The miR-19a/Cylindromatosis Axis Regulates Pituitary Adenoma Bone Invasion by Promoting Osteoclast Differentiation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fluorescence In Situ Hybridization (FISH)
2.2. Animal Experiments
2.3. Cell Culture and Reagents
2.4. Osteoclastogenesis Assay
2.5. F-Actin Ring Assay
2.6. Bone Resorption Assay
2.7. Quantitative Real-Time PCR (qRT–PCR)
2.8. Dual-Luciferase Reporter Gene Assay
2.9. Western Blotting (WB) and Co-Immunoprecipitation (Co-IP)
2.10. Electrophoretic Mobility Shift Assay (EMSA)
2.11. Statistical Analysis
3. Results
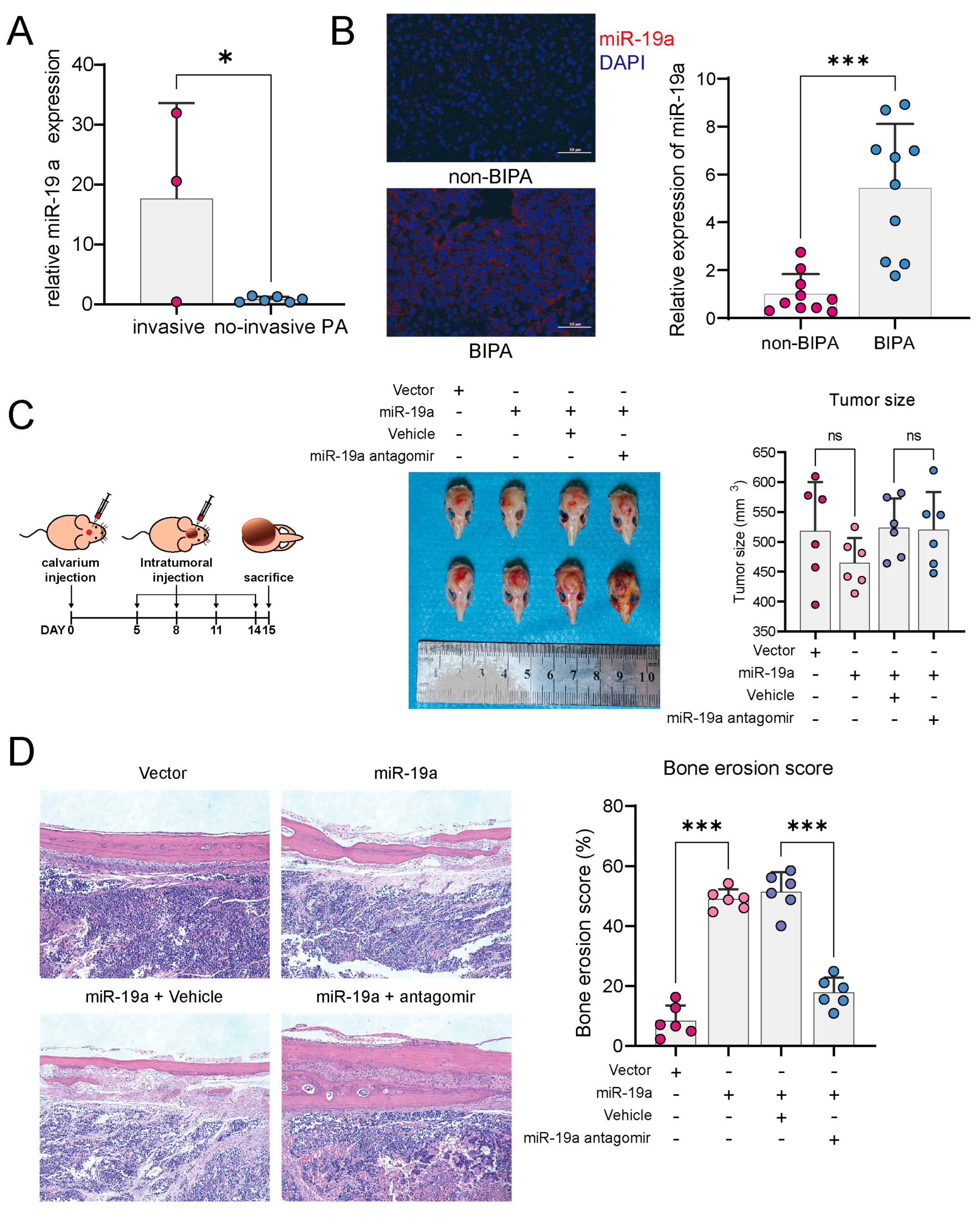
3.1. miR-19a Promotes PA Bone Invasion
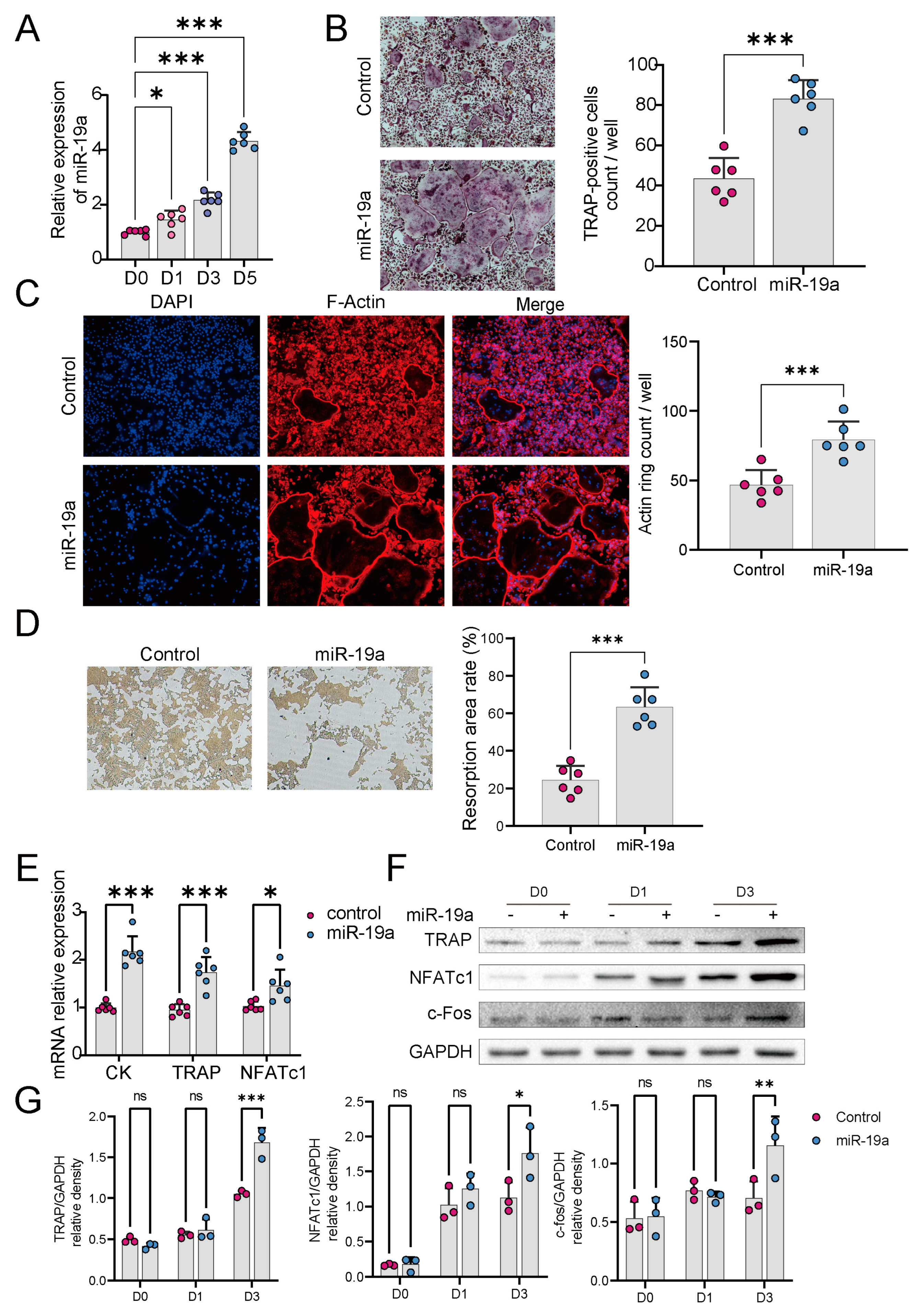
3.2. miR-19a Accelerates Osteoclast Differentiation and Function
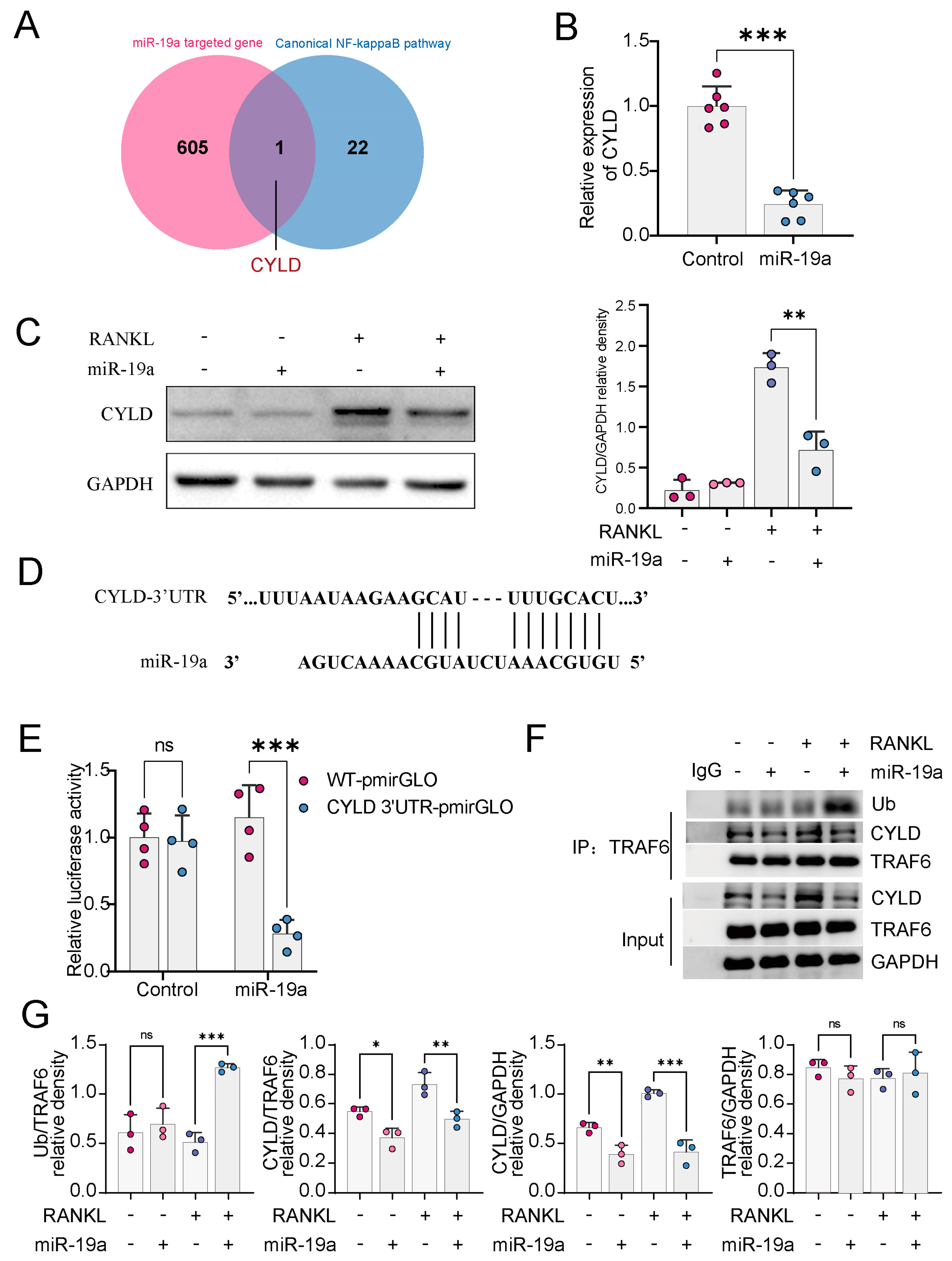
3.3. miR-19a Inhibits CYLD Expression and, Thus, Promotes TRAF6 Ubiquitination
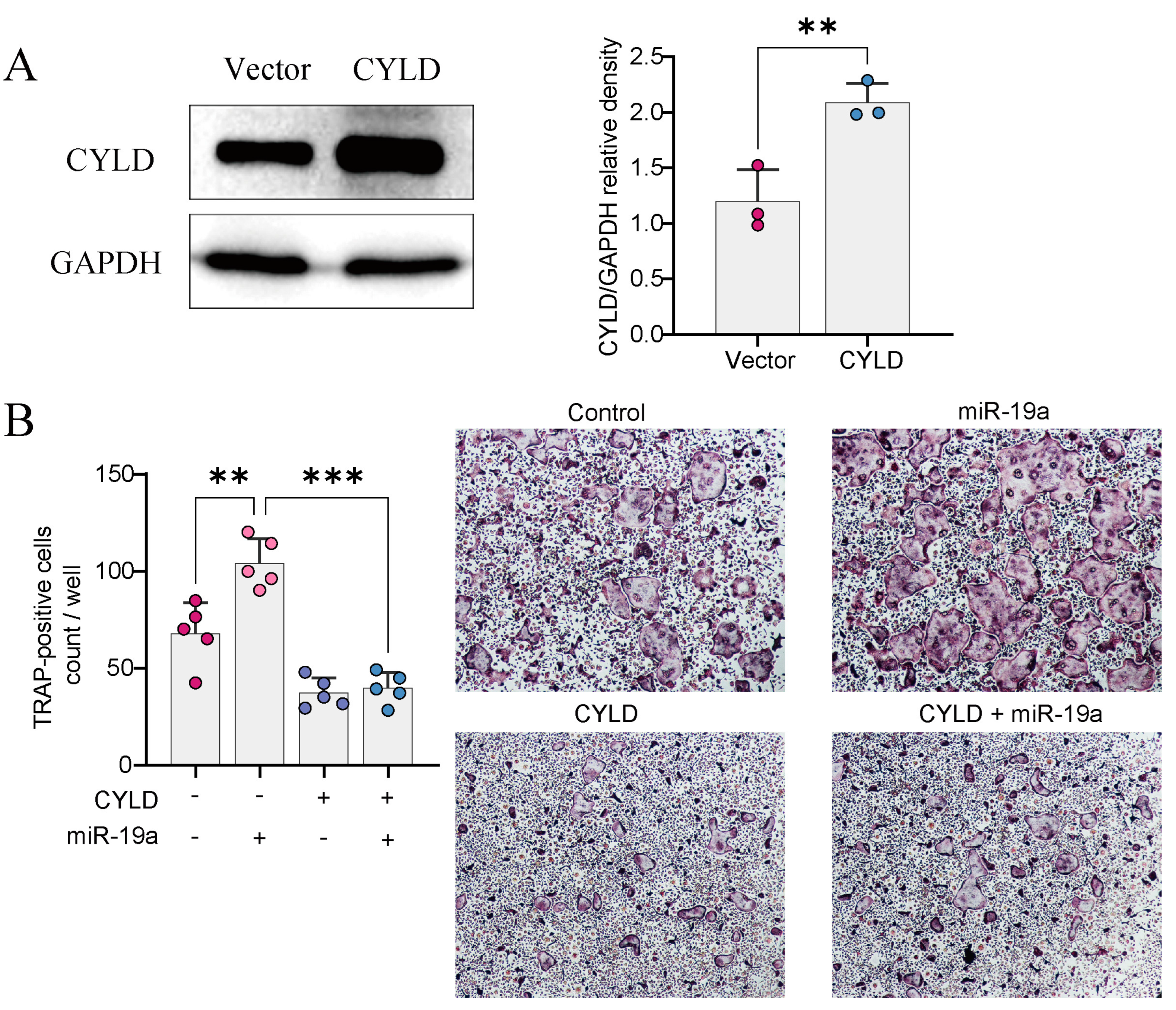
3.4. CYLD Is Required for miRNA-Mediated Regulation of Osteoclast Formation
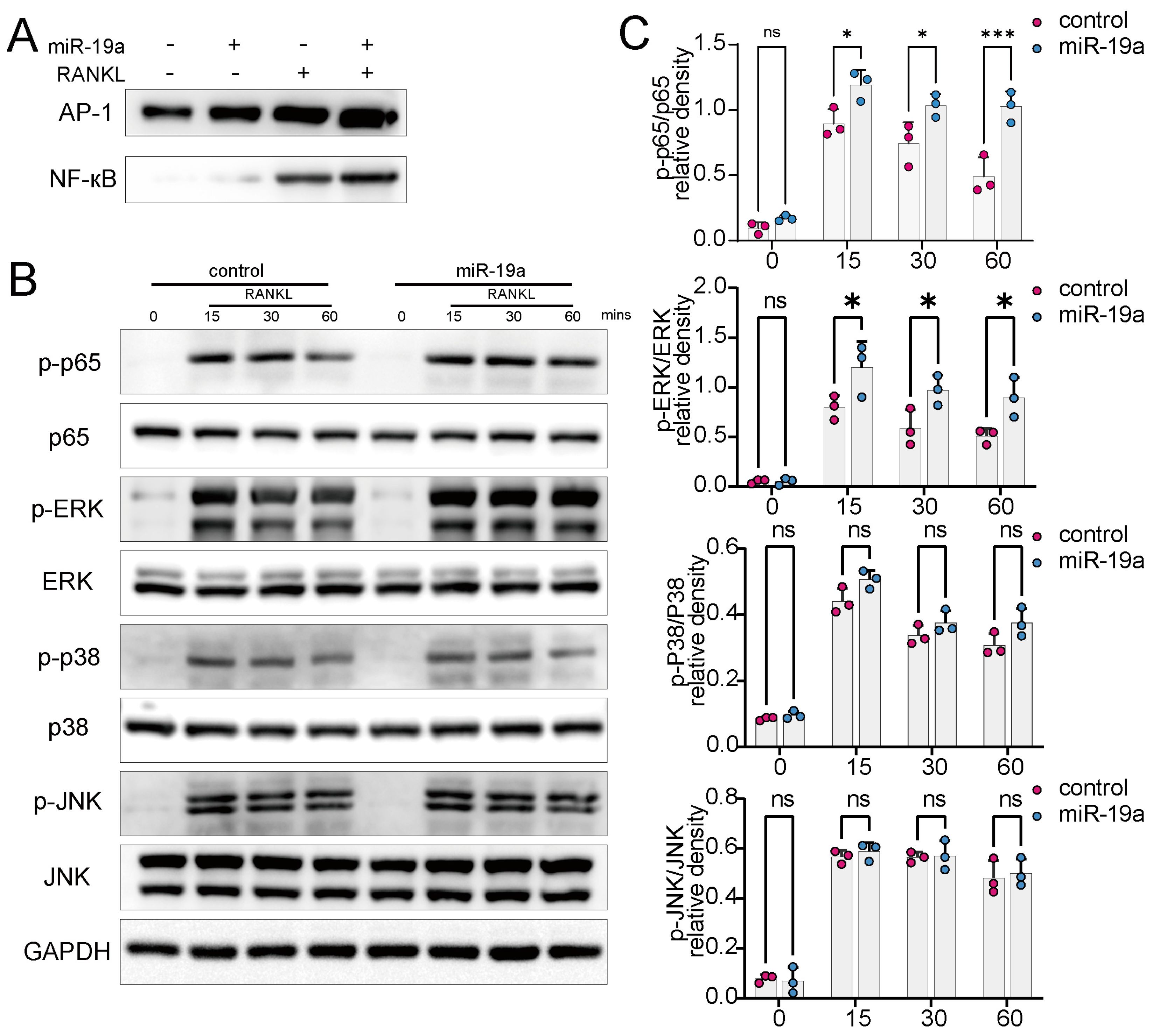
3.5. miR-19a Promotes RANKL-Induced NF-кB and p-ERK Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef]
- Ikebuchi, Y.; Aoki, S.; Honma, M.; Hayashi, M.; Sugamori, Y.; Khan, M.; Kariya, Y.; Kato, G.; Tabata, Y.; Penninger, J.M.; et al. Coupling of bone resorption and formation by RANKL reverse signalling. Nature 2018, 561, 195–200. [Google Scholar] [CrossRef]
