Shift in Tissue-Specific Immune Niches and CD137 Expression in Tuberculoma of Pembrolizumab-Treated Nasopharyngeal Carcinoma Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patient Characteristics
2.3. Tissue Collection
2.4. Image Capturing
2.5. Quantification and Multiplexed Analysis
2.6. PBMC Cell Isolation from Human Blood
2.7. PBMC Infection and Three-Dimensional In Vitro Granuloma Formation
2.8. Flow Cytometric Analysis
2.9. Whole-Blood and Tumor Tissue Transcriptomic Analysis
2.10. Statistical Analysis
2.11. Multiplexed Tissue Staining
2.12. RNA-Sequencing and Microarray Analysis of Data from Public Databases
2.13. Data Analysis, Bioinformatics and Statistical Considerations
3. Results
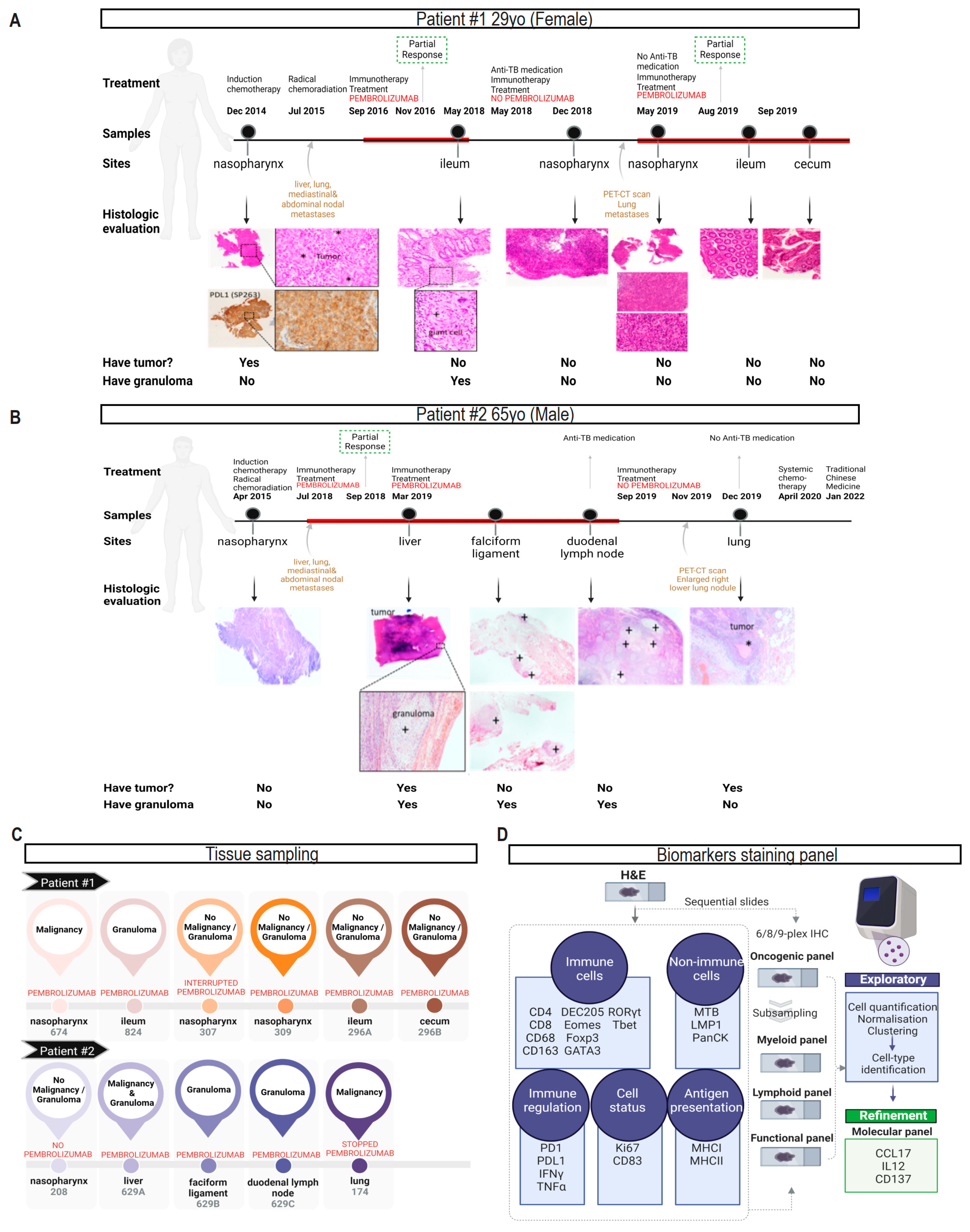
3.1. Characteristics of Two NPC Patients with Reactivation of Gastrointestinal TB after Pem-Brolizumab Treatment
3.2. Clustering Samples Based on Intrinsic Shared Biological Features of Benign, Malignant and Tuberculosis Tissue Lesions
3.3. Multi-Tissue mIHC Characterization of Immune Landscapes in Patient Tissues
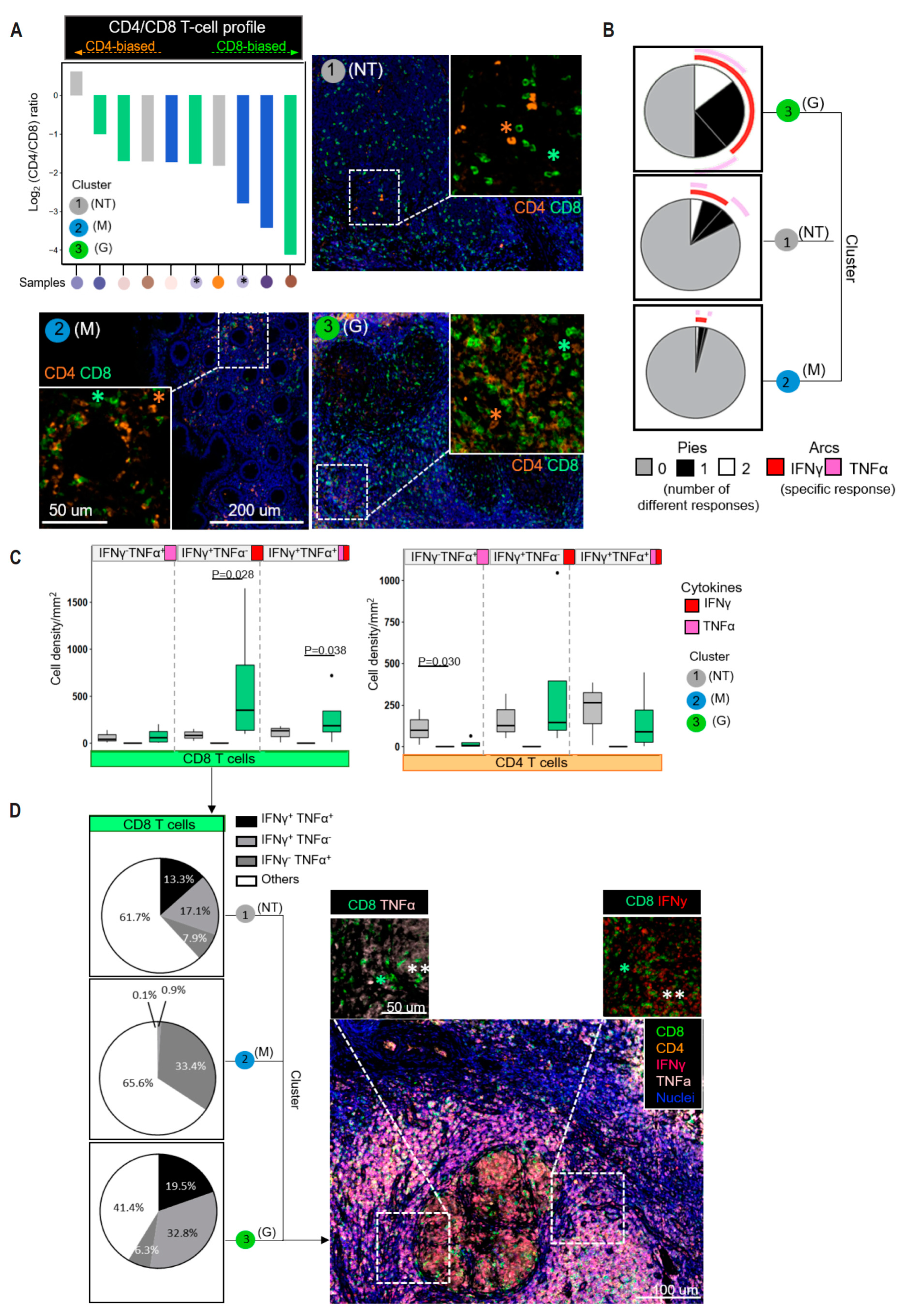
3.4. CD8+ T Cells Dominate the T-Cell Infiltrates in the Different Tissue Types
3.5. IFNγ-Producing CD8+ T Cells Significantly Accumulate in the Granulomatous Tissues
3.6. Infiltrating Immune Cells Are Phenotypically and Functionally Heterogeneous in Different Tissue Types
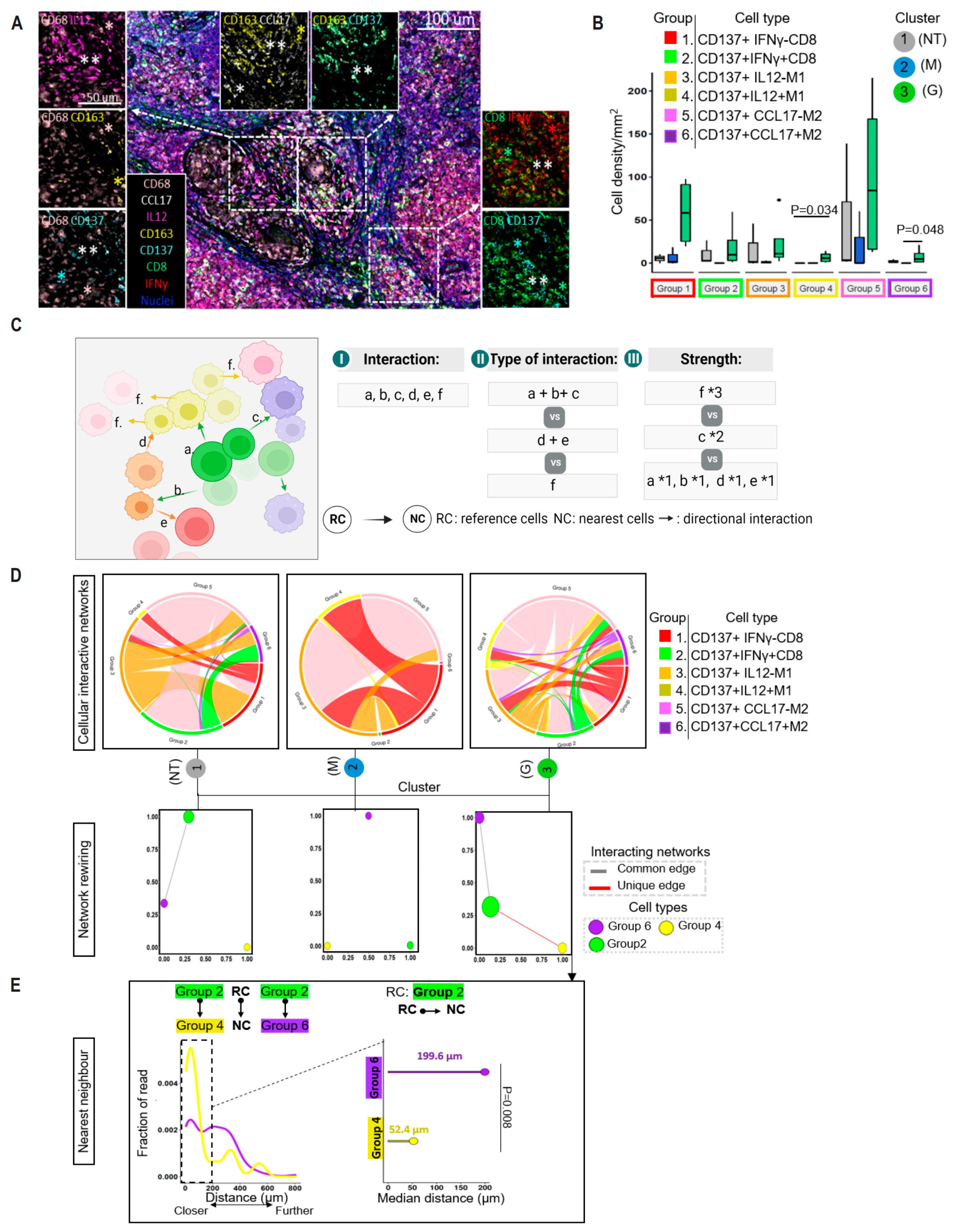
3.7. Enrichment of CD137 in Granuloma-Residing CD8+ T Cells
3.8. Spatial Proximity of CD137-Expressing Immune Cells in the Reactivation of TB Granuloma
3.9. CD137-Rich Cellular Microenviornment That Is Spatially Organized in Tumor Tissues in Response to ICI Is Absent in Ordinary TB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, A.W.; Sze, W.M.; Au, J.S.; Leung, S.F.; Leung, T.W.; Chua, D.T.; Zee, B.C.; Law, S.C.; Teo, P.M.; Tung, S.Y.; et al. Treatment results for nasopharyngeal carcinoma in the modern era: The Hong Kong experience. Int. J. Radiat. Oncol. Biol. Phys. 2005, 61, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Michot, J.M.; Bigenwald, C.; Champiat, S.; Collins, M.; Carbonnel, F.; Postel-Vinay, S.; Berdelou, A.; Varga, A.; Bahleda, R.; Hollebecque, A.; et