Bioinformatics Analysis of Human Papillomavirus 16 Integration in Cervical Cancer: Changes in MAGI-1 Expression in Premalignant Lesions and Invasive Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Analysis
2.2. Interaction Network Analysis
2.3. Cell Lines
2.4. Collection of Pap Smears, Cervical Tissue, and Informed Consent
2.5. Western Blot
2.6. RT-qPCR
2.7. Immunocytochemistry and Immunohistochemistry
2.8. Scoring Criteria
2.9. Immunofluorescence
2.10. TCGA Dataset
2.11. Statistical Analysis
3. Results
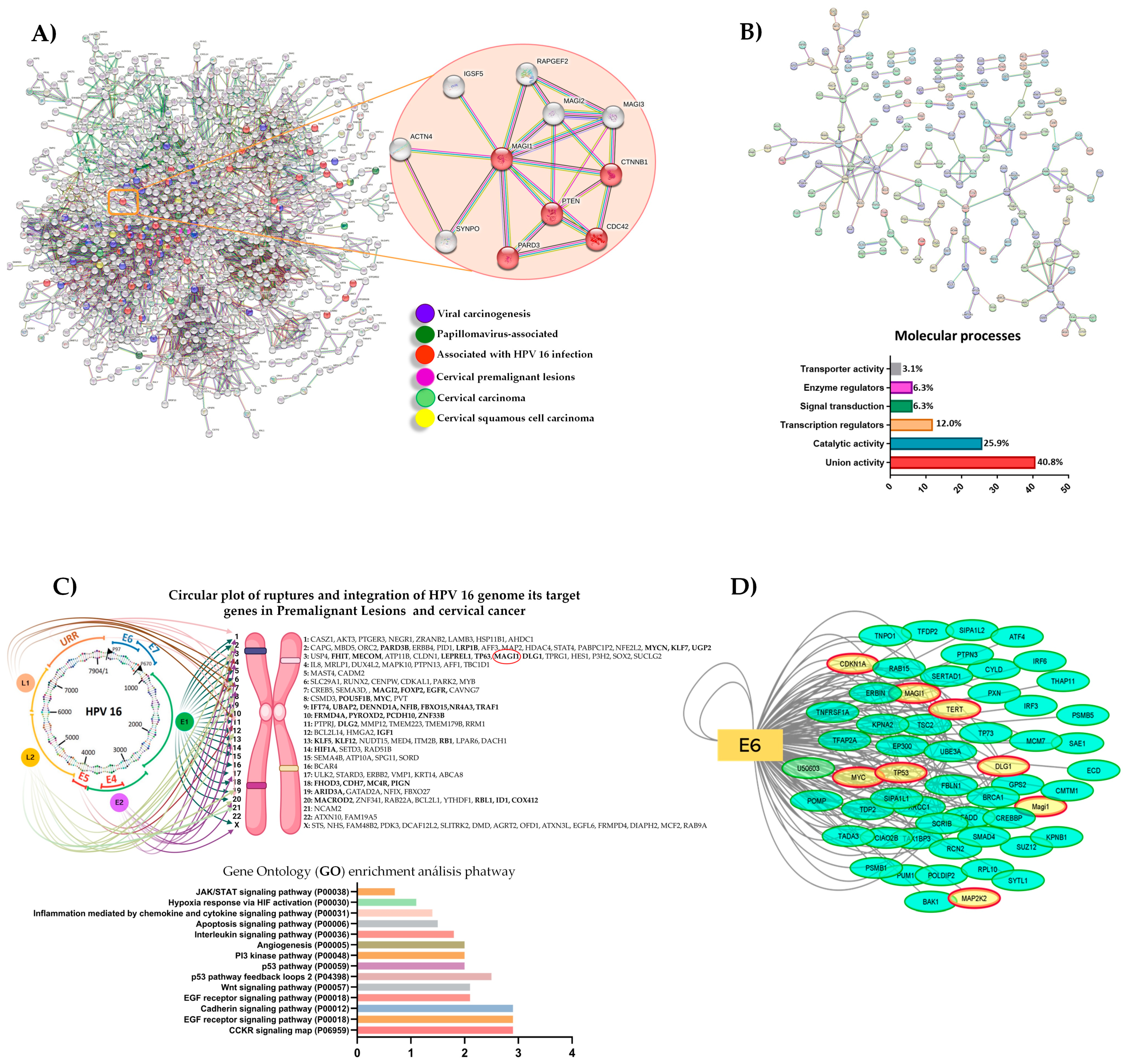
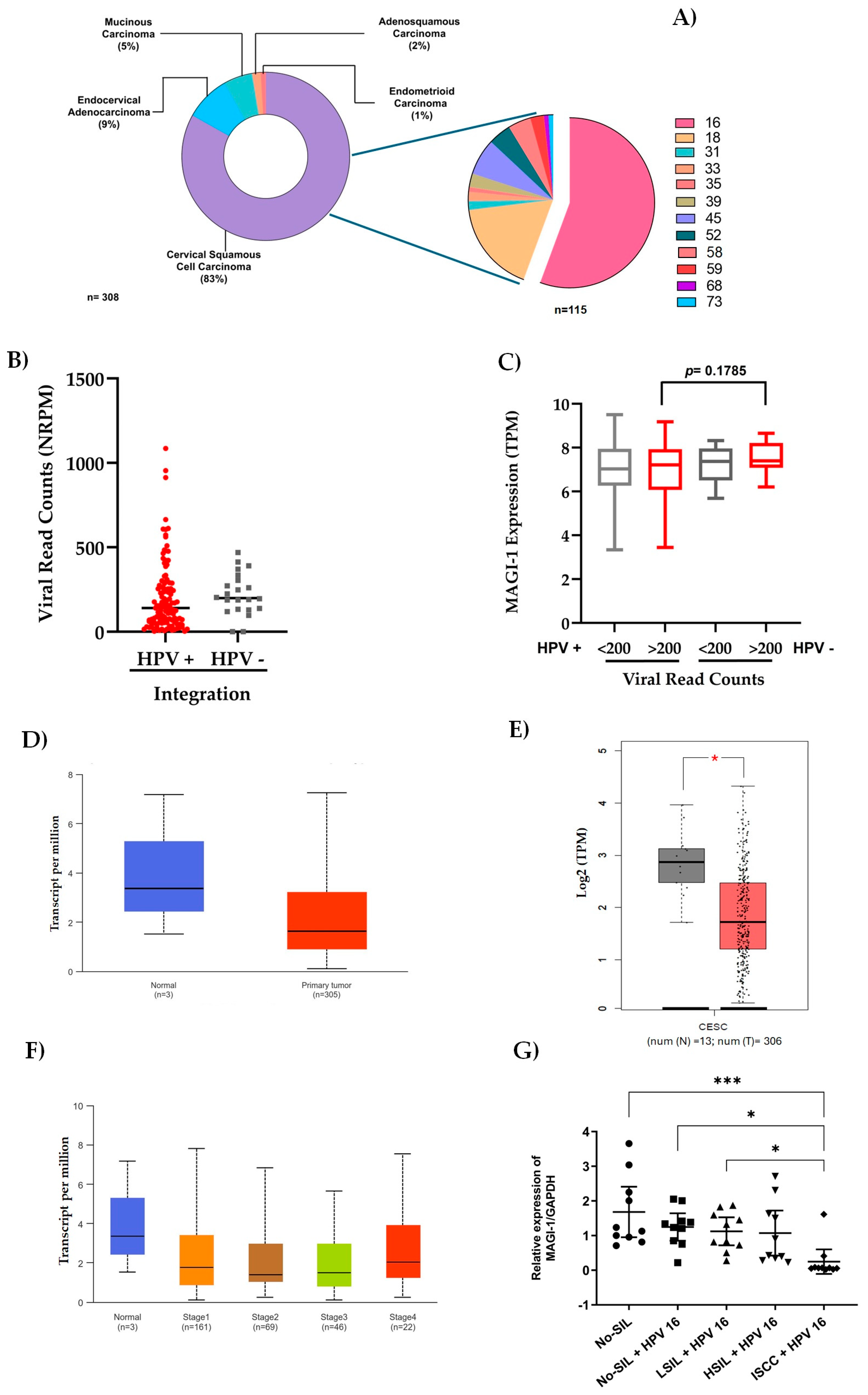
3.1. In Silico Analysis of Integrated HPV 16 Associated Proteins in Cervical Cancer
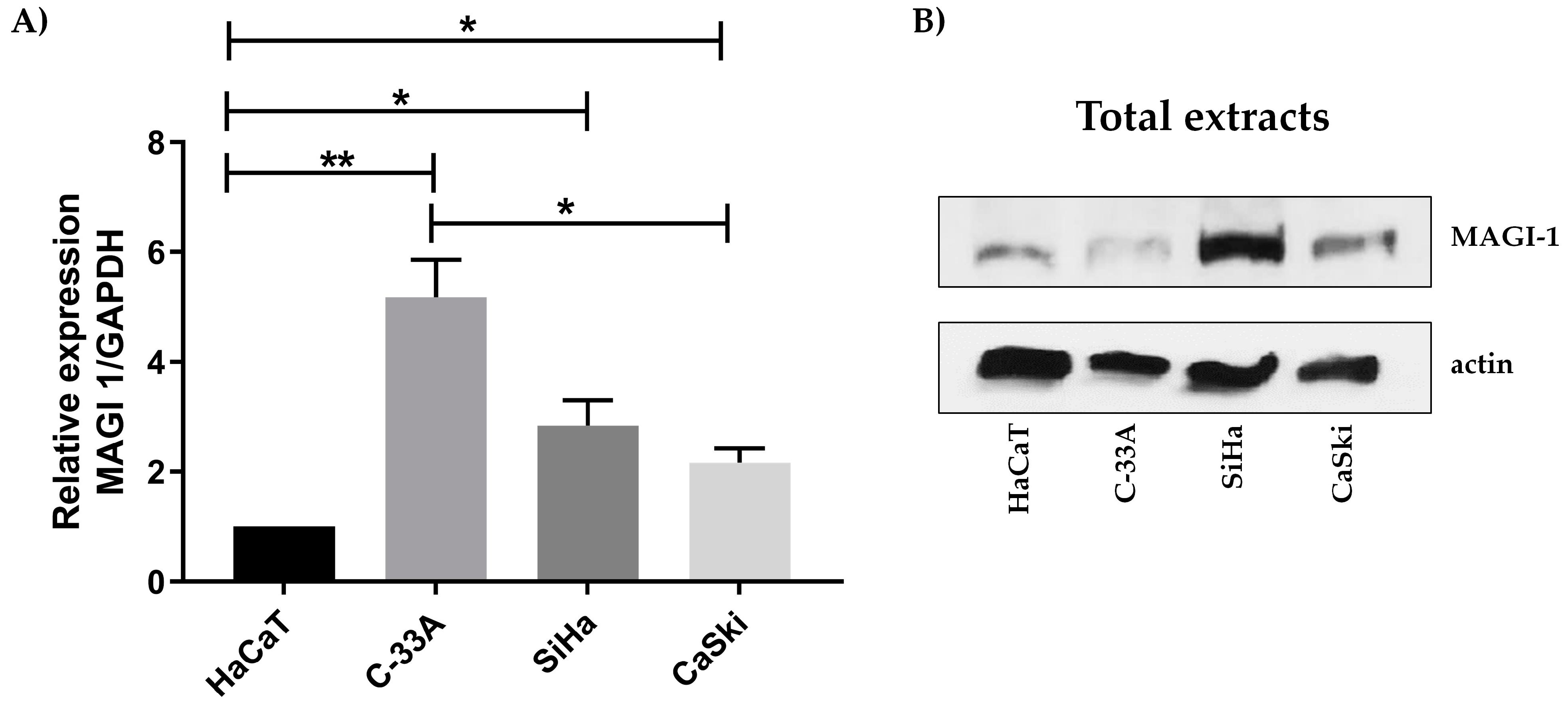
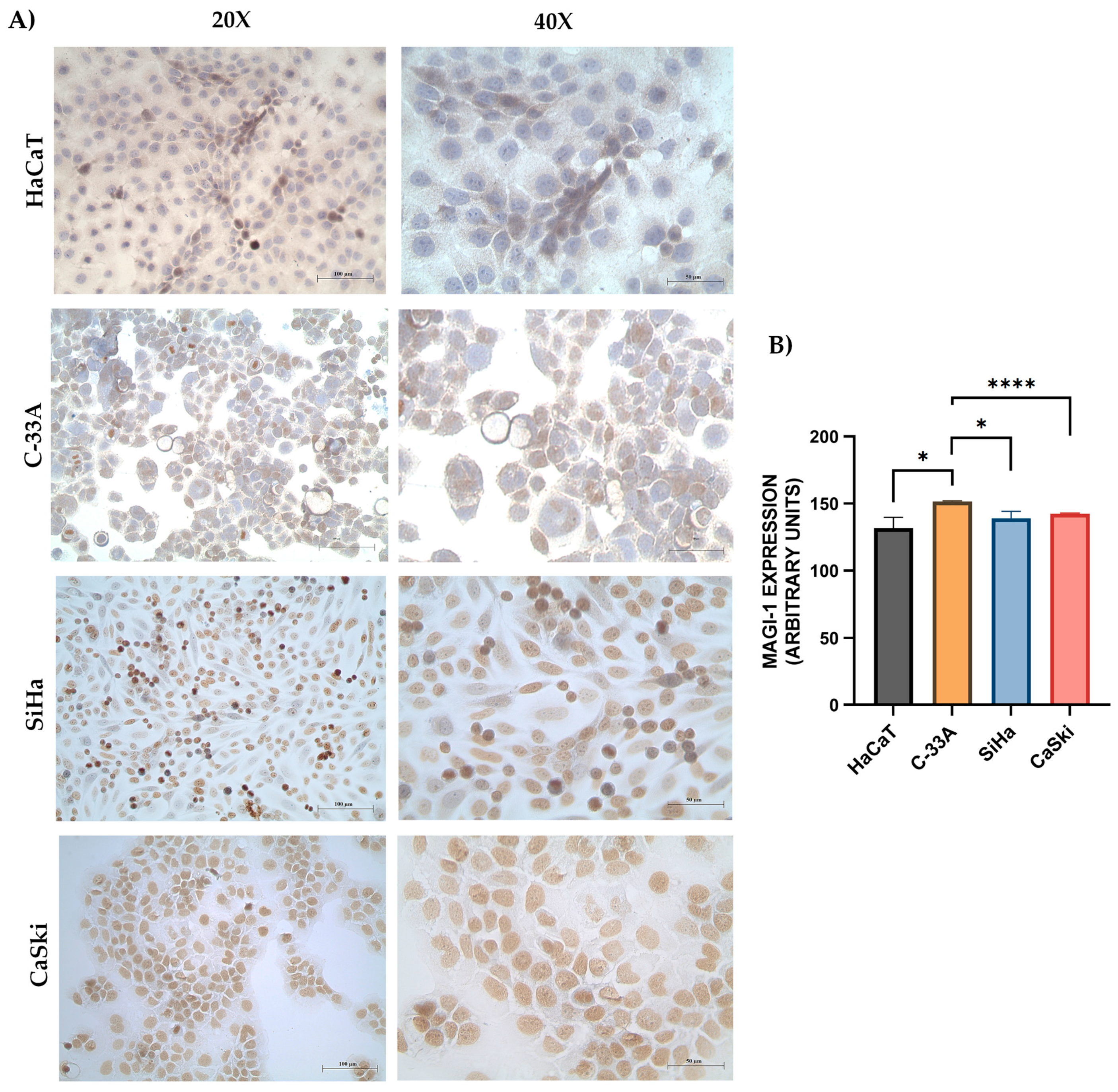
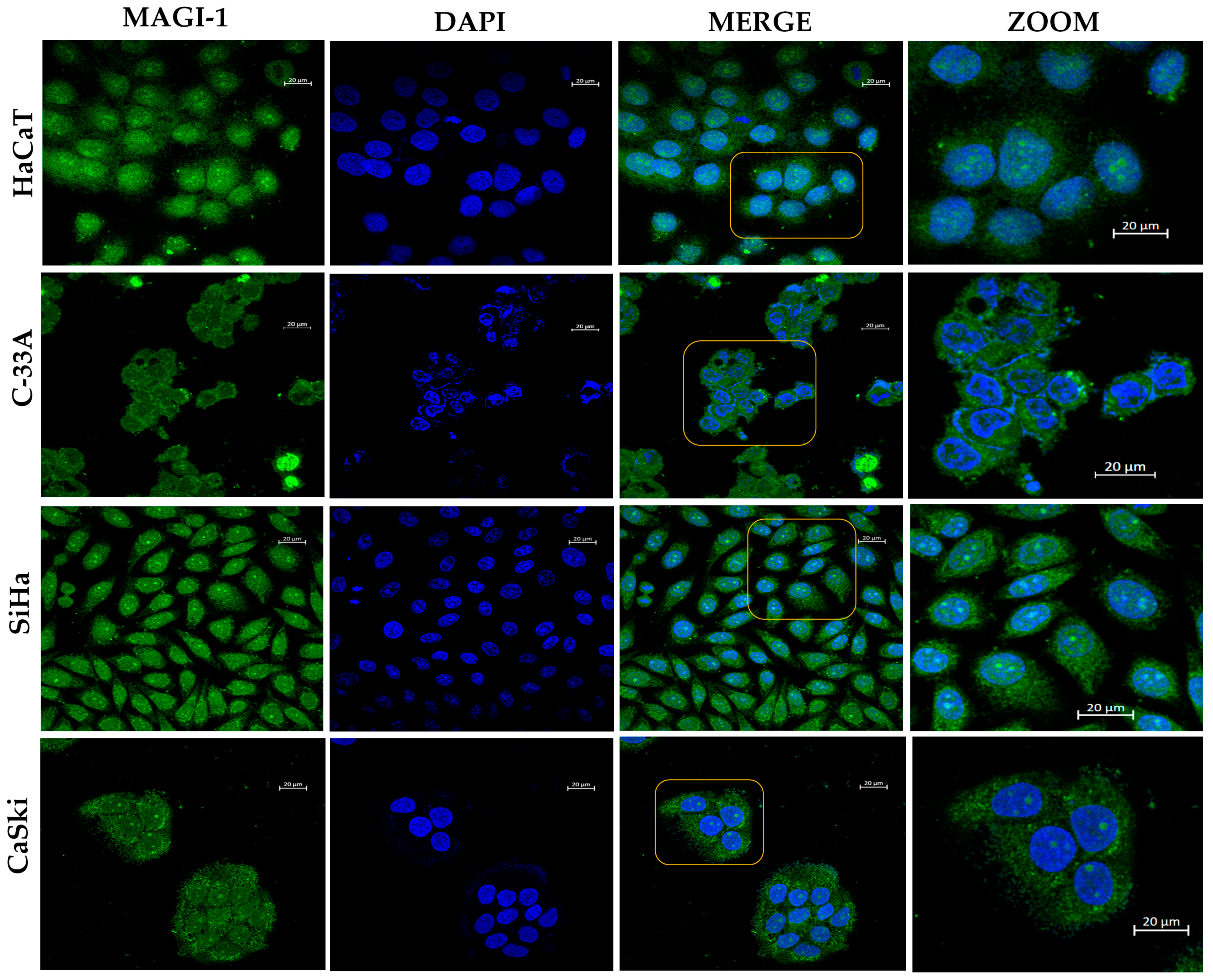
3.2. MAGI-1 Expression Profile in Cervical Cancer Cell Lines
3.3. Characteristics of the Study Population
3.4. Role of HPV Genotype, Integration, Viral Load, and MAGI-1 Expression
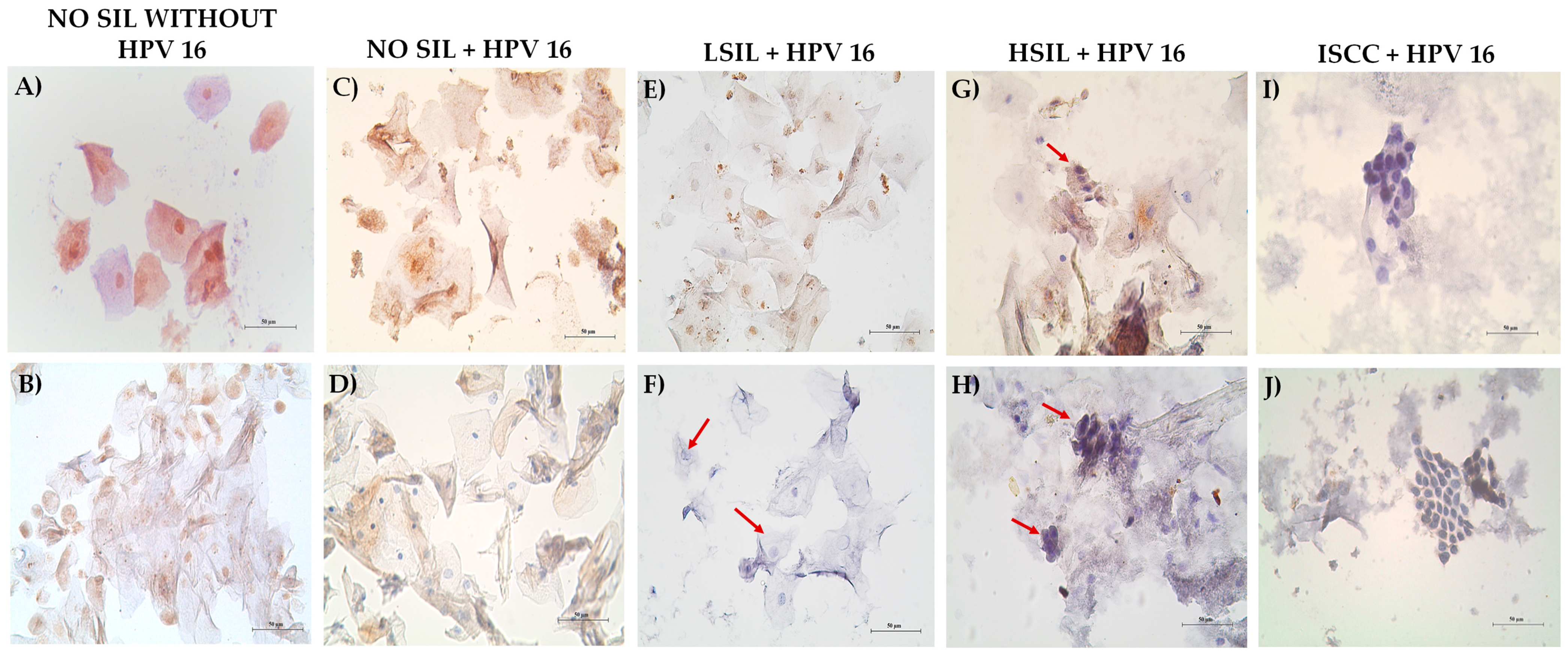
3.5. Decreased MAGI-1 Expression Is Associated with the Cervical Lesion Grade
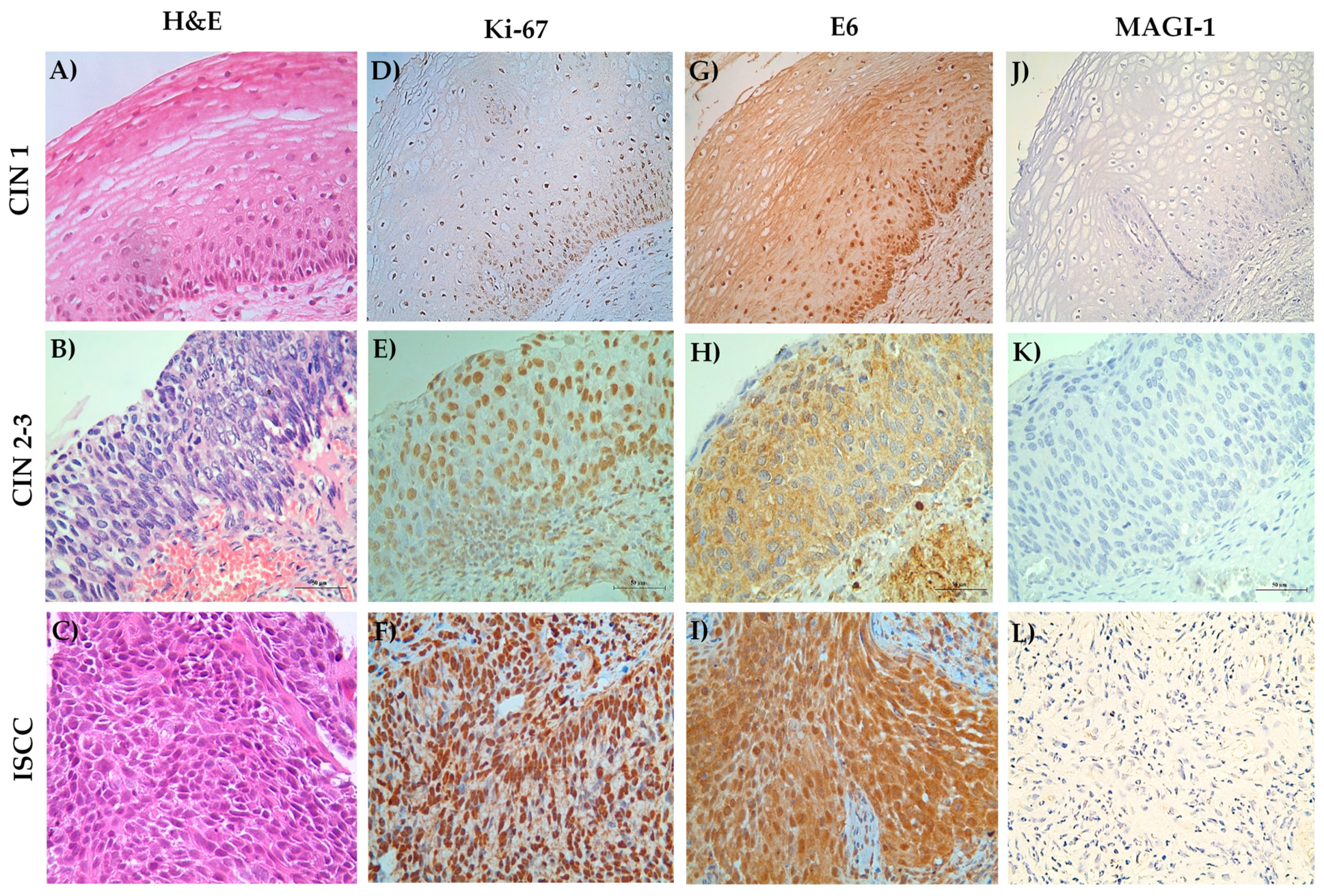
3.6. MAGI-1 Diminishing Expression Is Associated with Cell Proliferation and E6 Overexpression in CIN and ISCC HR-HPV
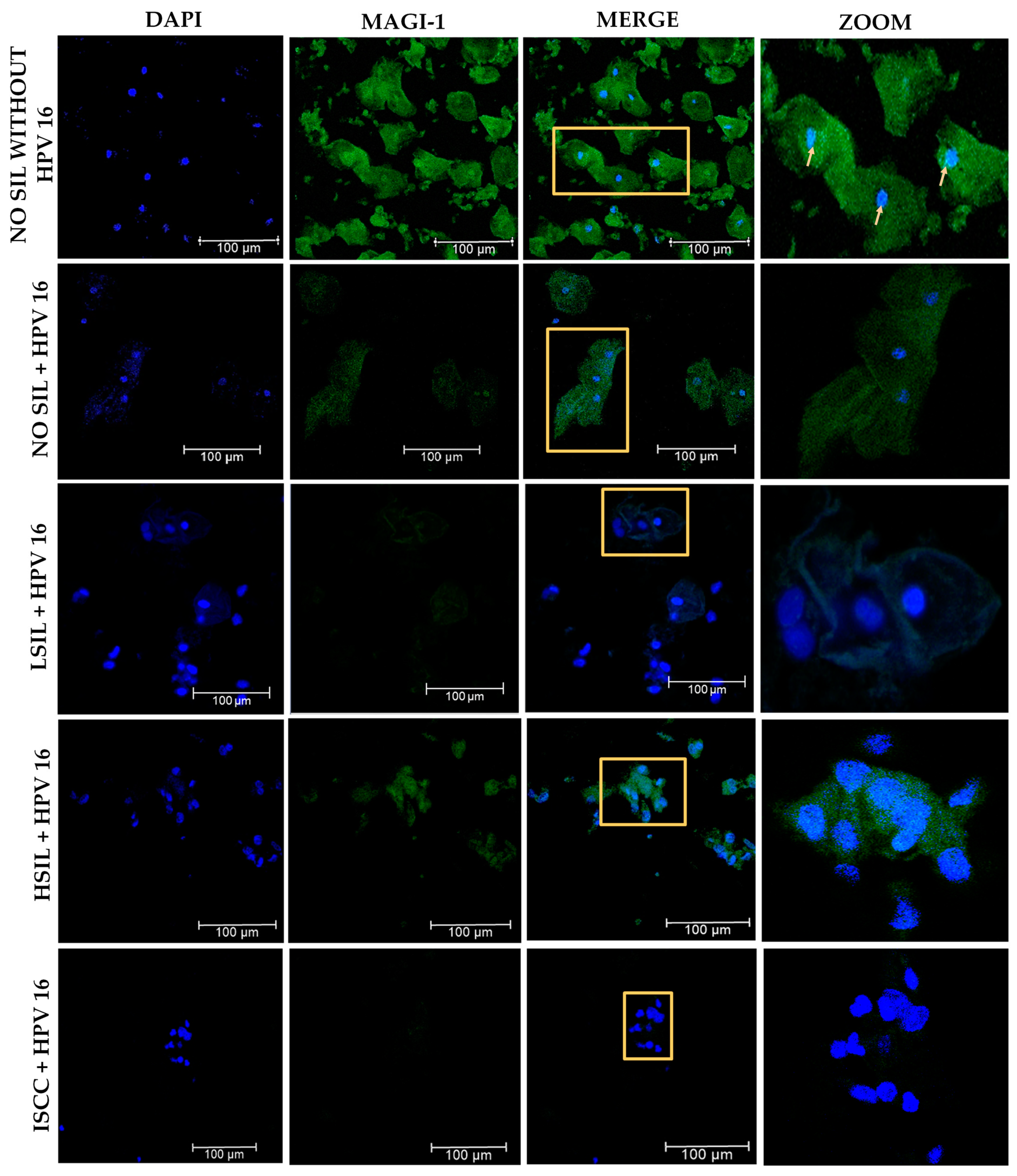
3.7. Changes in the Subcellular Localization of MAGI-1 in Premalignant Lesions and ISCC with HPV 16 Integrated
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, D.K.; Hashibe, M.; Kepka, D.; Maurer, K.A.; Werner, T.L. Too many women are dying from cervix cancer: Problems and solutions. Gynecol. Oncol. 2018, 151, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Senapati, R.; Senapati, N.N.; Dwibedi, B. Molecular mechanisms of HPV mediated neoplastic progression. Infect. Agent. Cancer. 2016, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Gilham, C.; Sargent, A.; Kitchener, H.C.; Peto, J. HPV testing compared with routine cytology in cervical screening: Long-term follow-up of ARTISTIC RCT. Health Technol. Assess. 2019, 23, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Hu, J.; Zeng, L.; Chen, G.; Cai, H.; Cao, H.; Ma, Q.; Wu, X. Characteristics of HPV integration in cervical adenocarcinoma and squamous carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 17973–17986. [Google Scholar] [CrossRef] [PubMed]
- Estêvão, D.; Costa, N.R.; Gil da Costa, R.M.; Medeiros, R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Andrake, M.D.; Skalka, A.M. Retroviral Integrase: Then and Now. Annu. Rev. Virol. 2015, 2, 241–264. [Google Scholar] [CrossRef] [PubMed]
