santo, C.; Marigo, I.; Colombo, M.P.; Basso, G.; Brombacher, F.; Borrello, I.; Zanovello, P.; et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J. Clin. Investig. 2006, 116, 2777–2790. [Google Scholar] [CrossRef]
- Roth, F.; De La Fuente, A.C.; Vella, J.L.; Zoso, A.; Inverardi, L.; Serafini, P. Aptamer-mediated blockade of IL4Rα triggers apoptosis of MDSCs and limits tumor progression. Cancer Res. 2012, 72, 1373–1383. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, Q.; Xu, M.; Wang, L.; Chen, X.; Feng, Y.; Li, Y.; Zhang, X.; Cui, W.; Jia, X. CCL2-CCR2 axis recruits tumor associated macrophages to induce immune evasion through PD-1 signaling in esophageal carcinogenesis. Mol. Cancer 2020, 19, 41. [Google Scholar] [CrossRef] [PubMed]
- Willems, J.J.L.P.; Arnold, B.P.; Gregory, C.D. Sinister Self-Sacrifice: The Contribution of Apoptosis to Malignancy. Front. Immunol. 2014, 5, 299. [Google Scholar] [CrossRef] [PubMed]
- Siveen, K.S.; Kuttan, G. Role of macrophages in tumour progression. Immunol. Lett. 2009, 123, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Kouketsu, A.; Sato, I.; Oikawa, M.; Shimizu, Y.; Saito, H.; Tashiro, K.; Yamashita, Y.; Takahashi, T.; Kumamoto, H. Regulatory T cells and M2-polarized tumour-associated macrophages are associated with the oncogenesis and progression of oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2019, 48, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Laviron, M.; Boissonnas, A. Ontogeny of Tumor-Associated Macrophages. Front. Immunol. 2019, 10, 1799. [Google Scholar] [CrossRef] [PubMed]
- El-Rouby, D.H. Association of macrophages with angiogenesis in oral verrucous and squamous cell carcinomas. J. Oral Pathol. Med. 2010, 39, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Rodriguez, P.C.; Hernandez, C.P.; Quiceno, D.; Dubinett, S.M.; Zabaleta, J.; Ochoa, J.B.; Gilbert, J.; Ochoa, A.C. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J. Exp. Med. 2005, 202, 931–939. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Takahashi, H.; Sakakura, K.; Kudo, T.; Toyoda, M.; Kaira, K.; Oyama, T.; Chikamatsu, K. Cancer-associated fibroblasts promote an immunosuppressive microenvironment through the induction and accumulation of protumoral macrophages. Oncotarget 2017, 8, 8633–8647. [Google Scholar] [CrossRef]
- Van Ginderachter, J.A.; Meerschaut, S.; Liu, Y.; Brys, L.; De Groeve, K.; Hassanzadeh Ghassabeh, G.; Raes, G.; De Baetselier, P. Peroxisome proliferator-activated receptor gamma (PPARgamma) ligands reverse CTL suppression by alternatively activated (M2) macrophages in cancer. Blood 2006, 108, 525–535. [Google Scholar] [CrossRef]
- Vaupel, P.; Multhoff, G. Hypoxia-/HIF-1α-Driven Factors of the Tumor Microenvironment Impeding Antitumor Immune Responses and Promoting Malignant Progression. Adv. Exp. Med. Biol. 2018, 1072, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Tang, Y.L.; Liang, X.H. Transforming growth factor-β signaling in head and neck squamous cell carcinoma: Insights into cellular responses. Oncol. Lett. 2018, 16, 4799–4806. [Google Scholar] [CrossRef] [PubMed]
- Rückert, M.; Deloch, L.; Fietkau, R.; Frey, B.; Hecht, M.; Gaipl, U.S. Immune modulatory effects of radiotherapy as basis for well-reasoned radioimmunotherapies. Strahlenther. Onkol. 2018, 194, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Larmonier, N.; Marron, M.; Zeng, Y.; Cantrell, J.; Romanoski, A.; Sepassi, M.; Thompson, S.; Chen, X.; Andreansky, S.; Katsanis, E. Tumor-derived CD4(+)CD25(+) regulatory T cell suppression of dendritic cell function involves TGF-beta and IL-10. Cancer Immunol. Immunother. 2007, 56, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Nakasone, E.S.; Werb, Z. Tumors as organs: Complex tissues that interface with the entire organism. Dev. Cell 2010, 18, 884–901. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Albelda, S.M. Tumor-associated neutrophils: Friend or foe? Carcinogenesis 2012, 33, 949–955. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Kaffenberger, W.; Clasen, B.P.; van Beuningen, D. The respiratory burst of neutrophils, a prognostic parameter in head and neck cancer? Clin. Immunol. Immunopathol. 1992, 64, 57–62. [Google Scholar] [CrossRef]