- Rodan, G.A.; Martin, T.J. Therapeutic approaches to bone diseases. Science 2000, 289, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Liu, X.; He, Z.; Han, X.; Yan, M.; Qu, X.; Li, X.; Yu, Z. Articular Cartilage Degradation and Aberrant Subchondral Bone Remodeling in Patients with Osteoarthritis and Osteoporosis. J. Bone Miner. Res. 2020, 35, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Emami, A.J.; Toupadakis, C.A.; Telek, S.M.; Fyhrie, D.P.; Yellowley, C.E.; Christiansen, B.A. Age Dependence of Systemic Bone Loss and Recovery Following Femur Fracture in Mice. J. Bone Miner. Res. 2019, 34, 157–170. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, Z.; Wang, Z.; Jiang, Q.; Zhang, Z.; Liu, X.; Xing, B.; Li, S.; Guo, X.; Liu, Y.; et al. PKCtheta Regulates Pituitary Adenoma Bone Invasion by Activating Osteoclast in NF-kappaB/IL-1beta-Dependent Manner. Cancers 2023, 15, 1624. [Google Scholar] [CrossRef]
- Lu, L.; Wan, X.; Xu, Y.; Chen, J.; Shu, K.; Lei, T. Classifying Pituitary Adenoma Invasiveness Based on Radiological, Surgical and Histological Features: A Retrospective Assessment of 903 Cases. J. Clin. Med. 2022, 11, 2464. [Google Scholar] [CrossRef]
- Kodama, J.; Kaito, T. Osteoclast Multinucleation: Review of Current Literature. Int. J. Mol. Sci. 2020, 21, 5685. [Google Scholar] [CrossRef]