al. Immune-related adverse events with immune checkpoint blockade: A comprehensive review. Eur. J. Cancer 2016, 54, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Sahu, G.; Burns, E.; Ensor, A.; Ensor, J.; Pingali, S.R.; Subbiah, V.; Iyer, S.P. Mycobacterial infections due to PD-1 and PD-L1 checkpoint inhibitors. ESMO Open 2020, 5, e000866. [Google Scholar] [CrossRef] [PubMed]
- Menardo, F.; Duchene, S.; Brites, D.; Gagneux, S. The molecular clock of Mycobacterium tuberculosis. PLoS Pathog. 2019, 15, e1008067. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.S.; Cheung, B.M.; Lam, K.O.; Chan, S.Y.; Lam, K.M.; Yeung, C.F.; Hung, I.F.; Kwong, D.L.; Tong, C.C.; Leung, T.W.; et al. Tuberculosis reactivation at ileum following immune checkpoint inhibition with pembrolizumab for metastatic nasopharyngeal carcinoma: A case report. BMC Infect. Dis. 2021, 21, 1148. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.L.; Sakai, S.; Kudchadkar, R.R.; Fling, S.P.; Day, T.A.; Vergara, J.A.; Ashkin, D.; Cheng, J.H.; Lundgren, L.M.; Raabe, V.N.; et al. Tuberculosis following PD-1 blockade for cancer immunotherapy. Sci. Transl. Med. 2019, 11, eaat2702. [Google Scholar] [CrossRef]
- Zaemes, J.; Kim, C. Immune checkpoint inhibitor use and tuberculosis: A systematic review of the literature. Eur. J. Cancer 2020, 132, 168–175. [Google Scholar] [CrossRef]
- Song, J.S.; Jeffery, C.C. Laryngeal Tuberculosis in a Patient on Avelumab for Metastatic Nasopharyngeal Carcinoma. J. Immunother. 2020, 43, 222–223. [Google Scholar] [CrossRef]
- Inthasot, V.; Bruyneel, M.; Muylle, I.; Ninane, V. Severe pulmonary infections complicating nivolumab treatment for lung cancer: A report of two cases. Acta Clin. Belg. 2020, 75, 308–310. [Google Scholar] [CrossRef]
- Tsai, C.C.; Chen, J.H.; Wang, Y.C.; Chang, F.Y. Re-activation of pulmonary tuberculosis during anti-programmed death-1 (PD-1) treatment. QJM 2019, 112, 41–42. [Google Scholar] [CrossRef] [PubMed]