- Gasparoto, T.H.; de Souza Malaspina, T.S.; Benevides, L.; de Melo, E.J., Jr.; Costa, M.R.; Damante, J.H.; Ikoma, M.R.; Garlet, G.P.; Cavassani, K.A.; da Silva, J.S.; et al. Patients with oral squamous cell carcinoma are characterized by increased frequency of suppressive regulatory T cells in the blood and tumor microenvironment. Cancer Immunol. Immunother. 2010, 59, 819–828. [Google Scholar] [CrossRef]
- Maggioni, D.; Pignataro, L.; Garavello, W. T-helper and T-regulatory cells modulation in head and neck squamous cell carcinoma. Oncoimmunology 2017, 6, e1325066. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; Van Waes, C.; Allen, C.T. Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and Tregs as mediators of the immunosuppressive microenvironment in head and neck cancer. Oral Oncol. 2016, 58, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Umansky, V.; Blattner, C.; Gebhardt, C.; Utikal, J. The Role of Myeloid-Derived Suppressor Cells (MDSC) in Cancer Progression. Vaccines 2016, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019, 19, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Poh, A.R.; Ernst, M. Targeting Macrophages in Cancer: From Bench to Bedside. Front. Oncol. 2018, 8, 49. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef]
- Geiger, R.; Rieckmann, J.C.; Wolf, T.; Basso, C.; Feng, Y.; Fuhrer, T.; Kogadeeva, M.; Picotti, P.; Meissner, F.; Mann, M.; et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 2016, 167, 829–842.e813. [Google Scholar] [CrossRef]
- Vasquez-Dunddel, D.; Pan, F.; Zeng, Q.; Gorbounov, M.; Albesiano, E.; Fu, J.; Blosser, R.L.; Tam, A.J.; Bruno, T.; Zhang, H.; et al. STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J. Clin. Investig. 2013, 123, 1580–1589. [Google Scholar] [CrossRef]
- Hyman, M.C.; Petrovic-Djergovic, D.; Visovatti, S.H.; Liao, H.; Yanamadala, S.; Bouïs, D.; Su, E.J.; Lawrence, D.A.; Broekman, M.J.; Marcus, A.J.; et al. Self-regulation of inflammatory cell trafficking in mice by the leukocyte surface apyrase CD39. J. Clin. Investig. 2009, 119, 1136–1149. [Google Scholar] [CrossRef]
- Knapp, K.; Zebisch, M.; Pippel, J.; El-Tayeb, A.; Müller, C.E.; Sträter, N. Crystal structure of the human ecto-5′-nucleotidase (CD73): Insights into the regulation of purinergic signaling. Structure 2012, 20, 2161–2173. [Google Scholar] [CrossRef]
- Mandapathil, M.; Hilldorfer, B.; Szczepanski, M.J.; Czystowska, M.; Szajnik, M.; Ren, J.; Lang, S.; Jackson, E.K.; Gorelik, E.; Whiteside, T.L. Generation and accumulation of immunosuppressive adenosine by human CD4+CD25highFOXP3+ regulatory T cells. J. Biol. Chem. 2010, 285, 7176–7186. [Google Scholar] [CrossRef]
- Sitkovsky, M.; Lukashev, D.; Deaglio, S.; Dwyer, K.; Robson, S.C.; Ohta, A. Adenosine A2A receptor antagonists: Blockade of adenosinergic effects and T regulatory cells. Br. J. Pharmacol. 2008, 153 (Suppl. S1), S457–S464. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Bravo, M.; Carvalho, J.L.; Saldanha-Araujo, F. Adenosine production: A common path for mesenchymal stem-cell and regulatory T-cell-mediated immunosuppression. Purinergic Signal 2016, 12, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Ohta, A.; Kini, R.; Ohta, A.; Subramanian, M.; Madasu, M.; Sitkovsky, M. The development and immunosuppressive functions of CD4(+) CD25(+) FoxP3(+) regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Front. Immunol. 2012, 3, 190. [Google Scholar] [CrossRef] [PubMed]
- Molon, B.; Ugel, S.; Del Pozzo, F.; Soldani, C.; Zilio, S.; Avella, D.; De Palma, A.; Mauri, P.; Monegal, A.; Rescigno, M.; et al. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J. Exp. Med. 2011, 208, 1949–1962. [Google Scholar] [CrossRef]
- Yarosz, E.L.; Chang, C.H. The Role of Reactive Oxygen Species in Regulating T Cell-mediated Immunity and Disease. Immune Netw. 2018, 18, e14. [Google Scholar] [CrossRef] [PubMed]
- Frossi, B.; De Carli, M.; Piemonte, M.; Pucillo, C. Oxidative microenvironment exerts an opposite regulatory effect on cytokine production by Th1 and Th2 cells. Mol. Immunol. 2008, 45, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, M.M.; Sauer, S.W.; Klemke, C.D.; Süss, D.; Okun, J.G.; Krammer, P.H.; Gülow, K. Mitochondrial reactive oxygen species control T cell activation by regulating IL-2 and IL-4 expression: Mechanism of ciprofloxacin-mediated immunosuppression. J. Immunol. 2010, 184, 4827–4841. [Google Scholar] [CrossRef]