- Reinson, T.; Toots, M.; Kadaja, M.; Pipitch, R.; Allik, M.; Ustav, E.; Ustav, M. Engagement of the ATR-Dependent DNA Damage Response at the Human Papillomavirus 18 Replication Centers during the Initial Amplification. J. Virol. 2013, 87, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Shen, K.; McBride, A.A. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014, 10, e1004117. [Google Scholar] [CrossRef] [PubMed]
- Dooley, K.E.; Warburton, A.; McBride, A.A. Tandemly Integrated HPV16 Can Form a Brd4-Dependent Super-Enhancer-Like Element That Drives Transcription of Viral Oncogenes. mBio 2016, 7, 10–1128. [Google Scholar] [CrossRef]
- Bristol, M.L.; Das, D.; Morgan, I.M. Why Human Papillomaviruses Activate the DNA Damage Response (DDR) and How Cellular and Viral Replication Persists in the Presence of DDR Signaling. Viruses 2017, 9, 268. [Google Scholar] [CrossRef]
- Hadami, K.; Saby, C.; Dakka, N.; Collin, G.; Attaleb, M.; Khyatti, M.; Filali-Maltouf, A.; Morjani, H.; El Mzibri, M. Degradation of p53 by HPV16-E6 variants isolated from cervical cancer specimens of Moroccan women. Gene 2021, 791, 145709. [Google Scholar] [CrossRef] [PubMed]
- Ganti, K.; Broniarczyk, J.; Manoubi, W.; Massimi, P.; Mittal, S.; Pim, D.; Szalmas, A.; Thatte, J.; Thomas, M.; Tomaić, V.; et al. The Human Papillomavirus E6 PDZ Binding Motif: From Life Cycle to Malignancy. Viruses 2015, 7, 3530–3551. [Google Scholar] [CrossRef] [PubMed]
- Pim, D.; Bergant, M.; Boon, S.S.; Ganti, K.; Kranjec, C.; Massimi, P.; Subbaiah, V.K.; Thomas, M.; Tomaić, V.; Banks, L. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J. 2012, 279, 3530–3537. [Google Scholar] [CrossRef] [PubMed]
- Sarabia-Vega, V.; Banks, L. Acquisition of a phospho-acceptor site enhances HPV E6 PDZ-binding motif functional promiscuity. J. Gen. Virol. 2020, 101, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Dizanzo, M.P.; Marziali, F.; Brunet Avalos, C.; Bugnon Valdano, M.; Leiva, S.; Cavatorta, A.L.; Gardiol, D. HPV E6 and E7 oncoproteins cooperatively alter the expression of Disc Large 1 polarity protein in epithelial cells. BMC Cancer 2020, 20, 293. [Google Scholar] [CrossRef] [PubMed]
- Araujo-Arcos, L.E.; Montaño, S.; Bello-Rios, C.; Garibay-Cerdenares, O.L.; Leyva-Vázquez, M.A.; Illades-Aguiar, B. Molecular insights into the interaction of HPV-16 E6 variants against MAGI-1 PDZ1 domain. Sci. Rep. 2022, 12, 1898. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Yun, H.-Y.; Lee, E.-W.; Shin, H.-C.; Kim, S.J.; Ku, B. Structural and biochemical analysis of the PTPN4 PDZ domain bound to the C-terminal tail of the human papillomavirus E6 oncoprotein. J. Microbiol. 2022, 60, 395–401. [Google Scholar] [CrossRef]