- Takata, S.; Koh, G.; Han, Y.; Yoshida, H.; Shiroyama, T.; Takada, H.; Masuhiro, K.; Nasu, S.; Morita, S.; Tanaka, A.; et al. Paradoxical response in a patient with non-small cell lung cancer who received nivolumab followed by anti-Mycobacterium tuberculosis agents. J. Infect. Chemother. 2019, 25, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.D.; Sakai, S.; Lora, N.E.; Namasivayam, S.; Baker, P.J.; Kamenyeva, O.; Foreman, T.W.; Nelson, C.E.; Oliveira-de-Souza, D.; Vinhaes, C.L.; et al. PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci. Immunol. 2021, 6, eabf3861. [Google Scholar] [CrossRef] [PubMed]
- Gideon, H.P.; Phuah, J.; Junecko, B.A.; Mattila, J.T. Neutrophils express pro- and anti-inflammatory cytokines in granulomas from Mycobacterium tuberculosis-infected cynomolgus macaques. Mucosal Immunol. 2019, 12, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- McCaffrey, E.F.; Donato, M.; Keren, L.; Chen, Z.; Delmastro, A.; Fitzpatrick, M.B.; Gupta, S.; Greenwald, N.F.; Baranski, A.; Graf, W.; et al. The immunoregulatory landscape of human tuberculosis granulomas. Nat. Immunol. 2022, 23, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Hughes, T.K.; Teles, R.M.B.; Andrade, P.R.; de Andrade Silva, B.J.; Plazyo, O.; Tsoi, L.C.; Do, T.; Wadsworth, M.H., 2nd; Oulee, A.; et al. The cellular architecture of the antimicrobial response network in human leprosy granulomas. Nat. Immunol. 2021, 22, 839–850. [Google Scholar] [CrossRef]
- Jimenez-Sanchez, A.; Memon, D.; Pourpe, S.; Veeraraghavan, H.; Li, Y.; Vargas, H.A.; Gill, M.B.; Park, K.J.; Zivanovic, O.; Konner, J.; et al. Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell 2017, 170, 927–938.e920. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, J.; Hong, S.; Zhan, J.; Chen, N.; Qin, T.; Tang, Y.; Zhang, Y.; Kang, S.; Zhou, T.; et al. EBV-driven LMP1 and IFN-gamma up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget 2014, 5, 12189–12202. [Google Scholar] [CrossRef]
- Green, A.M.; Difazio, R.; Flynn, J.L. IFN-gamma from CD4 T cells is essential for host survival and enhances CD8 T cell function during Mycobacterium tuberculosis infection. J. Immunol. 2013, 190, 270–277. [Google Scholar] [CrossRef]
- Winje, B.A.; White, R.; Syre, H.; Skutlaberg, D.H.; Oftung, F.; Mengshoel, A.T.; Blix, H.S.; Brantsaeter, A.B.; Holter, E.K.; Handal, N.; et al. Stratification by interferon-gamma release assay level predicts risk of incident TB. Thorax 2018, 73, 652–661. [Google Scholar] [CrossRef]
