Testing Adaptive Therapy Protocols Using Gemcitabine and Capecitabine in a Preclinical Model of Endocrine-Resistant Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. In Vitro Experiments
Cell Culture
Preparing Endocrine-Resistant Cell Lines
Drug Dose–Response Curve Analysis of Resistant Cell Lines
2.3. In Vivo Experiment
2.3.1. Xenograft Model of Human Breast Cancer
2.3.2. Tumor Burden Measurement
2.3.3. Starting Therapy and Drug Dosing
Standard Therapy Protocol
Dose Modulation Adaptive Therapy Protocol
Intermittent Adaptive Therapy Protocol
Ping-Pong Dose Modulation Adaptive Therapy Protocol
Ping-Pong Intermittent Adaptive Therapy Protocol
Tandem Dose Modulation Adaptive Therapy Protocol
Tandem Intermittent Adaptive Therapy Protocol
Endpoints
2.4. Ex Vivo Experiments
2.4.1. Derivation of Cancer Cell Lines from Mice
2.4.2. Histological Analysis and Immunohistochemistry
2.5. Statistical Methods
2.6. Computational Modeling
3. Results
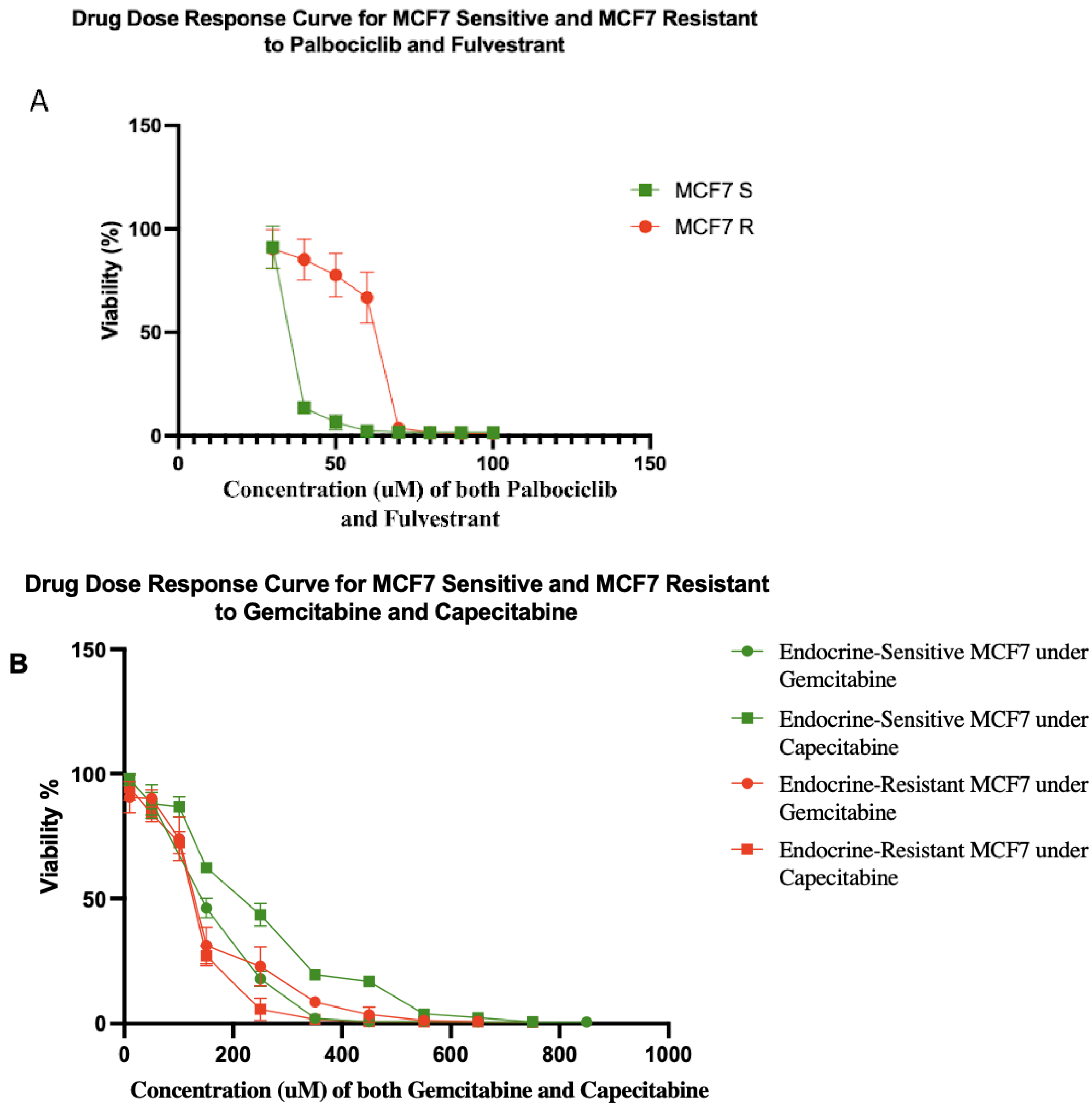
3.1. Endocrine-Resistant MCF7 Cell Line
3.2. Tumor Growth Control after Therapy Cessation
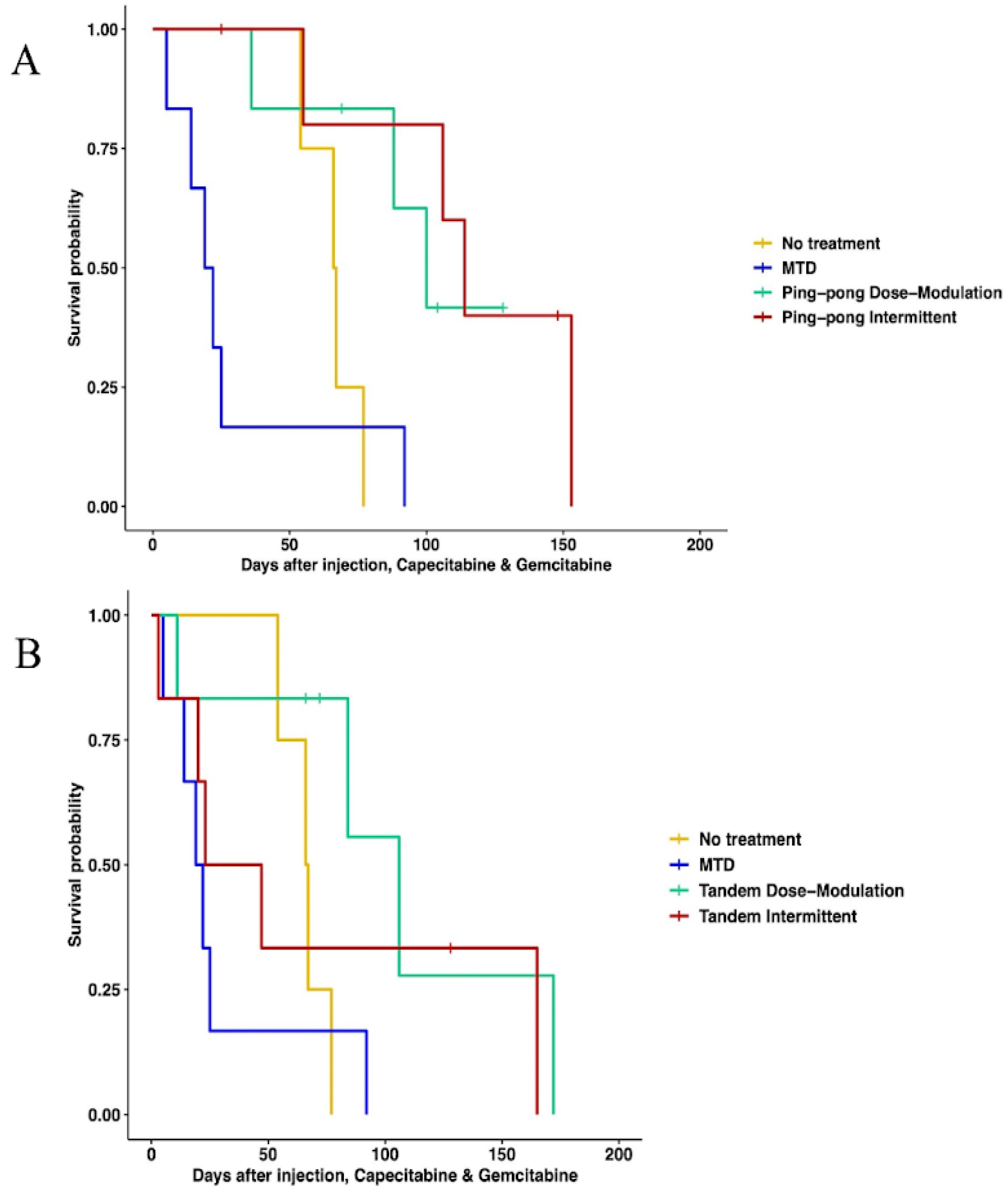
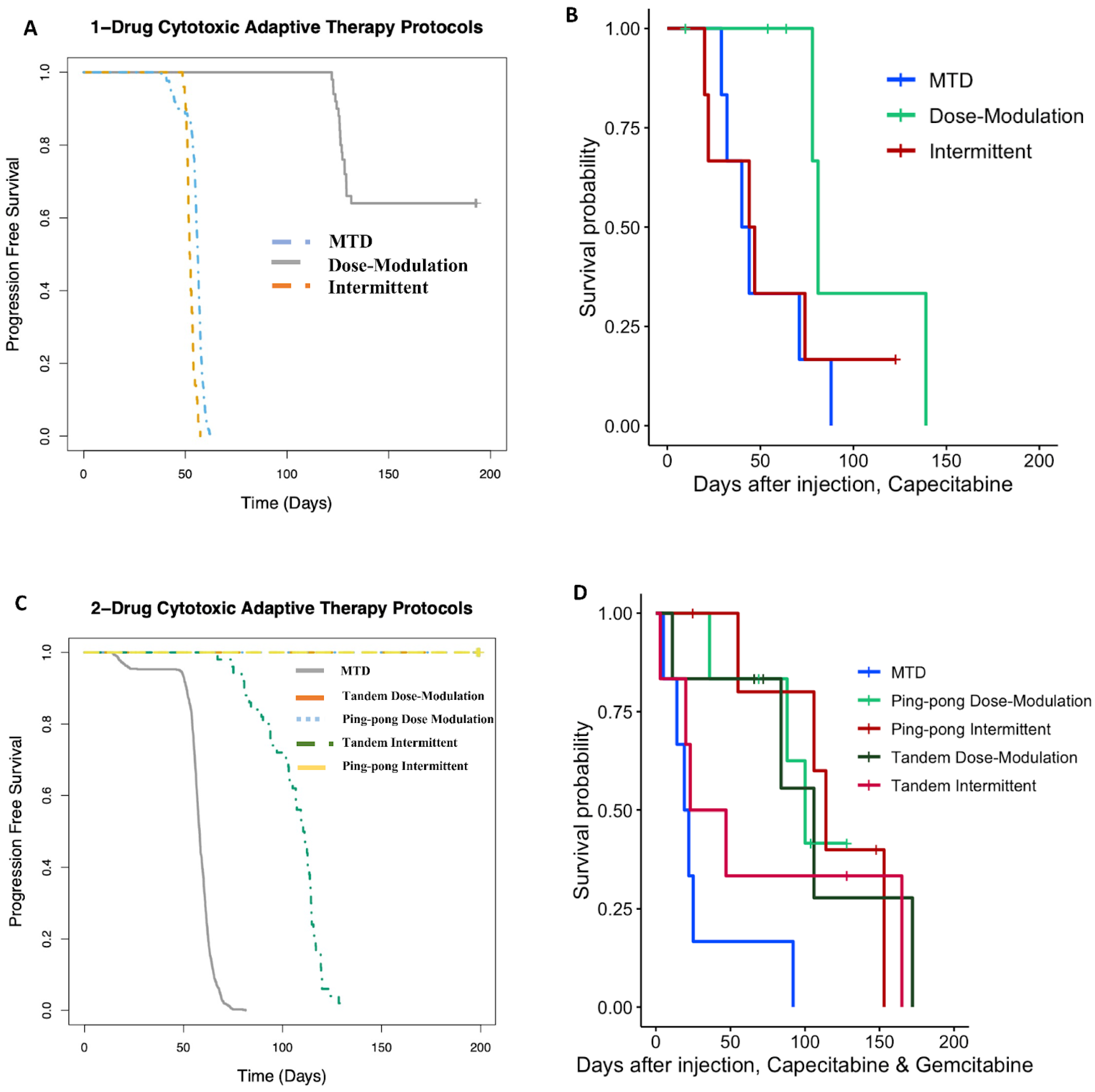
3.3. Prolonged Survival Benefit of Adaptive Therapy
3.3.1. Single-Drug Therapy
3.3.2. Multidrug Therapy
3.4. A Strong Correlation between the Percentage of Maximum Tolerated Drug Dose Used with Survival Time
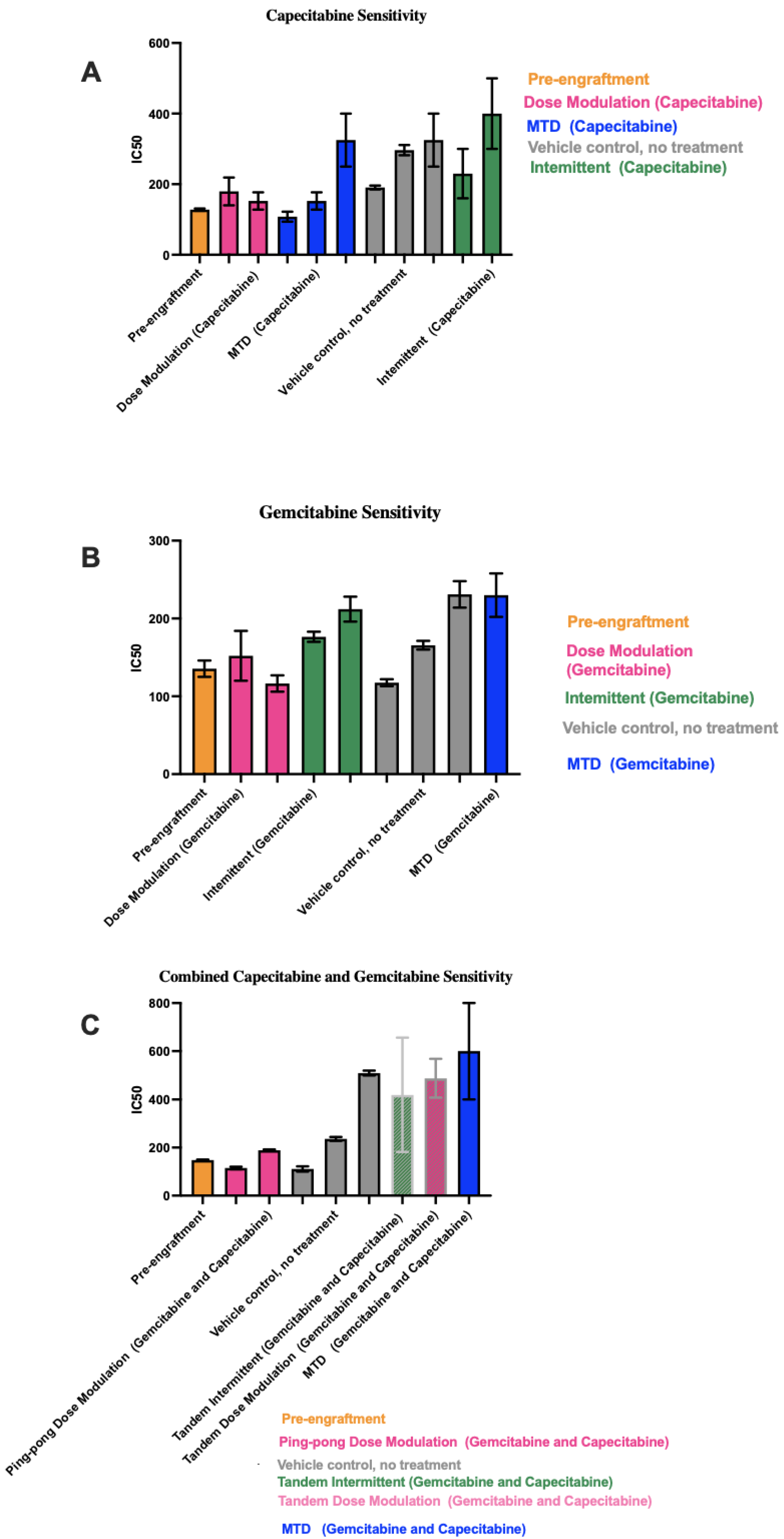
3.5. Different Chemosensitivity in Cell Lines Retrieved from Different Treatment Groups
3.6. Correlation between IC50 Values with Both Tumor Burden and Drug Dose
3.7. Immunohistochemistry and Histological Analysis
3.8. Computational Simulations Match the Rank Orders for the Different Adaptive Therapy Protocols
4. Discussion
4.1. Caveats