- Lyford-Pike, S.; Peng, S.; Young, G.D.; Taube, J.M.; Westra, W.H.; Akpeng, B.; Bruno, T.C.; Richmon, J.D.; Wang, H.; Bishop, J.A.; et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013, 73, 1733–1741. [Google Scholar] [CrossRef]
- Parry, R.V.; Chemnitz, J.M.; Frauwirth, K.A.; Lanfranco, A.R.; Braunstein, I.; Kobayashi, S.V.; Linsley, P.S.; Thompson, C.B.; Riley, J.L. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell Biol. 2005, 25, 9543–9553. [Google Scholar] [CrossRef]
- Kubota, K.; Moriyama, M.; Furukawa, S.; Rafiul, H.; Maruse, Y.; Jinno, T.; Tanaka, A.; Ohta, M.; Ishiguro, N.; Yamauchi, M.; et al. CD163(+)CD204(+) tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci. Rep. 2017, 7, 1755. [Google Scholar] [CrossRef]
- Bergmann, C.; Strauss, L.; Wang, Y.; Szczepanski, M.J.; Lang, S.; Johnson, J.T.; Whiteside, T.L. T regulatory type 1 cells in squamous cell carcinoma of the head and neck: Mechanisms of suppression and expansion in advanced disease. Clin. Cancer Res. 2008, 14, 3706–3715. [Google Scholar] [CrossRef]
- Strauss, L.; Bergmann, C.; Szczepanski, M.; Gooding, W.; Johnson, J.T.; Whiteside, T.L. A unique subset of CD4+CD25highFoxp3+ T cells secreting interleukin-10 and transforming growth factor-beta1 mediates suppression in the tumor microenvironment. Clin. Cancer Res. 2007, 13, 4345–4354. [Google Scholar] [CrossRef]
- Ralainirina, N.; Poli, A.; Michel, T.; Poos, L.; Andrès, E.; Hentges, F.; Zimmer, J. Control of NK cell functions by CD4+CD25+ regulatory T cells. J. Leukoc. Biol. 2007, 81, 144–153. [Google Scholar] [CrossRef]
- Viguier, M.; Lemaître, F.; Verola, O.; Cho, M.S.; Gorochov, G.; Dubertret, L.; Bachelez, H.; Kourilsky, P.; Ferradini, L. Foxp3 expressing CD4+CD25(high) regulatory T cells are overrepresented in human metastatic melanoma lymph nodes and inhibit the function of infiltrating T cells. J. Immunol. 2004, 173, 1444–1453. [Google Scholar] [CrossRef]
- Valzasina, B.; Piconese, S.; Guiducci, C.; Colombo, M.P. Tumor-induced expansion of regulatory T cells by conversion of CD4+CD25- lymphocytes is thymus and proliferation independent. Cancer Res. 2006, 66, 4488–4495. [Google Scholar] [CrossRef]
- Zhou, G.; Levitsky, H.I. Natural regulatory T cells and de novo-induced regulatory T cells contribute independently to tumor-specific tolerance. J. Immunol. 2007, 178, 2155–2162. [Google Scholar] [CrossRef]
- Jie, H.B.; Gildener-Leapman, N.; Li, J.; Srivastava, R.M.; Gibson, S.P.; Whiteside, T.L.; Ferris, R.L. Intratumoral regulatory T cells upregulate immunosuppressive molecules in head and neck cancer patients. Br. J. Cancer 2013, 109, 2629–2635. [Google Scholar] [CrossRef]
- Facciabene, A.; Peng, X.; Hagemann, I.S.; Balint, K.; Barchetti, A.; Wang, L.P.; Gimotty, P.A.; Gilks, C.B.; Lal, P.; Zhang, L.; et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature 2011, 475, 226–230. [Google Scholar] [CrossRef]
- Lippitz, B.E. Cytokine patterns in patients with cancer: A systematic review. Lancet Oncol. 2013, 14, e218–e228. [Google Scholar] [CrossRef]
- Nishikawa, H.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Curr. Opin. Immunol. 2014, 27, 1–7. [Google Scholar] [CrossRef]
- Zou, L.; Barnett, B.; Safah, H.; Larussa, V.F.; Evdemon-Hogan, M.; Mottram, P.; Wei, S.; David, O.; Curiel, T.J.; Zou, W. Bone marrow is a reservoir for CD4+CD25+ regulatory T cells that traffic through CXCL12/CXCR4 signals. Cancer Res. 2004, 64, 8451–8455. [Google Scholar] [CrossRef]
- Pallandre, J.R.; Brillard, E.; Créhange, G.; Radlovic, A.; Remy-Martin, J.P.; Saas, P.; Rohrlich, P.S.; Pivot, X.; Ling, X.; Tiberghien, P.; et al. Role of STAT3 in CD4+CD25+FOXP3+ regulatory lymphocyte generation: Implications in graft-versus-host disease and antitumor immunity. J. Immunol. 2007, 179, 7593–7604. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Liang, J.; Liu, J.; Hao, J.; Wan, Q.; Liu, J.; Luo, C.; Lu, Z. CircRNA has_circ_0069313 induced OSCC immunity escape by miR-325-3p-Foxp3 axes in both OSCC cells and Treg cells. Aging 2022, 14, 4376–4389. [Google Scholar] [CrossRef]
- Chikamatsu, K.; Sakakura, K.; Whiteside, T.L.; Furuya, N. Relationships between regulatory T cells and CD8+ effector populations in patients with squamous cell carcinoma of the head and neck. Head. Neck 2007, 29, 120–127. [Google Scholar] [CrossRef]