- Barber, D.L.; Mayer-Barber, K.D.; Feng, C.G.; Sharpe, A.H.; Sher, A. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J. Immunol. 2011, 186, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Tousif, S.; Singh, Y.; Prasad, D.V.; Sharma, P.; Van Kaer, L.; Das, G. T cells from Programmed Death-1 deficient mice respond poorly to Mycobacterium tuberculosis infection. PLoS ONE 2011, 6, e19864. [Google Scholar] [CrossRef] [PubMed]
- Clay, H.; Davis, J.M.; Beery, D.; Huttenlocher, A.; Lyons, S.E.; Ramakrishnan, L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe 2007, 2, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Putra, I.; Astuti, P.A.S.; Suarjana, I.K.; Mulyawan, K.H.; Duana, I.M.K.; Kurniasari, N.M.D.; Putra, I. Factors Associated with Participation in Pulmonary Tuberculosis Screening Using Chest X-Ray among Diabetes Mellitus Type II Patients in Denpasar, Bali, Indonesia. Tuberc. Res. Treat. 2018, 2018, 9285195. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.M.; Jenner, R.G. Transcription factor interplay in T helper cell differentiation. Brief. Funct. Genom. 2013, 12, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yu, D.; Sarnaik, A.A.; Yu, B.; Hall, M.; Morelli, D.; Zhang, Y.; Zhao, X.; Weber, J.S. Biomarkers on melanoma patient T cells associated with ipilimumab treatment. J. Transl. Med. 2012, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Postow, M.A.; Orlowski, R.J.; Mick, R.; Bengsch, B.; Manne, S.; Xu, W.; Harmon, S.; Giles, J.R.; Wenz, B.; et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017, 545, 60–65. [Google Scholar] [CrossRef] [PubMed]
- O’Garra, A.; Redford, P.S.; McNab, F.W.; Bloom, C.I.; Wilkinson, R.J.; Berry, M.P. The immune response in tuberculosis. Annu. Rev. Immunol. 2013, 31, 475–527. [Google Scholar] [CrossRef]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef]
- Henry, C.J.; Ornelles, D.A.; Mitchell, L.M.; Brzoza-Lewis, K.L.; Hiltbold, E.M. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J. Immunol. 2008, 181, 8576–8584. [Google Scholar] [CrossRef]