- Genera, M.; Samson, D.; Raynal, B.; Haouz, A.; Baron, B.; Simenel, C.; Guerois, R.; Wolff, N.; Caillet-Saguy, C. Structural and functional characterization of the PDZ domain of the human phosphatase PTPN3 and its interaction with the human papillomavirus E6 oncoprotein. Sci. Rep. 2019, 9, 7438. [Google Scholar] [CrossRef]
- Ramírez, J.; Recht, R.; Charbonnier, S.; Ennifar, E.; Atkinson, R.A.; Travé, G.; Nominé, Y.; Kieffer, B. Disorder-To-Order Transition of MAGI-1 PDZ1 C-Terminal Extension upon Peptide Binding: Thermodynamic and Dynamic Insights. Biochemistry 2015, 54, 1327–1337. [Google Scholar] [CrossRef]
- Excoffon, K.J.D.A.; Avila, C.L.; Alghamri, M.S.; Kolawole, A.O. The magic of MAGI-1: A scaffolding protein with multi signalosomes and functional plasticity. Biol. Cell. 2022, 114, 185–198. [Google Scholar] [CrossRef]
- Kozakai, T.; Takahashi, M.; Higuchi, M.; Hara, T.; Saito, K.; Tanaka, Y.; Masuko, M.; Takizawa, J.; Sone, H.; Fujii, M. MAGI-1 expression is decreased in several types of human T-cell leukemia cell lines, including adult T-cell leukemia. Int. J. Hematol. 2018, 107, 337–344. [Google Scholar] [CrossRef]
- Zaric, J.; Joseph, J.M.; Tercier, S.; Sengstag, T.; Ponsonnet, L.; Delorenzi, M.; Rüegg, C. Identification of MAGI1 as a tumor-suppressor protein induced by cyclooxygenase-2 inhibitors in colorectal cancer cells. Oncogene 2012, 31, 48–59. [Google Scholar] [CrossRef]
- Alday-Parejo, B.; Richard, F.; Wörthmüller, J.; Rau, T.; Galván, J.A.; Desmedt, C.; Santamaria-Martinez, A.; Rüegg, C. MAGI1, a New Potential Tumor Suppressor Gene in Estrogen Receptor Positive Breast Cancer. Cancers 2020, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Lu, J.; Qu, T.; Feng, Y.; Wang, X.; Liu, C.; Ji, J. MAGI1 inhibits migration and invasion via blocking MAPK/ERK signaling pathway in gastric cancer. Chin. J. Cancer Res. 2017, 29, 25–35. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, T.; Wang, Z. Downregulation of MAGI1 Associates with Poor Prognosis of Hepatocellular Carcinoma. J. Investig. Surg. 2012, 25, 93–99. [Google Scholar] [CrossRef]
- Kranjec, C.; Massimi, P.; Banks, L. Restoration of MAGI-1 Expression in Human Papillomavirus-Positive Tumor Cells Induces Cell Growth Arrest and Apoptosis. J. Virol. 2014, 88, 7155–7169. [Google Scholar] [CrossRef] [PubMed]
- Wörthmüller, J.; Rüegg, C. MAGI1, a Scaffold Protein with Tumor Suppressive and Vascular Functions. Cells 2021, 10, 1494. [Google Scholar] [CrossRef]
- Meissner, J.D. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J. General. Virol. 1999, 80, 1725–1733. [Google Scholar] [CrossRef]