- Jarnicki, A.G.; Lysaght, J.; Todryk, S.; Mills, K.H. Suppression of antitumor immunity by IL-10 and TGF-beta-producing T cells infiltrating the growing tumor: Influence of tumor environment on the induction of CD4+ and CD8+ regulatory T cells. J. Immunol. 2006, 177, 896–904. [Google Scholar] [CrossRef]
- Togashi, Y.; Shitara, K.; Nishikawa, H. Regulatory T cells in cancer immunosuppression—Implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019, 16, 356–371. [Google Scholar] [CrossRef]
- Mandapathil, M.; Szczepanski, M.J.; Szajnik, M.; Ren, J.; Lenzner, D.E.; Jackson, E.K.; Gorelik, E.; Lang, S.; Johnson, J.T.; Whiteside, T.L. Increased ectonucleotidase expression and activity in regulatory T cells of patients with head and neck cancer. Clin. Cancer Res. 2009, 15, 6348–6357. [Google Scholar] [CrossRef]
- Perri, F.; Ionna, F.; Longo, F.; Della Vittoria Scarpati, G.; De Angelis, C.; Ottaiano, A.; Botti, G.; Caponigro, F. Immune Response Against Head and Neck Cancer: Biological Mechanisms and Implication on Therapy. Transl. Oncol. 2020, 13, 262–274. [Google Scholar] [CrossRef]
- Grosso, J.F.; Kelleher, C.C.; Harris, T.J.; Maris, C.H.; Hipkiss, E.L.; De Marzo, A.; Anders, R.; Netto, G.; Getnet, D.; Bruno, T.C.; et al. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J. Clin. Investig. 2007, 117, 3383–3392. [Google Scholar] [CrossRef]
- Mishra, A.K.; Kadoishi, T.; Wang, X.; Driver, E.; Chen, Z.; Wang, X.J.; Wang, J.H. Squamous cell carcinomas escape immune surveillance via inducing chronic activation and exhaustion of CD8+ T Cells co-expressing PD-1 and LAG-3 inhibitory receptors. Oncotarget 2016, 7, 81341–81356. [Google Scholar] [CrossRef]
- Collison, L.W.; Chaturvedi, V.; Henderson, A.L.; Giacomin, P.R.; Guy, C.; Bankoti, J.; Finkelstein, D.; Forbes, K.; Workman, C.J.; Brown, S.A.; et al. IL-35-mediated induction of a potent regulatory T cell population. Nat. Immunol. 2010, 11, 1093–1101. [Google Scholar] [CrossRef]
- Collison, L.W.; Workman, C.J.; Kuo, T.T.; Boyd, K.; Wang, Y.; Vignali, K.M.; Cross, R.; Sehy, D.; Blumberg, R.S.; Vignali, D.A.A. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 2007, 450, 566–569. [Google Scholar] [CrossRef]
- Olson, B.; Sullivan, J.; Burlingham, W. Interleukin 35: A Key Mediator of Suppression and the Propagation of Infectious Tolerance. Front. Immunol. 2013, 4, 315. [Google Scholar] [CrossRef]
- Olson, B.M.; Jankowska-Gan, E.; Becker, J.T.; Vignali, D.A.A.; Burlingham, W.J.; McNeel, D.G. Human Prostate Tumor Antigen–Specific CD8+ Regulatory T Cells Are Inhibited by CTLA-4 or IL-35 Blockade. J. Immunol. 2012, 189, 5590–5601. [Google Scholar] [CrossRef]
- Schmidt, A.; Oberle, N.; Krammer, P.H. Molecular mechanisms of treg-mediated T cell suppression. Front. Immunol. 2012, 3, 51. [Google Scholar] [CrossRef]
- Zou, W. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 2006, 6, 295–307. [Google Scholar] [CrossRef]
- Banerjee, H.; Nieves-Rosado, H.; Kulkarni, A.; Murter, B.; McGrath, K.V.; Chandran, U.R.; Chang, A.; Szymczak-Workman, A.L.; Vujanovic, L.; Delgoffe, G.M.; et al. Expression of Tim-3 drives phenotypic and functional changes in Treg cells in secondary lymphoid organs and the tumor microenvironment. Cell Rep. 2021, 36, 109699. [Google Scholar] [CrossRef]
- Sheng, C.C.; Han, F.Y. Immunoregulation effects of TIM-3 on tumors. Neoplasma 2019, 66, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhang, H.; Sun, S.; Sun, S.; Li, L. The effects of Tim-3 activation on T-cells in gastric cancer progression. Oncol. Lett. 2019, 17, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Cai, S.F.; Fehniger, T.A.; Song, J.; Collins, L.I.; Piwnica-Worms, D.R.; Ley, T.J. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 2007, 27, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Strauss, L.; Bergmann, C.; Whiteside, T.L. Human circulating CD4+CD25highFoxp3+ regulatory T cells kill autologous CD8+ but not CD4+ responder cells by Fas-mediated apoptosis. J. Immunol. 2009, 182, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Kuropkat, C.; Duenne, A.A.; Herz, U.; Renz, H.; Werner, J.A. Significant correlation of matrix metalloproteinase and macrophage colony-stimulating factor serum concentrations in patients with head and neck cancer. Neoplasma 2004, 51, 375–378. [Google Scholar] [PubMed]