- Jakubzick, C.; Wen, H.; Matsukawa, A.; Keller, M.; Kunkel, S.L.; Hogaboam, C.M. Role of CCR4 ligands, CCL17 and CCL22, during Schistosoma mansoni egg-induced pulmonary granuloma formation in mice. Am. J. Pathol. 2004, 165, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Tezera, L.B.; Bielecka, M.K.; Ogongo, P.; Walker, N.F.; Ellis, M.; Garay-Baquero, D.J.; Thomas, K.; Reichmann, M.T.; Johnston, D.A.; Wilkinson, K.A.; et al. Anti-PD-1 immunotherapy leads to tuberculosis reactivation via dysregulation of TNF-alpha. eLife 2020, 9, e52668. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yan, Z.; Hao, J.; Yang, B.; Wang, J.; Yi, L.; Wang, X.; Li, S.; Zhang, H.; Zhang, S. CD137 ligand feedback upregulates PD-L1 expression on lung cancer via T cell production of IFN-gamma. Thorac. Cancer 2019, 10, 2225–2235. [Google Scholar] [CrossRef]
- Kim, S.H.; Singh, R.; Han, C.; Cho, E.; Kim, Y.I.; Lee, D.G.; Kim, Y.H.; Kim, S.S.; Shin, D.H.; You, H.J.; et al. Chronic activation of 4-1BB signaling induces granuloma development in tumor-draining lymph nodes that is detrimental to subsequent CD8(+) T cell responses. Cell. Mol. Immunol. 2021, 18, 1956–1968. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.; Rice, S.; Jiang, L.; Liu, X.; Gragnoli, C.; Belani, C.P.; Wu, R. A rewiring model of intratumoral interaction networks. Comput. Struct. Biotechnol. J. 2020, 18, 45–51. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Day, C.L.; Abrahams, D.A.; Lerumo, L.; Janse van Rensburg, E.; Stone, L.; O’Rie, T.; Pienaar, B.; de Kock, M.; Kaplan, G.; Mahomed, H.; et al. Functional capacity of Mycobacterium tuberculosis-specific T cell responses in humans is associated with mycobacterial load. J. Immunol. 2011, 187, 2222–2232. [Google Scholar] [CrossRef]
- Harari, A.; Rozot, V.; Bellutti Enders, F.; Perreau, M.; Stalder, J.M.; Nicod, L.P.; Cavassini, M.; Calandra, T.; Blanchet, C.L.; Jaton, K.; et al. Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat. Med. 2011, 17, 372–376. [Google Scholar] [CrossRef]
- Marin, N.D.; Paris, S.C.; Rojas, M.; Garcia, L.F. Functional profile of CD4+ and CD8+ T cells in latently infected individuals and patients with active TB. Tuberculosis 2013, 93, 155–166. [Google Scholar] [CrossRef]