- Kobayashi, N.; Kadono, Y.; Naito, A.; Matsumoto, K.; Yamamoto, T.; Tanaka, S.; Inoue, J. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 2001, 20, 1271–1280. [Google Scholar] [CrossRef]
- Jin, W.; Chang, M.; Paul, E.M.; Babu, G.; Lee, A.J.; Reiley, W.; Wright, A.; Zhang, M.; You, J.; Sun, S.C. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J. Clin. Investig. 2008, 118, 1858–1866. [Google Scholar] [CrossRef]
- Lee, S.E.; Woo, K.M.; Kim, S.Y.; Kim, H.M.; Kwack, K.; Lee, Z.H.; Kim, H.H. The phosphatidylinositol 3-kinase, p38, and extracellular signal-regulated kinase pathways are involved in osteoclast differentiation. Bone 2002, 30, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Lee, Z.H.; Kim, H.H. Signal transduction by receptor activator of nuclear factor kappa B in osteoclasts. Biochem. Biophys. Res. Commun. 2003, 305, 211–214. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, N. Signaling Pathways in Osteoclast Differentiation. Chonnam Med. J. 2016, 52, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Asagiri, M.; Sato, K.; Usami, T.; Ochi, S.; Nishina, H.; Yoshida, H.; Morita, I.; Wagner, E.F.; Mak, T.W.; Serfling, E.; et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 2005, 202, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, F.; Nishimura, R.; Matsubara, T.; Tanaka, S.; Inoue, J.; Reddy, S.V.; Hata, K.; Yamashita, K.; Hiraga, T.; Watanabe, T.; et al. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J. Clin. Investig. 2004, 114, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Taipaleenmaki, H.; Saito, H.; Schroder, S.; Maeda, M.; Mettler, R.; Ring, M.; Rollmann, E.; Gasser, A.; Haasper, C.; Gehrke, T.; et al. Antagonizing microRNA-19a/b augments PTH anabolic action and restores bone mass in osteoporosis in mice. EMBO Mol. Med. 2022, 14, e13617. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wang, B.; Qiu, M.; Huang, Y. MiR-19b-3p accelerates bone loss after spinal cord injury by suppressing osteogenesis via regulating PTEN/Akt/mTOR signalling. J. Cell Mol. Med. 2021, 25, 990–1000. [Google Scholar] [CrossRef]
- Jiang, Q.; Lei, Z.; Wang, Z.; Wang, Q.; Zhang, Z.; Liu, X.; Xing, B.; Li, S.; Guo, X.; Liu, Y.; et al. Tumor-Associated Fibroblast-Derived Exosomal circDennd1b Promotes Pituitary Adenoma Progression by Modulating the miR-145-5p/ONECUT2 Axis and Activating the MAPK Pathway. Cancers 2023, 15, 3375. [Google Scholar] [CrossRef]
- Zhang, Z.; Schäfer, A.; Voellger, B.; Wang, J.-W.; Lei, T.; Nimsky, C.; Bartsch, J.W. MicroRNA-149 Regulates Proliferation, Migration, and Invasion of Pituitary Adenoma Cells by Targeting ADAM12 and MMP14. Curr. Med. Sci. 2022, 42, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Han, Y.C.; Betel, D.; Yao, E.; Squatrito, M.; Ogrodowski, P.; de Stanchina, E.; D’Andrea, A.; Sander, C.; Ventura, A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes. Dev. 2009, 23, 2806–2811. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, M.; Guo, Q.; Lou, J.; Wang, L. Non-small cell lung cancer cell-derived exosomal miR-17-5p promotes osteoclast differentiation by targeting PTEN. Exp. Cell Res. 2021, 408, 112834. [Google Scholar] [CrossRef]
- Wu, K.; Feng, J.; Lyu, F.; Xing, F.; Sharma, S.; Liu, Y.; Wu, S.Y.; Zhao, D.; Tyagi, A.; Deshpande, R.P.; et al. Exosomal miR-19a and IBSP cooperate to induce osteolytic bone metastasis of estrogen receptor-positive breast cancer. Nat. Commun. 2021, 12, 5196. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fang, Z.; Song, C.; Kang, H.; Guo, Q.; Dong, Y.; Zhang, Y.; Peng, R.; Guan, H.; Li, F. Schisandrin B Inhibits Osteoclastogenesis and Protects Against Ovariectomy-Induced Bone Loss. Front. Pharmacol. 2020, 11, 1175. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Tan, P.; Zhang, Z.; Wu, W.; Dong, Y.; Zhao, L.; Liu, H.; Guan, H.; Li, F. REV-ERB agonism suppresses osteoclastogenesis and prevents ovariectomy-induced bone loss partially via FABP4 upregulation. FASEB J. 2018, 32, 3215–3228. [Google Scholar] [CrossRef]
- Lei, Z.; Wang, J.; Sun, W.; Chen, X.; Jiao, W.; Zhang, H.; Lei, T.; Li, F. PKCdelta reveals a tumor promoter function by promoting cell proliferation and migration in somatotropinomas. Int. J. Clin. Exp. Pathol. 2018, 11, 208–215. [Google Scholar]