- Torres--Rojas, F.I.; Alarcón--Romero, L.d.C.; Leyva--Vázquez, M.A.; Ortiz--Ortiz, J.; Mendoza--Catalán, M.Á.; Hernández--Sotelo, D.; Del Moral--Hernández, O.; Rodríguez--Ruiz, H.A.; Leyva--Illades, D.; Flores-Alfaro, E.; et al. Methylation of the L1 gene and integration of human papillomavirus 16 and 18 in cervical carcinoma and premalignant lesions. Oncol. Lett. 2018, 15, 2278–2286. [Google Scholar] [CrossRef] [PubMed]
- Cambruzzi, E.; Zettler, C.G.; Alexandre, C.O. Expression of Ki-67 and squamous intraepithelial lesions are related with HPV in endocervical adenocarcinoma. Pathol. Oncol. Res. 2005, 11, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Portari, E.A.; Russomano, F.B.; de Camargo, M.J.; Machado Gayer, C.R.; da Rocha Guillobel, H.C.; Santos-Rebouças, C.B.; Brito Macedo, J.M. Immunohistochemical Expression of Cyclin D1, p16Ink4a, p21WAF1, and Ki-67 Correlates With the Severity of Cervical Neoplasia. Int. J. Gynecol. Pathol. 2013, 32, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.D.; Chen, Z.; Saller, C.; Tarvin, K.; Carvalho, A.L.; Scapulatempo-Neto, C.; Silveira, H.C.; Fregnani, J.H.; Creighton, C.J.; Anderson, M.L.; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e814. [Google Scholar] [CrossRef] [PubMed]
- Bodelon, C.; Untereiner, M.E.; Machiela, M.J.; Vinokurova, S.; Wentzensen, N. Genomic characterization of viral integration sites in HPV-related cancers. Int. J. Cancer 2016, 139, 2001–2011. [Google Scholar] [CrossRef] [PubMed]
- Vats, A.; Trejo-Cerro, O.; Thomas, M.; Banks, L. Human papillomavirus E6 and E7: What remains? Tumour Virus Res. 2021, 11, 200213. [Google Scholar] [CrossRef] [PubMed]
- Warburton, A.; Markowitz, T.E.; Katz, J.P.; Pipas, J.M.; McBride, A.A. Recurrent integration of human papillomavirus genomes at transcriptional regulatory hubs. NPJ Genom. Med. 2021, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.; Lameiras, S.; Jeannot, E.; Marie, Y.; Castera, L.; Sastre-Garau, X.; Nicolas, A. Mechanistic signatures of HPV insertions in cervical carcinomas. NPJ Genom. Med. 2016, 1, 16004. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhu, D.; Wang, W.; Li, W.; Jia, W.; Zeng, X.; Ding, W.; Yu, L.; Wang, X.; Wang, L.; et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015, 47, 158–163. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014, 24, 185–199. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Zhang, R.; Cai, Y.; Yang, X.; Wang, Z.; Li, Y.; Cheng, X.; Ye, X.; Xiang, Y.; et al. Preferential sites for the integration and disruption of human papillomavirus 16 in cervical lesions. J. Clin. Virol. 2013, 56, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Qiu, Q.; Zhou, Q.; Li, J.; Yu, M.; Li, K.; Xu, L.; Ke, X.; Xu, H.; Lu, B.; et al. Long-read sequencing unveils high-resolution HPV integration and its oncogenic progression in cervical cancer. Nat. Commun. 