4.2. Future Challenges and Opportunities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Enriquez-Navas, P.M.; Kam, Y.; Das, T.; Hassan, S.; Silva, A.; Foroutan, P.; Ruiz, E.; Martinez, G.; Minton, S.; Gillies, R.J.; et al. Exploiting evolutionary principles to prolong tumor control in preclinical models of breast cancer. Sci. Transl. Med. 2016, 8, 327ra24. [Google Scholar] [CrossRef] [PubMed]
- Barzman, M.; Bàrberi, P.; Birch, A.N.E.; Boonekamp, P.; Dachbrodt-Saaydeh, S.; Graf, B.; Hommel, B.; Jensen, J.E.; Kiss, J.; Kudsk, P.; et al. Eight principles of integrated pest management. Agron. Sustain. Dev. 2015, 35, 1199–1215. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Silva, A.S.; Gillies, R.J.; Frieden, B.R. Adaptive therapy. Cancer Res. 2009, 69, 4894–4903. [Google Scholar] [CrossRef]
- Seyedi, S.; Harris, V.K.; Kapsetaki, S.E.; Saha, D.; Compton, Z.; Yousefi, R.; May, A.; Fakir, E.; Boddy, A.M.; Gerlinger, M.; et al. Resistance Management for Cancer: Lessons from Farmers. arXiv 2023. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Brown, J.S. The Evolution and Ecology of Resistance in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2020, 10, a040972. [Google Scholar] [CrossRef]
- Thomas, E.A.; Zaman, A.; Sloggett, K.J.; Steinke, S.; Grau, L.; Catenacci, V.A.; Cornier, M.-A.; Rynders, C.A. Early time-restricted eating compared with daily caloric restriction: A randomized trial in adults with obesity. Obesity 2022, 30, 1027–1038. [Google Scholar] [CrossRef]
- Zhang, J.; Cunningham, J.J.; Brown, J.S.; Gatenby, R.A. Abstract 5562: Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer: A pilot multidisciplinary study. Cancer Res. 2017, 77, 5562. [Google Scholar] [CrossRef]
- Zhang, J.; Gallaher, J.; Cunningham, J.J.; Choi, J.W.; Ionescu, F.; Chatwal, M.S.; Jain, R.; Kim, Y.; Wang, L.; Brown, J.S.; et al. A Phase 1b Adaptive Androgen Deprivation Therapy Trial in Metastatic Castration Sensitive Prostate Cancer. Cancers 2022, 14, 5225. [Google Scholar] [CrossRef]