- Caccamo, N.; Guggino, G.; Joosten, S.A.; Gelsomino, G.; Di Carlo, P.; Titone, L.; Galati, D.; Bocchino, M.; Matarese, A.; Salerno, A.; et al. Multifunctional CD4(+) T cells correlate with active Mycobacterium tuberculosis infection. Eur. J. Immunol. 2010, 40, 2211–2220. [Google Scholar] [CrossRef]
- Sutherland, J.S.; Adetifa, I.M.; Hill, P.C.; Adegbola, R.A.; Ota, M.O. Pattern and diversity of cytokine production differentiates between Mycobacterium tuberculosis infection and disease. Eur. J. Immunol. 2009, 39, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Lazar-Molnar, E.; Chen, B.; Sweeney, K.A.; Wang, E.J.; Liu, W.; Lin, J.; Porcelli, S.A.; Almo, S.C.; Nathenson, S.G.; Jacobs, W.R., Jr. Programmed death-1 (PD-1)-deficient mice are extraordinarily sensitive to tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 13402–13407. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Song, D.G.; Poussin, M.; Yamamoto, T.; Best, A.; Li, C.; Coukos, G.; Powell, D.J., Jr. CD137 accurately identifies and enriches for naturally occurring tumor-reactive T cells in tumor. Clin. Cancer Res. 2014, 20, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Zizzari, I.G.; Di Filippo, A.; Botticelli, A.; Strigari, L.; Pernazza, A.; Rullo, E.; Pignataro, M.G.; Ugolini, A.; Scirocchi, F.; Di Pietro, F.R.; et al. Circulating CD137+ T Cells Correlate with Improved Response to Anti-PD1 Immunotherapy in Patients with Cancer. Clin. Cancer Res. 2022, 28, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Ugolini, A.; Zizzari, I.G.; Ceccarelli, F.; Botticelli, A.; Colasanti, T.; Strigari, L.; Rughetti, A.; Rahimi, H.; Conti, F.; Valesini, G.; et al. IgM-Rheumatoid factor confers primary resistance to anti-PD-1 immunotherapies in NSCLC patients by reducing CD137(+)T-cells. EBioMedicine 2020, 62, 103098. [Google Scholar] [CrossRef] [PubMed]
- Zizzari, I.G.; Napoletano, C.; Di Filippo, A.; Botticelli, A.; Gelibter, A.; Calabro, F.; Rossi, E.; Schinzari, G.; Urbano, F.; Pomati, G.; et al. Exploratory Pilot Study of Circulating Biomarkers in Metastatic Renal Cell Carcinoma. Cancers 2020, 12, 2620. [Google Scholar] [CrossRef] [PubMed]