2022, 13, 2563. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jia, W.; Wang, M.; Liu, J.; Zhou, X.; Liang, Z.; Zhang, Q.; Long, S.; Quzhen, S.; Li, X.; et al. Human papillomavirus integration perspective in small cell cervical carcinoma. Nat. Commun. 2022, 13, 5968. [Google Scholar] [CrossRef] [PubMed]
- Moberg, M.; Gustavsson, I.; Wilander, E.; Gyllensten, U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br. J. Cancer 2005, 92, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Adcock, R.; Cuzick, J.; Hunt, W.C.; McDonald, R.M.; Wheeler, C.M.; Joste, N.E.; Kinney, W.; Wheeler, C.M.; Hunt, W.C.; McDonald, R.M.; et al. Role of HPV Genotype, Multiple Infections, and Viral Load on the Risk of High-Grade Cervical Neoplasia. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1816–1824. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shi, X.; Liu, J.; Zhang, L. Correlation between human papillomavirus viral load and cervical lesions classification: A review of current research. Front. Med. 2023, 10, 1111269. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Mehta Kavi, P.; Laimins Laimonis, A. Suppression of STAT-1 Expression by Human Papillomaviruses Is Necessary for Differentiation-Dependent Genome Amplification and Plasmid Maintenance. J. Virol. 2011, 85, 9486–9494. [Google Scholar] [CrossRef] [PubMed]
- Sakuta, K.; Sasaki, Y.; Abe, Y.; Sato, H.; Shoji, M.; Yaoita, T.; Yagi, M.; Mizumoto, N.; Onozato, Y.; Kon, T.; et al. Somatic alterations and mutational burden are potential predictive factors for metachronous development of early gastric cancer. Sci. Rep. 2020, 10, 22071. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, X.; Luo, J.; Liang, B.; Peng, J.; Chen, C.; Guo, H.; Wang, Q.; Xing, X.; Deng, Q.; et al. MiR-486-5p-directed MAGI1/Rap1/RASSF5 signaling pathway contributes to hydroquinone-induced inhibition of erythroid differentiation in K562 cells. Toxicol. Vitro. 2020, 66, 104830. [Google Scholar] [CrossRef]
- Wang, W.; Yang, Y.; Chen, X.; Shao, S.; Hu, S.; Zhang, T. MAGI1 mediates tumor metastasis through c-Myb/miR-520h/MAGI1 signaling pathway in renal cell carcinoma. Apoptosis 2019, 24, 837–848. [Google Scholar] [CrossRef]
- Ravi, N.; Yang, M.; Mylona, N.; Wennerberg, J.; Paulsson, K. Global RNA Expression and DNA Methylation Patterns in Primary Anaplastic Thyroid Cancer. Cancers 2020, 12, 680. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.Q.; Tong, W.G.; Yang, H.; Lin, W.; Lee, M.K.; Fang, Z.H.; Wei, Y.; Jelinek, J.; Issa, J.P.; Garcia-Manero, G. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia 2008, 22, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.; Driesch, C.; Jansen, L.; Runnebaum, I.B.; Dürst, M. Non-Random Integration of the HPV Genome in Cervical Cancer. PLoS ONE 2012, 7, e39632. [Google Scholar] [CrossRef] [PubMed]