- Gallaher, J.; Strobl, M.; West, J.; Zhang, J.; Gatenby, R.; Robertson-Tessi, M.; Anderson, A.R. The sum and the parts: Dynamics of multiple and individual metastases during adaptive therapy. bioRxiv 2022. [Google Scholar] [CrossRef]
- Hockings, H.; Lakatos, E.; Huang, W.; Mossner, M.; Khan, M.A.; Metcalf, S.; Nicolini, F.; Smith, K.; Baker, A.-M.; Graham, T.A.; et al. Adaptive therapy achieves long-term control of chemotherapy resistance in high grade ovarian cancer. bioRxiv 2023. [Google Scholar] [CrossRef]
- Gluzman, M.; Scott, J.G.; Vladimirsky, A. Optimizing adaptive cancer therapy: Dynamic programming and evolutionary game theory. Proc. Biol. Sci. 2020, 287, 20192454. [Google Scholar] [CrossRef] [PubMed]
- Strobl, M.A.R.; West, J.; Viossat, Y.; Damaghi, M.; Robertson-Tessi, M.; Brown, J.S.; Gatenby, R.A.; Maini, P.K.; Anderson, A.R. Turnover Modulates the Need for a Cost of Resistance in Adaptive Therapy. Cancer Res. 2021, 81, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Brown, J.S.; Eroglu, Z.; Anderson, A.R.A. Adaptive Therapy for Metastatic Melanoma: Predictions from Patient Calibrated Mathematical Models. Cancers 2021, 13, 823. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.S.; Cisneros, L.H.; Anderson, A.R.A.; Maley, C.C. In Silico Investigations of Multi-Drug Adaptive Therapy Protocols. Cancers 2022, 14, 2699. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Cavalcante, L.; Monteiro, G. Gemcitabine: Metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur. J. Pharmacol. 2014, 741, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Walko, C.M.; Lindley, C. Capecitabine: A review. Clin. Ther. 2005, 27, 23–44. [Google Scholar] [CrossRef]
- Kurata, N.; Fujita, H.; Ohuchida, K.; Mizumoto, K.; Mahawithitwong, P.; Sakai, H.; Manabe, T.; Ohtsuka, T.; Tanaka, M. Predicting the chemosensitivity of pancreatic cancer cells by quantifying the expression levels of genes associated with the metabolism of gemcitabine and 5-fluorouracil. Int. J. Oncol. 2011, 39, 473–482. [Google Scholar]