- Liu, L.; Jin, R.; Duan, J.; Yang, L.; Cai, Z.; Zhu, W.; Nie, Y.; He, J.; Xia, C.; Gong, Q.; et al. Bioactive iron oxide nanoparticles suppress osteoclastogenesis and ovariectomy-induced bone loss through regulating the TRAF6-p62-CYLD signaling complex. Acta Biomater. 2020, 103, 281–292. [Google Scholar] [CrossRef]
- Lin, F.T.; Lin, V.Y.; Lin, V.T.; Lin, W.C. TRIP6 antagonizes the recruitment of A20 and CYLD to TRAF6 to promote the LPA2 receptor-mediated TRAF6 activation. Cell Discov. 2016, 2, 15048. [Google Scholar] [CrossRef]
- Sun, K.T.; Chen, M.Y.; Tu, M.G.; Wang, I.K.; Chang, S.S.; Li, C.Y. MicroRNA-20a regulates autophagy related protein-ATG16L1 in hypoxia-induced osteoclast differentiation. Bone 2015, 73, 145–153. [Google Scholar] [CrossRef]
- Zhao, M.; Dong, J.; Liao, Y.; Lu, G.; Pan, W.; Zhou, H.; Zuo, X.; Shan, B. MicroRNA miR-18a-3p promotes osteoporosis and possibly contributes to spinal fracture by inhibiting the glutamate AMPA receptor subunit 1 gene (GRIA1). Bioengineered 2021, 13, 370–382. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Y.; Feng, S.; He, P.; Sheng, B.; Ni, J. miR-19b enhances osteogenic differentiation of mesenchymal stem cells and promotes fracture healing through the WWP1/Smurf2-mediated KLF5/beta-catenin signaling pathway. Exp. Mol. Med. 2021, 53, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Xiaoling, G.; Shuaibin, L.; Kailu, L. MicroRNA-19b-3p promotes cell proliferation and osteogenic differentiation of BMSCs by interacting with lncRNA H19. BMC Med. Genet. 2020, 21, 11. [Google Scholar] [CrossRef] [PubMed]
- Wa, Q.; Li, L.; Lin, H.; Peng, X.; Ren, D.; Huang, Y.; He, P.; Huang, S. Downregulation of miR-19a-3p promotes invasion, migration and bone metastasis via activating TGF-beta signaling in prostate cancer. Oncol. Rep. 2018, 39, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Brummelkamp, T.R.; Nijman, S.M.; Dirac, A.M.; Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003, 424, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Herlyn, M.; Yang, X. TRIM15 and CYLD regulate ERK activation via lysine-63-linked polyubiquitination. Nat. Cell Biol. 2021, 23, 978–991. [Google Scholar] [CrossRef]
- Nguyen, J.; Massoumi, R.; Alliston, T. CYLD, a mechanosensitive deubiquitinase, regulates TGFbeta signaling in load-induced bone formation. Bone 2020, 131, 115148. [Google Scholar] [CrossRef]
- Ye, H.; Liu, X.; Lv, M.; Wu, Y.; Kuang, S.; Gong, J.; Yuan, P.; Zhong, Z.; Li, Q.; Jia, H.; et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012, 40, 5201–5214. [Google Scholar] [CrossRef]