- Xuemin, F.; Shuqin, J.I.A.; Tracey, A.M.; Wen, G.J. Regulation and Involvement in Cancer and Pathological Conditions of MAGI1, a Tight Junction Protein. Anticancer. Res. 2014, 34, 3251–3256. [Google Scholar]
- Kranjec, C.; Banks, L. A Systematic Analysis of Human Papillomavirus (HPV) E6 PDZ Substrates Identifies MAGI-1 as a Major Target of HPV Type 16 (HPV-16) and HPV-18 Whose Loss Accompanies Disruption of Tight Junctions. J. Virol. 2011, 85, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Knights, A.J.; Funnell, A.P.; Crossley, M.; Pearson, R.C. Holding Tight: Cell Junctions and Cancer Spread. Trends Cancer Res. 2012, 8, 61–69. [Google Scholar] [PubMed]
- Kotelevets, L.; Chastre, E. A New Story of the Three Magi: Scaffolding Proteins and lncRNA Suppressors of Cancer. Cancers 2021, 13, 4264. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, S.; Repnik, U.; Banks, L.; Turk, V.; Turk, B. Cellular localization of MAGI-1 caspase cleavage products and their role in apoptosis. Biol. Chem. 2007, 388, 1195–1198. [Google Scholar] [CrossRef]
- Abe, J.-I.; Ko, K.A.; Kotla, S.; Wang, Y.; Paez-Mayorga, J.; Shin, I.J.; Imanishi, M.; Vu, H.T.; Tao, Y.; Leiva-Juarez, M.M.; et al. MAGI1 as a link between endothelial activation and ER stress drives atherosclerosis. JCI Insight 2019, 4, e125570. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, Z. MAGI1 inhibits cancer cell migration and invasion of hepatocellular carcinoma via regulating PTEN. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2011, 36, 381–385. [Google Scholar] [PubMed]
- Fournane, S.; Charbonnier, S.; Chapelle, A.; Kieffer, B.; Orfanoudakis, G.; Travé, G.; Masson, M.; Nominé, Y. Surface plasmon resonance analysis of the binding of high-risk mucosal HPV E6 oncoproteins to the PDZ1 domain of the tight junction protein MAGI-1. J. Mol. Recognit. 2011, 24, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Boldt, K.; van Reeuwijk, J.; Lu, Q.; Koutroumpas, K.; Nguyen, T.-M.T.; Texier, Y.; van Beersum, S.E.C.; Horn, N.; Willer, J.R.; Mans, D.A.; et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat. Commun. 2016, 7, 11491. [Google Scholar] [CrossRef] [PubMed]
- Cavatorta, A.L.; Fumero, G.; Chouhy, D.; Aguirre, R.; Nocito, A.L.; Giri, A.A.; Banks, L.; Gardiol, D. Differential expression of the human homologue of drosophila discs large oncosuppressor in histologic samples from human papillomavirus–associated lesions as a marker for progression to malignancy. Int. J. Cancer. 2004, 111, 373–380. [Google Scholar] [CrossRef]