- Bland, K.I.; Copeland, E.M.; Suzanne Klimberg, V.; Gradishar, W.J. The Breast-E-Book: Comprehensive Management of Benign and Malignant Diseases; Elsevier Health Sciences: Philadelphia, PA, USA, 2023. [Google Scholar]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Cambridge, MA, USA, 2016. [Google Scholar]
- Mullen, P. The Use of Matrigel to Facilitate the Establishment of Human Cancer Cell Lines as Xenografts. In Cancer Cell Culture: Methods and Protocols; Langdon, S.P., Ed.; Humana Press: Totowa, NJ, USA, 2004; pp. 287–292. [Google Scholar]
- Lippman, M.E.; Osborne, C.K.; Knazek, R.; Young, N. In vitro model systems for the study of hormone-dependent human breast cancer. N. Engl. J. Med. 1977, 296, 154–159. [Google Scholar]
- Dall, G.; Vieusseux, J.; Unsworth, A.; Anderson, R.; Britt, K. Low Dose, Low Cost Estradiol Pellets Can Support MCF-7 Tumour Growth in Nude Mice without Bladder Symptoms. J. Cancer 2015, 6, 1331–1336. [Google Scholar] [CrossRef]
- Shipley, L.A.; Brown, T.J.; Cornpropst, J.D.; Hamilton, M.; Daniels, W.D.; Culp, H.W. Metabolism and disposition of gemcitabine, and oncolytic deoxycytidine analog, in mice, rats, and dogs. Drug Metab. Dispos. 1992, 20, 849–855. [Google Scholar]
- Sahin, A.; Zhang, H. Invasive Breast Carcinoma. In Pathobiology of Human Disease; Elsevier: Amsterdam, The Netherlands, 2014; pp. 934–951. [Google Scholar] [CrossRef]
- Duan, W.R.; Garner, D.S.; Williams, S.D.; Funckes-Shippy, C.L.; Spath, I.S.; Blomme, E.A. Comparison of immunohistochemistry for activated caspase-3 and cleaved cytokeratin 18 with the TUNEL method for quantification of apoptosis in histological sections of PC-3 subcutaneous xenografts. J. Pathol. 2003, 199, 221–228. [Google Scholar] [CrossRef]