- Yu, J.; Yun, H.; Shin, B.; Kim, Y.; Park, E.S.; Choi, S.; Yu, J.; Amarasekara, D.S.; Kim, S.; Inoue, J.; et al. Interaction of Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) and Vav3 in the Receptor Activator of Nuclear Factor kappaB (RANK) Signaling Complex Enhances Osteoclastogenesis. J. Biol. Chem. 2016, 291, 20643–20660. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, Z.; Wang, Q.; Jiang, Q.; Liu, H.; Xu, L.; Kang, H.; Li, F.; Huang, Y.; Lei, T. The miR-19a/Cylindromatosis Axis Regulates Pituitary Adenoma Bone Invasion by Promoting Osteoclast Differentiation. Cancers 2024, 16, 302. https://doi.org/10.3390/cancers16020302
Lei Z, Wang Q, Jiang Q, Liu H, Xu L, Kang H, Li F, Huang Y, Lei T. The miR-19a/Cylindromatosis Axis Regulates Pituitary Adenoma Bone Invasion by Promoting Osteoclast Differentiation. Cancers. 2024; 16(2):302. https://doi.org/10.3390/cancers16020302
Chicago/Turabian StyleLei, Zhuowei, Quanji Wang, Qian Jiang, Huiyong Liu, Linpeng Xu, Honglei Kang, Feng Li, Yimin Huang, and Ting Lei. 2024. "The miR-19a/Cylindromatosis Axis Regulates Pituitary Adenoma Bone Invasion by Promoting Osteoclast Differentiation" Cancers 16, no. 2: 302. https://doi.org/10.3390/cancers16020302
APA StyleLei, Z., Wang, Q., Jiang, Q., Liu, H., Xu, L., Kang, H., Li, F., Huang, Y., & Lei, T. (2024). The miR-19a/Cylindromatosis Axis Regulates Pituitary Adenoma Bone Invasion by Promoting Osteoclast Differentiation. Cancers, 16(2), 302. https://doi.org/10.3390/cancers16020302