- McDermott, R.S.; Deneux, L.; Mosseri, V.; Védrenne, J.; Clough, K.; Fourquet, A.; Rodriguez, J.; Cosset, J.M.; Sastre, X.; Beuzeboc, P.; et al. Circulating macrophage colony stimulating factor as a marker of tumour progression. Eur. Cytokine Netw. 2002, 13, 121–127. [Google Scholar]
- Itoh, S.; Matsui, K.; Furuta, I.; Takano, Y. Immunohistochemical study on overexpression of cyclooxygenase-2 in squamous cell carcinoma of the oral cavity: Its importance as a prognostic predictor. Oral Oncol. 2003, 39, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.O.; Leskov, I.L.; Lin, M.; Abreo, F.W.; Shi, R.; Hartman, G.H.; Glass, J. COX-2 expression in dysplasia of the head and neck: Correlation with elF4E. Cancer 2001, 92, 1888–1895. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Wang, D.; Daikoku, T.; Sun, H.; Dey, S.K.; Dubois, R.N. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 2013, 24, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Knolhoff, B.L.; Meyer, M.A.; Nywening, T.M.; West, B.L.; Luo, J.; Wang-Gillam, A.; Goedegebuure, S.P.; Linehan, D.C.; DeNardo, D.G. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014, 74, 5057–5069. [Google Scholar] [CrossRef]
- Hamilton, J.A. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008, 8, 533–544. [Google Scholar] [CrossRef]
- Priceman, S.J.; Sung, J.L.; Shaposhnik, Z.; Burton, J.B.; Torres-Collado, A.X.; Moughon, D.L.; Johnson, M.; Lusis, A.J.; Cohen, D.A.; Iruela-Arispe, M.L.; et al. Targeting distinct tumor-infiltrating myeloid cells by inhibiting CSF-1 receptor: Combating tumor evasion of antiangiogenic therapy. Blood 2010, 115, 1461–1471. [Google Scholar] [CrossRef]
- Xu, J.; Escamilla, J.; Mok, S.; David, J.; Priceman, S.; West, B.; Bollag, G.; McBride, W.; Wu, L. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer Res. 2013, 73, 2782–2794. [Google Scholar] [CrossRef] [PubMed]
- Serafini, P. Editorial: PGE2-producing MDSC: A role in tumor progression? J. Leukoc. Biol. 2010, 88, 827–829. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S.; Sinha, P. Myeloid-derived suppressor cells: Linking inflammation and cancer. J. Immunol. 2009, 182, 4499–4506. [Google Scholar] [CrossRef]
- Serafini, P.; Meckel, K.; Kelso, M.; Noonan, K.; Califano, J.; Koch, W.; Dolcetti, L.; Bronte, V.; Borrello, I. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J. Exp. Med. 2006, 203, 2691–2702. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef] [PubMed]
- Rapidis, A.D.; Wolf, G.T. Immunotherapy of head and neck cancer: Current and future considerations. J. Oncol. 2009, 2009, 346345. [Google Scholar] [CrossRef] [PubMed]
- Braza, M.S.; Conde, P.; Garcia, M.; Cortegano, I.; Brahmachary, M.; Pothula, V.; Fay, F.; Boros, P.; Werner, S.A.; Ginhoux, F.; et al. Neutrophil derived CSF1 induces macrophage polarization and promotes transplantation tolerance. Am. J. Transplant. 2018, 18, 1247–1255. [Google Scholar] [CrossRef]
- Condamine, T.; Dominguez, G.A.; Youn, J.I.; Kossenkov, A.V.; Mony, S.; Alicea-Torres, K.; Tcyganov, E.; Hashimoto, A.; Nefedova, Y.; Lin, C.; et al. Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 2016, 1, aaf8943. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S. Immune surveillance: A balance between protumor and antitumor immunity. Curr. Opin. Genet. Dev. 2008, 18, 11–18. [Google Scholar] [CrossRef]
- Rodriguez, P.C.; Ernstoff, M.S.; Hernandez, C.; Atkins, M.; Zabaleta, J.; Sierra, R.; Ochoa, A.C. Arginase I-producing myeloid-derived suppressor cells in renal cell carcinoma are a subpopulation of activated granulocytes. Cancer Res. 2009, 69, 1553–1560. [Google Scholar] [CrossRef]
- Rotondo, R.; Barisione, G.; Mastracci, L.; Grossi, F.; Orengo, A.M.; Costa, R.; Truini, M.; Fabbi, M.; Ferrini, S.; Barbieri, O. IL-8 induces exocytosis of arginase 1 by neutrophil polymorphonuclears in nonsmall cell lung cancer. Int. J. Cancer 2009, 125, 887–893. [Google Scholar] [CrossRef]
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336 Pt 1, 1–17. [Google Scholar] [CrossRef]
- Yu, J.; Du, W.; Yan, F.; Wang, Y.; Li, H.; Cao, S.; Yu, W.; Shen, C.; Liu, J.; Ren, X. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J. Immunol. 2013, 190, 3783–3797. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef]