- Krishna Subbaiah, V.; Massimi, P.; Boon, S.S.; Myers, M.P.; Sharek, L.; Garcia-Mata, R.; Banks, L. The Invasive Capacity of HPV Transformed Cells Requires the hDlg-Dependent Enhancement of SGEF/RhoG Activity. PLoS Pathog. 2012, 8, e1002543. [Google Scholar] [CrossRef]

| Variable | No-SIL without HPV n (%) | No-SIL + HPV 16 n (%) | LSIL+ HPV 16 n (%) | HSIL + HPV 16 n (%) | ISCC + HPV 16 n (%) | Total n = 50 | p |
|---|---|---|---|---|---|---|---|
| Age groups | |||||||
| 20–37 | 6 (60) | 2 (20) | 5 (50) | 6 (60) | 2 (20) | 21 (43) | |
| 38–49 | 2 (20) | 4 (40) | 2 (20) | 2 (20) | 3 (30) | 13 (24) | 0.093 |
| 50–86 | 2 (20) | 4 (40) | 3 (30) | 2 (20) | 5 (50) | 16 (33) | |
| * SD | |||||||
| 12–16 | 1 (10) | 5 (50) | 2 (20) | 5 (50) | 5 (50) | 18 (36) | |
| 17–18 | 3 (30) | 1 (10) | 2 (20) | 4 (40) | 3 (30) | 13 (26) | 0.037 |
| 19–30 | 6 (60) | 4 (40) | 6 (60) | 1 (10) | 2 (20) | 19 (38) | |
| Menarche | |||||||
| 11–12 | 3 (30) | 6 (60) | 5 (50) | 3 (30) | 2 (20) | 19 (38) | |
| 13–14 | 6 (60) | 3 (30) | 3 (30) | 5 (50) | 5 (50) | 22 (44) | 0.697 |
| 15–16 | 1 (10) | 1 (10) | 2 (20) | 2 (20) | 3 (30) | 9 (18) | |
| Number of sex partners | |||||||
| 1 | 7 (70) | 4 (40) | 6 (60) | 8 (80) | 6 (60) | 31 (62) | 0.987 |
| >2 | 3 (30) | 6 (60) | 4 (40) | 2 (20) | 4 (40) | 19 (38) | |
| Giving birth | |||||||
| 0 | 5 (50) | 4 (40) | 2 (20) | 3 (30) | 0 | 14 (28) | 0.054 |
| >1 | 5 (50) | 6 (60) | 8 (80) | 7 (70) | 10 (100) | 36 (72) |
| Viral Physical State | No-SIL + HPV 16 n = 10 | LSIL + HPV 16 n = 10 | HSIL + HPV 16 n = 10 | ISCC + HPV 16 n = 10 | Total n = 40 | p |
|---|---|---|---|---|---|---|
| Episomal | 0 | 0 | 0 | 0 | 0 | 0.170 |
| Mixed | 2 (20) | 3 (30) | 4 (40) | 0 | 9 (22.5) | |
| Integrated | 8 (80) | 7 (70) | 6 (60) | 10 (100) | 31 (77.5) |
| Assessment Criteria | No SIL/without HPV n = 10 | No-SIL + HPV 16 n = 10 | LSIL + HPV 16 n = 10 | HSIL + HPV 16 n = 10 | ISCC + HPV 16 n = 10 | p |
|---|---|---|---|---|---|---|
| MAGI-1 | ||||||
| Level of expression | ||||||
| Negative | 0 | 3 (30) | 3 (30) | 6 (60) | 10 (100) | <0.001 |
| Mild (1–10% of positive cells) | 0 | 4 (40) | 3 (30) | 4 (40) | 0 | |
| Moderate (11–50% of positive cells) | 2 (20) | 3 (30) | 4 (40) | 0 | 0 | |
| Intense (>50% positive cells) | 8 (80) | 0 | 0 | 0 | 0 | |
| Subcellular localization | ||||||
| * Negative | 0 | 3 (30) | 3 (30) | 6 (56) | 10 (100) | |
| Nucleus | 0 | 0 | 1 (10) | 2 (22) | 0 | <0.001 |
| Cytoplasm | 1 (10) | 3 (30) | 2 (20) | 0 | 0 | |
| Nucleus–Cytoplasm | 9 (90) | 4 (40) | 4 (40) | 2 (22) | 0 |