- Silva, M.N.; Leite, J.S.; Mello, M.F.; Silva, K.V.; Corgozinho, K.B.; de Souza, H.J.; Cunha, S.C.S.; Ferreira, A.M.R. Histologic evaluation of Ki-67 and cleaved caspase-3 expression in feline mammary carcinoma. J. Feline Med. Surg. 2017, 19, 440–445. [Google Scholar] [CrossRef]
- Bravo, R.R.; Baratchart, E.; West, J.; Schenck, R.O.; Miller, A.K.; Gallaher, J.; Gatenbee, C.D.; Basanta, D.; Robertson-Tessi, M.; Anderson, A.R.A. Hybrid Automata Library: A flexible platform for hybrid modeling with real-time visualization. PLoS Comput. Biol. 2020, 16, e1007635. [Google Scholar] [CrossRef]
- McDermott, M.; Eustace, A.J.; Busschots, S.; Breen, L.; Crown, J.; Clynes, M.; O’Donovan, N.; Stordal, B. In vitro Development of Chemotherapy and Targeted Therapy Drug-Resistant Cancer Cell Lines: A Practical Guide with Case Studies. Front. Oncol. 2014, 4, 40. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Moiseyenko, V.M.; Tjulandin, S.; Majlis, A.; Constenla, M.; Boni, C.; Adriano, R.; Miguel, F.; Yee, C.; Edouard, V.; et al. Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: A report of the V325 Study Group. J. Clin. Oncol. 2006, 24, 4991–4997. [Google Scholar] [CrossRef]
- Wagner, A.D.; Grothe, W.; Haerting, J.; Kleber, G.; Grothey, A.; Fleig, W.E. Chemotherapy in advanced gastric cancer: A systematic review and meta-analysis based on aggregate data. J. Clin. Oncol. 2006, 24, 2903–2909. [Google Scholar] [CrossRef]
- Carrick, S.; Parker, S.; Thornton, C.E.; Ghersi, D.; Simes, J.; Wilcken, N. Single agent versus combination chemotherapy for metastatic breast cancer. Cochrane Database Syst. Rev. 2009, 2009, CD003372. [Google Scholar] [PubMed]
- Lanfear, R.; Kokko, H.; Eyre-Walker, A. Population size and the rate of evolution. Trends Ecol. Evol. 2014, 29, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.K.; Anderson, A.R.A.; Cisneros, L.; Maley, C.C. In Silico Investigations of Adaptive Therapy Using a Single Cytotoxic or a Single Cytostatic Drug. bioRxiv 2023. [Google Scholar] [CrossRef]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Delitala, M.; Ferraro, M. Is the Allee effect relevant in cancer evolution and therapy? AIMS Math. 2020, 5, 7648–7659. [Google Scholar] [CrossRef]