- Weber, R.; Fleming, V.; Hu, X.; Nagibin, V.; Groth, C.; Altevogt, P.; Utikal, J.; Umansky, V. Myeloid-Derived Suppressor Cells Hinder the Anti-Cancer Activity of Immune Checkpoint Inhibitors. Front. Immunol. 2018, 9, 1310. [Google Scholar] [CrossRef]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Rosbe, K.W.; Prazma, J.; Petrusz, P.; Mims, W.; Ball, S.S.; Weissler, M.C. Immunohistochemical characterization of nitric oxide synthase activity in squamous cell carcinoma of the head and neck. Otolaryngol. Head Neck Surg. 1995, 113, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Pan, P.Y.; Li, Q.; Sato, A.I.; Levy, D.E.; Bromberg, J.; Divino, C.M.; Chen, S.H. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006, 66, 1123–1131. [Google Scholar] [CrossRef]
- Sinha, P.; Clements, V.K.; Bunt, S.K.; Albelda, S.M.; Ostrand-Rosenberg, S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J. Immunol. 2007, 179, 977–983. [Google Scholar] [CrossRef]
- Srivastava, M.K.; Sinha, P.; Clements, V.K.; Rodriguez, P.; Ostrand-Rosenberg, S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010, 70, 68–77. [Google Scholar] [CrossRef]
- Greene, S.; Robbins, Y.; Mydlarz, W.K.; Huynh, A.P.; Schmitt, N.C.; Friedman, J.; Horn, L.A.; Palena, C.; Schlom, J.; Maeda, D.Y.; et al. Inhibition of MDSC Trafficking with SX-682, a CXCR1/2 Inhibitor, Enhances NK-Cell Immunotherapy in Head and Neck Cancer Models. Clin. Cancer Res. 2020, 26, 1420–1431. [Google Scholar] [CrossRef]
- Pak, A.S.; Wright, M.A.; Matthews, J.P.; Collins, S.L.; Petruzzelli, G.J.; Young, M.R. Mechanisms of immune suppression in patients with head and neck cancer: Presence of CD34(+) cells which suppress immune functions within cancers that secrete granulocyte-macrophage colony-stimulating factor. Clin. Cancer Res. 1995, 1, 95–103. [Google Scholar]
- Parker, K.H.; Beury, D.W.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv. Cancer Res. 2015, 128, 95–139. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.C.; Quiceno, D.G.; Zabaleta, J.; Ortiz, B.; Zea, A.H.; Piazuelo, M.B.; Delgado, A.; Correa, P.; Brayer, J.; Sotomayor, E.M.; et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004, 64, 5839–5849. [Google Scholar] [CrossRef]
- Rodriguez, P.C.; Quiceno, D.G.; Ochoa, A.C. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 2007, 109, 1568–1573. [Google Scholar] [CrossRef] [PubMed]
- Harari, O.; Liao, J.K. Inhibition of MHC II gene transcription by nitric oxide and antioxidants. Curr. Pharm. Des. 2004, 10, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Stiff, A.; Trikha, P.; Mundy-Bosse, B.; McMichael, E.; Mace, T.A.; Benner, B.; Kendra, K.; Campbell, A.; Gautam, S.; Abood, D.; et al. Nitric Oxide Production by Myeloid-Derived Suppressor Cells Plays a Role in Impairing Fc Receptor-Mediated Natural Killer Cell Function. Clin. Cancer Res. 2018, 24, 1891–1904. [Google Scholar] [CrossRef]
- Mazzoni, A.; Bronte, V.; Visintin, A.; Spitzer, J.H.; Apolloni, E.; Serafini, P.; Zanovello, P.; Segal, D.M. Myeloid suppressor lines inhibit T cell responses by an NO-dependent mechanism. J. Immunol. 2002, 168, 689–695. [Google Scholar] [CrossRef]
- Bingisser, R.M.; Tilbrook, P.A.; Holt, P.G.; Kees, U.R. Macrophage-derived nitric oxide regulates T cell activation via reversible disruption of the Jak3/STAT5 signaling pathway. J. Immunol. 1998, 160, 5729–5734. [Google Scholar] [CrossRef]
- Liu, C.; Yu, S.; Kappes, J.; Wang, J.; Grizzle, W.E.; Zinn, K.R.; Zhang, H.G. Expansion of spleen myeloid suppressor cells represses NK cell cytotoxicity in tumor-bearing host. Blood 2007, 109, 4336–4342. [Google Scholar] [CrossRef]
- Bentz, B.G.; Haines, G.K., 3rd; Radosevich, J.A. Increased protein nitrosylation in head and neck squamous cell carcinogenesis. Head. Neck 2000, 22, 64–70. [Google Scholar] [CrossRef]
- Bronte, V.; Serafini, P.; De Santo, C.; Marigo, I.; Tosello, V.; Mazzoni, A.; Segal, D.M.; Staib, C.; Lowel, M.; Sutter, G.; et al. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J. Immunol. 2003, 170, 270–278. [Google Scholar] [CrossRef]