- Fernandez Do Porto, D.A.; Jurado, J.O.; Pasquinelli, V.; Alvarez, I.B.; Aspera, R.H.; Musella, R.M.; Garcia, V.E. CD137 differentially regulates innate and adaptive immunity against Mycobacterium tuberculosis. Immunol. Cell Biol. 2012, 90, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.H.W.; Wong, N.S.; Leung, C.C.; Chan, C.K.; Lau, A.K.H.; Tian, L.; Lee, S.S. Seasonality of tuberculosis in intermediate endemicity setting dominated by reactivation diseases in Hong Kong. Sci. Rep. 2021, 11, 20259. [Google Scholar] [CrossRef]
- Olmo-Fontanez, A.M.; Turner, J. Tuberculosis in an Aging World. Pathogens 2022, 11, 1101. [Google Scholar] [CrossRef]
- Lopez-Cuevas, P.; Cross, S.J.; Martin, P. Modulating the Inflammatory Response to Wounds and Cancer Through Infection. Front. Cell Dev. Biol. 2021, 9, 676193. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kam, N.W.; Lo, A.W.I.; Hung, D.T.Y.; Ko, H.; Wu, K.C.; Kwong, D.L.W.; Lam, K.O.; Leung, T.W.; Che, C.M.; Lee, V.H.F. Shift in Tissue-Specific Immune Niches and CD137 Expression in Tuberculoma of Pembrolizumab-Treated Nasopharyngeal Carcinoma Patients. Cancers 2024, 16, 268. https://doi.org/10.3390/cancers16020268
Kam NW, Lo AWI, Hung DTY, Ko H, Wu KC, Kwong DLW, Lam KO, Leung TW, Che CM, Lee VHF. Shift in Tissue-Specific Immune Niches and CD137 Expression in Tuberculoma of Pembrolizumab-Treated Nasopharyngeal Carcinoma Patients. Cancers. 2024; 16(2):268. https://doi.org/10.3390/cancers16020268
Chicago/Turabian StyleKam, Ngar Woon, Anthony Wing Ip Lo, Desmond Tae Yang Hung, Ho Ko, Ka Chun Wu, Dora Lai Wan Kwong, Ka On Lam, To Wai Leung, Chi Ming Che, and Victor Ho Fun Lee. 2024. "Shift in Tissue-Specific Immune Niches and CD137 Expression in Tuberculoma of Pembrolizumab-Treated Nasopharyngeal Carcinoma Patients" Cancers 16, no. 2: 268. https://doi.org/10.3390/cancers16020268
APA StyleKam, N. W., Lo, A. W. I., Hung, D. T. Y., Ko, H., Wu, K. C., Kwong, D. L. W., Lam, K. O., Leung, T. W., Che, C. M., & Lee, V. H. F. (2024). Shift in Tissue-Specific Immune Niches and CD137 Expression in Tuberculoma of Pembrolizumab-Treated Nasopharyngeal Carcinoma Patients. Cancers, 16(2), 268. https://doi.org/10.3390/cancers16020268