- Azimzade, Y.; Saberi, A.A.; Gatenby, R.A. Superlinear growth reveals the Allee effect in tumors. Phys. Rev. E 2021, 103, 042405. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Zhang, J.; Brown, J.S. First Strike–Second Strike Strategies in Metastatic Cancer: Lessons from the Evolutionary Dynamics of Extinction. Cancer Res. 2019, 79, 3174–3177. [Google Scholar] [CrossRef]
- Frequently Asked NSGTM Questions. In The Jackson Laboratory. Available online: https://www.jax.org/jax-mice-and-services/find-and-order-jax-mice/nsg-portfolio/frequently-asked-nsg-questions (accessed on 5 September 2023).
- Aktipis, C.A.; Boddy, A.M.; Gatenby, R.A.; Brown, J.S.; Maley, C.C. Life history trade-offs in cancer evolution. Nat. Rev. Cancer 2013, 13, 883–892. [Google Scholar] [CrossRef]
- Jeschke, J.M.; Kokko, H. The roles of body size and phylogeny in fast and slow life histories. Evol. Ecol. 2009, 23, 867–878. [Google Scholar] [CrossRef]
- Del Monte, U. Does the cell number 109 still really fit one gram of tumor tissue? Cell Cycle 2009, 8, 505–506. [Google Scholar] [CrossRef]
- Nishijima, T.F.; Suzuki, M.; Muss, H.B. A comparison of toxicity profiles between the lower and standard dose capecitabine in breast cancer: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2016, 156, 227–236. [Google Scholar] [CrossRef]
- de Gregorio, A.; Häberle, L.; Fasching, P.A.; Müller, V.; Schrader, I.; Lorenz, R.; Forstbauer, H.; Friedl, T.W.P.; Bauer, E.; de Gregorio, N.; et al. Gemcitabine as adjuvant chemotherapy in patients with high-risk early breast cancer-results from the randomized phase III SUCCESS—A trial. Breast Cancer Res. 2020, 22, 111. [Google Scholar] [CrossRef]





| Treatment and Control Groups |
|---|
|
|
|
|
|
|
|