- Nagaraj, S.; Gupta, K.; Pisarev, V.; Kinarsky, L.; Sherman, S.; Kang, L.; Herber, D.L.; Schneck, J.; Gabrilovich, D.I. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat. Med. 2007, 13, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Schrum, A.G.; Cho, H.I.; Celis, E.; Gabrilovich, D.I. Mechanism of T cell tolerance induced by myeloid-derived suppressor cells. J. Immunol. 2010, 184, 3106–3116. [Google Scholar] [CrossRef]
- Trellakis, S.; Bruderek, K.; Dumitru, C.A.; Gholaman, H.; Gu, X.; Bankfalvi, A.; Scherag, A.; Hütte, J.; Dominas, N.; Lehnerdt, G.F.; et al. Polymorphonuclear granulocytes in human head and neck cancer: Enhanced inflammatory activity, modulation by cancer cells and expansion in advanced disease. Int. J. Cancer 2011, 129, 2183–2193. [Google Scholar] [CrossRef]
- Haqqani, A.S.; Sandhu, J.K.; Birnboim, H.C. Expression of interleukin-8 promotes neutrophil infiltration and genetic instability in mutatect tumors. Neoplasia 2000, 2, 561–568. [Google Scholar] [CrossRef]
- Castell, S.D.; Harman, M.F.; Morón, G.; Maletto, B.A.; Pistoresi-Palencia, M.C. Neutrophils Which Migrate to Lymph Nodes Modulate CD4(+) T Cell Response by a PD-L1 Dependent Mechanism. Front. Immunol. 2019, 10, 105. [Google Scholar] [CrossRef]
- Sun, R.; Xiong, Y.; Liu, H.; Gao, C.; Su, L.; Weng, J.; Yuan, X.; Zhang, D.; Feng, J. Tumor-associated neutrophils suppress antitumor immunity of NK cells through the PD-L1/PD-1 axis. Transl. Oncol. 2020, 13, 100825. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, J.; Shaul, M.E.; Mishalian, I.; Hovav, A.H.; Levy, L.; Zolotriov, L.; Granot, Z.; Fridlender, Z.G. Tumor-associated neutrophils induce apoptosis of non-activated CD8 T-cells in a TNFα and NO-dependent mechanism, promoting a tumor-supportive environment. Oncoimmunology 2017, 6, e1356965. [Google Scholar] [CrossRef]
- Tecchio, C.; Cassatella, M.A. Neutrophil-derived chemokines on the road to immunity. Semin. Immunol. 2016, 28, 119–128. [Google Scholar] [CrossRef]
- Odobasic, D.; Kitching, A.R.; Yang, Y.; O’Sullivan, K.M.; Muljadi, R.C.; Edgtton, K.L.; Tan, D.S.; Summers, S.A.; Morand, E.F.; Holdsworth, S.R. Neutrophil myeloperoxidase regulates T-cell-driven tissue inflammation in mice by inhibiting dendritic cell function. Blood 2013, 121, 4195–4204. [Google Scholar] [CrossRef]
- Monteran, L.; Erez, N. The Dark Side of Fibroblasts: Cancer-Associated Fibroblasts as Mediators of Immunosuppression in the Tumor Microenvironment. Front. Immunol. 2019, 10, 1835. [Google Scholar] [CrossRef] [PubMed]
- New, J.; Arnold, L.; Ananth, M.; Alvi, S.; Thornton, M.; Werner, L.; Tawfik, O.; Dai, H.; Shnayder, Y.; Kakarala, K.; et al. Secretory Autophagy in Cancer-Associated Fibroblasts Promotes Head and Neck Cancer Progression and Offers a Novel Therapeutic Target. Cancer Res. 2017, 77, 6679–6691. [Google Scholar] [CrossRef]
- Yu, B.; Wu, K.; Wang, X.; Zhang, J.; Wang, L.; Jiang, Y.; Zhu, X.; Chen, W.; Yan, M. Periostin secreted by cancer-associated fibroblasts promotes cancer stemness in head and neck cancer by activating protein tyrosine kinase 7. Cell Death Dis. 2018, 9, 1082. [Google Scholar] [CrossRef]
- Kratochwil, C.; Flechsig, P.; Lindner, T.; Abderrahim, L.; Altmann, A.; Mier, W.; Adeberg, S.; Rathke, H.; Röhrich, M.; Winter, H.; et al. (68)Ga-FAPI PET/CT: Tracer Uptake in 28 Different Kinds of Cancer. J. Nucl. Med. 2019, 60, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Gleich, L.L.; Srivastava, L.; Gluckman, J.L. Plasma Platelet-Derived Growth Factor: Preliminary Study of a Potential Marker in Head and Neck Cancer. Ann. Otol. Rhinol. Laryngol. 1996, 105, 710–712. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yan, M.; Wang, X.; Xu, Q.; Wang, X.; Zhu, X.; Shi, J.; Li, Z.; Zhang, J.; Chen, W. Cancer-associated Fibroblast-derived IL-6 Promotes Head and Neck Cancer Progression via the Osteopontin-NF-kappa B Signaling Pathway. Theranostics 2018, 8, 921–940. [Google Scholar] [CrossRef]
- Bae, J.Y.; Kim, E.K.; Yang, D.H.; Zhang, X.; Park, Y.J.; Lee, D.Y.; Che, C.M.; Kim, J. Reciprocal interaction between carcinoma-associated fibroblasts and squamous carcinoma cells through interleukin-1α induces cancer progression. Neoplasia 2014, 16, 928–938. [Google Scholar] [CrossRef]
- Bello, I.O.; Vered, M.; Dayan, D.; Dobriyan, A.; Yahalom, R.; Alanen, K.; Nieminen, P.; Kantola, S.; Läärä, E.; Salo, T. Cancer-associated fibroblasts, a parameter of the tumor microenvironment, overcomes carcinoma-associated parameters in the prognosis of patients with mobile tongue cancer. Oral Oncol. 2011, 47, 33–38. [Google Scholar] [CrossRef]