| Histological Diagnosis | ||||
|---|---|---|---|---|
| Expression Level | CIN 1 n (%) | CIN 2–3 n (%) | ISCC n (%) | p |
| Ki-67 | ||||
| Negative | 0 | 0 | 0 | |
| 1+ | 11 (79) | 1 (5) | 0 | |
| 2+ | 2 (14) | 6 (30) | 0 | <0.001 |
| 3+ | 1 (7) | 13 (65) | 0 | |
| Tumor nests in the stroma | 0 | 0 | 4 (100) | |
| E6 | ||||
| Negative | 0 | 0 | 0 | |
| 1+ | 1 (7) | 0 | 0 | |
| 2+ | 2 (14) | 2 (10) | 0 | <0.001 |
| 3+ | 11 (79) | 18 (90) | 0 | |
| Tumor nests in the stroma | 0 | 0 | 4 (100) | |
| MAGI-1 | ||||
| Negative | 11 (79) | 17 (85) | 4 (100) | |
| 1+ | 2 (14) | 2 (10) | 0 | |
| 2+ | 1 (7) | 1 (5) | 0 | 0.089 |
| 3+ | 0 | 0 | 0 | |
| Tumor nests in the stroma | 0 | 0 | 0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catalán-Castorena, O.; Garibay-Cerdenares, O.L.; Illades-Aguiar, B.; Castillo-Sánchez, R.; Zubillaga-Guerrero, M.I.; Leyva-Vazquez, M.A.; Encarnacion-Guevara, S.; Flores-Alfaro, E.; Ramirez-Ruano, M.; del Carmen Alarcón-Romero, L. Bioinformatics Analysis of Human Papillomavirus 16 Integration in Cervical Cancer: Changes in MAGI-1 Expression in Premalignant Lesions and Invasive Carcinoma. Cancers 2024, 16, 2225. https://doi.org/10.3390/cancers16122225
Catalán-Castorena O, Garibay-Cerdenares OL, Illades-Aguiar B, Castillo-Sánchez R, Zubillaga-Guerrero MI, Leyva-Vazquez MA, Encarnacion-Guevara S, Flores-Alfaro E, Ramirez-Ruano M, del Carmen Alarcón-Romero L. Bioinformatics Analysis of Human Papillomavirus 16 Integration in Cervical Cancer: Changes in MAGI-1 Expression in Premalignant Lesions and Invasive Carcinoma. Cancers. 2024; 16(12):2225. https://doi.org/10.3390/cancers16122225
Chicago/Turabian StyleCatalán-Castorena, Oscar, Olga Lilia Garibay-Cerdenares, Berenice Illades-Aguiar, Rocio Castillo-Sánchez, Ma. Isabel Zubillaga-Guerrero, Marco Antonio Leyva-Vazquez, Sergio Encarnacion-Guevara, Eugenia Flores-Alfaro, Mónica Ramirez-Ruano, and Luz del Carmen Alarcón-Romero. 2024. "Bioinformatics Analysis of Human Papillomavirus 16 Integration in Cervical Cancer: Changes in MAGI-1 Expression in Premalignant Lesions and Invasive Carcinoma" Cancers 16, no. 12: 2225. https://doi.org/10.3390/cancers16122225
APA StyleCatalán-Castorena, O., Garibay-Cerdenares, O. L., Illades-Aguiar, B., Castillo-Sánchez, R., Zubillaga-Guerrero, M. I., Leyva-Vazquez, M. A., Encarnacion-Guevara, S., Flores-Alfaro, E., Ramirez-Ruano, M., & del Carmen Alarcón-Romero, L. (2024). Bioinformatics Analysis of Human Papillomavirus 16 Integration in Cervical Cancer: Changes in MAGI-1 Expression in Premalignant Lesions and Invasive Carcinoma. Cancers, 16(12), 2225. https://doi.org/10.3390/cancers16122225