|
|
|
|
|
| A | |
|---|---|
| Parameter | Value |
| Cell division rate: sensitive | 0.06 per hour |
| Cell division rate: resistant | 0.02 per hour |
| Background death rate | 0.01 per hour |
| Replacement probability | 1.0 |
| Delta Tumor | 10% |
| Delta Dose | 50% |
| Probability of death due to drug potency (Ψ) | 0.04 per unit drug concentration |
| Maximum tolerated dose (MTD) | 2.5 units |
| Minimum drug dose | 0.5 units |
| Drug on time | 1 h |
| Frequency of drug application | Once every 24 h |
| Check tumor burden | Every 3 days |
| Drug decay | 10% per hour |
| Drug diffusion rate | 2.0 |
| Tumor size triggering treatment | Tumor burden is 50% or more of the carrying capacity |
| Mutation rate | 1 × 10−3 per cell division |
| Measurement noise standard deviation (SD) | 5 cells |
| Total grid size | 100 by 100 |
| Duration of simulation | 5000 h |
| Stop dosing/initiate treatment vacation when (DM protocols only) | Tumor burden is less than or equal to 25% of carrying capacity |
| Doubling time of sensitive cells | 13.86 h |
| Doubling time of resistant cells | 69.3 h |
| B | |
| Parameter | Value |
| Cell division rate: doubly sensitive | 0.10 per hour |
| Cell division rate: singly resistant | 0.06 per hour |
| Cell division rate: doubly resistant | 0.02 per hour |
| Background death rate | 0.01 per hour |
| Replacement probability | 1.0 |
| Delta Tumor | 10% |
| Delta Dose | 50% |
| Probability of death due to drug potency (Ψ) | 0.04 per unit drug concentration |
| Maximum tolerated dose (MTD): Drug 1 | 2.5 units |
| Maximum tolerated dose (MTD): Drug 2 | 2.5 units |
| Minimum drug dose | 0.5 units |
| Drug on time | 1 h |
| Frequency of drug application | Once every 24 h |
| Check tumor burden | Every 3 days |
| Drug decay | 10% per hour |
| Drug diffusion rate | 2.0 |
| Tumor size triggering treatment | Tumor burden is 50% or more of the carrying capacity |