- Lotti, F.; Jarrar, A.M.; Pai, R.K.; Hitomi, M.; Lathia, J.; Mace, A.; Gantt, G.A., Jr.; Sukhdeo, K.; DeVecchio, J.; Vasanji, A.; et al. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. J. Exp. Med. 2013, 210, 2851–2872. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Sakakura, K.; Kawabata-Iwakawa, R.; Rokudai, S.; Toyoda, M.; Nishiyama, M.; Chikamatsu, K. Immunosuppressive activity of cancer-associated fibroblasts in head and neck squamous cell carcinoma. Cancer Immunol. Immunother. 2015, 64, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Fearon, D.T. The carcinoma-associated fibroblast expressing fibroblast activation protein and escape from immune surveillance. Cancer Immunol. Res. 2014, 2, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef]
- Li, X.; Bu, W.; Meng, L.; Liu, X.; Wang, S.; Jiang, L.; Ren, M.; Fan, Y.; Sun, H. CXCL12/CXCR4 pathway orchestrates CSC-like properties by CAF recruited tumor associated macrophage in OSCC. Exp. Cell Res. 2019, 378, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; He, J.; Pan, Q.Z.; Yang, J.; Zhao, J.; Zhang, Y.J.; Huang, Y.; Tang, Y.; Wang, Q.; He, J.; et al. Cancer-Associated Fibroblast-Mediated Cellular Crosstalk Supports Hepatocellular Carcinoma Progression. Hepatology 2021, 73, 1717–1735. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Li, H.; Deng, Y.; Tai, Y.; Zeng, K.; Zhang, Y.; Liu, W.; Zhang, Q.; Yang, Y. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 422. [Google Scholar] [CrossRef]
- Huang, T.X.; Fu, L. The immune landscape of esophageal cancer. Cancer Commun. 2019, 39, 79. [Google Scholar] [CrossRef]
- Jenkins, R.W.; Barbie, D.A.; Flaherty, K.T. Mechanisms of resistance to immune checkpoint inhibitors. Br. J. Cancer 2018, 118, 9–16. [Google Scholar] [CrossRef]
- Karasarides, M.; Cogdill, A.P.; Robbins, P.B.; Bowden, M.; Burton, E.M.; Butterfield, L.H.; Cesano, A.; Hammer, C.; Haymaker, C.L.; Horak, C.E.; et al. Hallmarks of Resistance to Immune-Checkpoint Inhibitors. Cancer Immunol. Res. 2022, 10, 372–383. [Google Scholar] [CrossRef]
- Marei, H.E.; Hasan, A.; Pozzoli, G.; Cenciarelli, C. Cancer immunotherapy with immune checkpoint inhibitors (ICIs): Potential, mechanisms of resistance, and strategies for reinvigorating T cell responsiveness when resistance is acquired. Cancer Cell Int. 2023, 23, 64. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Han, Y.; Zhang, Y.; Zhao, Q.; Wang, H.; Wei, J.; Meng, L.; Xin, Y.; Jiang, X. Overcoming acquired resistance to cancer immune checkpoint therapy: Potential strategies based on molecular mechanisms. Cell Biosci. 2023, 13, 120. [Google Scholar] [CrossRef] [PubMed]
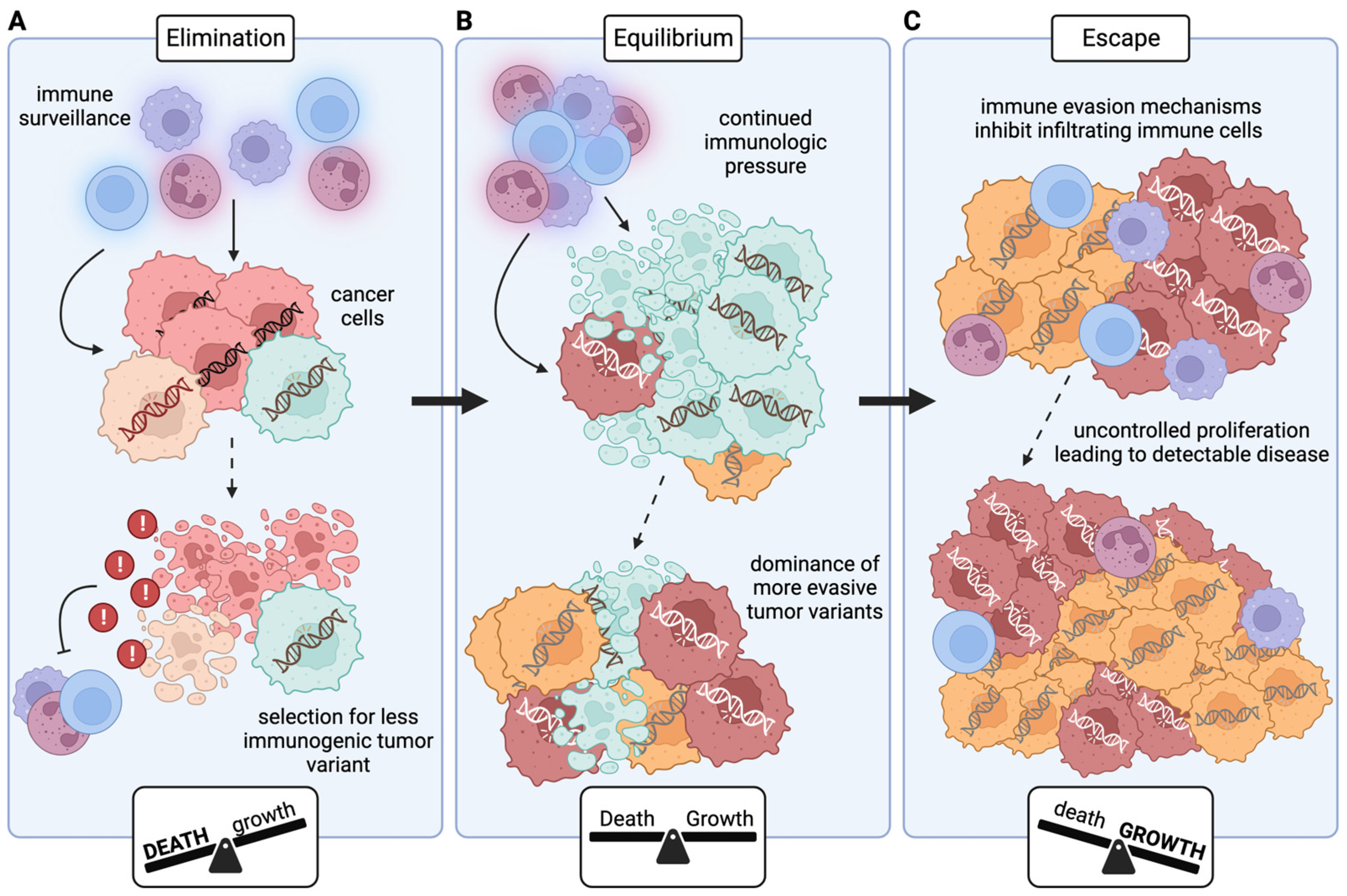
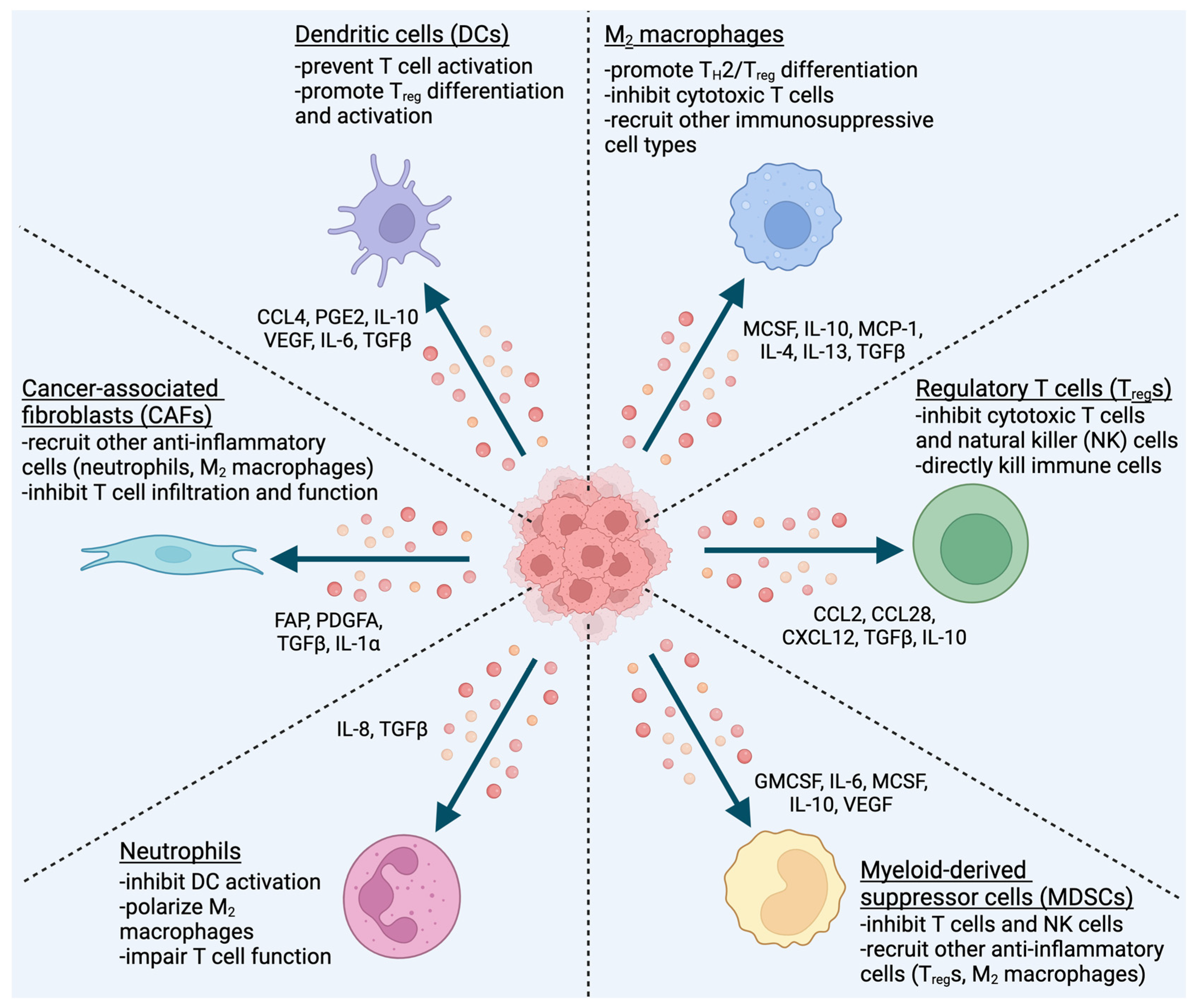
| AP Component | HNCs That Downregulate | References |
|---|---|---|
| TAP1 | 34–71% | [64,68,69,70,71] |
| TAP2 | 64–88% | [64,68,70] |
| Tapasin | 40–79% | [64,68,69,71] |
| LMP2 | 78–88% | [64,70,71] |
| LMP7 | ~60% | [64,70] |
| β2M | 60–87% | [64,71] |
| Calnexin | ~34% | [71] |
| Calreticulin | ~19% | [71] |
| ERp57 | ~26% | [71] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kostecki, K.L.; Iida, M.; Crossman, B.E.; Salgia, R.; Harari, P.M.; Bruce, J.Y.; Wheeler, D.L. Immune Escape Strategies in Head and Neck Cancer: Evade, Resist, Inhibit, Recruit. Cancers 2024, 16, 312. https://doi.org/10.3390/cancers16020312
Kostecki KL, Iida M, Crossman BE, Salgia R, Harari PM, Bruce JY, Wheeler DL. Immune Escape Strategies in Head and Neck Cancer: Evade, Resist, Inhibit, Recruit. Cancers. 2024; 16(2):312. https://doi.org/10.3390/cancers16020312
Chicago/Turabian StyleKostecki, Kourtney L., Mari Iida, Bridget E. Crossman, Ravi Salgia, Paul M. Harari, Justine Y. Bruce, and Deric L. Wheeler. 2024. "Immune Escape Strategies in Head and Neck Cancer: Evade, Resist, Inhibit, Recruit" Cancers 16, no. 2: 312. https://doi.org/10.3390/cancers16020312
APA StyleKostecki, K. L., Iida, M., Crossman, B. E., Salgia, R., Harari, P. M., Bruce, J. Y., & Wheeler, D. L. (2024). Immune Escape Strategies in Head and Neck Cancer: Evade, Resist, Inhibit, Recruit. Cancers, 16(2), 312. https://doi.org/10.3390/cancers16020312