| Mutation rate | 1 × 10−3 per cell division |
| Measurement noise standard deviation (SD) | 5 cells |
| Total grid size | 100 by 100 |
| Duration of simulation | 5000 h |
| Stop dosing/initiate treatment vacation when (DM protocols only): | Tumor burden is less than or equal to 25% of carrying capacity |
| Doubling time of doubly sensitive cells | 7.7 h |
| Doubling time of doubly resistant cells | 69.3 h |
| Doubling time of singly resistant cells | 13.86 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seyedi, S.; Teo, R.; Foster, L.; Saha, D.; Mina, L.; Northfelt, D.; Anderson, K.S.; Shibata, D.; Gatenby, R.; Cisneros, L.H.; et al. Testing Adaptive Therapy Protocols Using Gemcitabine and Capecitabine in a Preclinical Model of Endocrine-Resistant Breast Cancer. Cancers 2024, 16, 257. https://doi.org/10.3390/cancers16020257
Seyedi S, Teo R, Foster L, Saha D, Mina L, Northfelt D, Anderson KS, Shibata D, Gatenby R, Cisneros LH, et al. Testing Adaptive Therapy Protocols Using Gemcitabine and Capecitabine in a Preclinical Model of Endocrine-Resistant Breast Cancer. Cancers. 2024; 16(2):257. https://doi.org/10.3390/cancers16020257
Chicago/Turabian StyleSeyedi, Sareh, Ruthanne Teo, Luke Foster, Daniel Saha, Lida Mina, Donald Northfelt, Karen S. Anderson, Darryl Shibata, Robert Gatenby, Luis H. Cisneros, and et al. 2024. "Testing Adaptive Therapy Protocols Using Gemcitabine and Capecitabine in a Preclinical Model of Endocrine-Resistant Breast Cancer" Cancers 16, no. 2: 257. https://doi.org/10.3390/cancers16020257
APA StyleSeyedi, S., Teo, R., Foster, L., Saha, D., Mina, L., Northfelt, D., Anderson, K. S., Shibata, D., Gatenby, R., Cisneros, L. H., Troan, B., Anderson, A. R. A., & Maley, C. C. (2024). Testing Adaptive Therapy Protocols Using Gemcitabine and Capecitabine in a Preclinical Model of Endocrine-Resistant Breast Cancer. Cancers, 16(2), 257. https://doi.org/10.3390/cancers16020257








