Preoperative Classification of Peripheral Nerve Sheath Tumors on MRI Using Radiomics
Abstract
Simple Summary
Abstract
1. Introduction
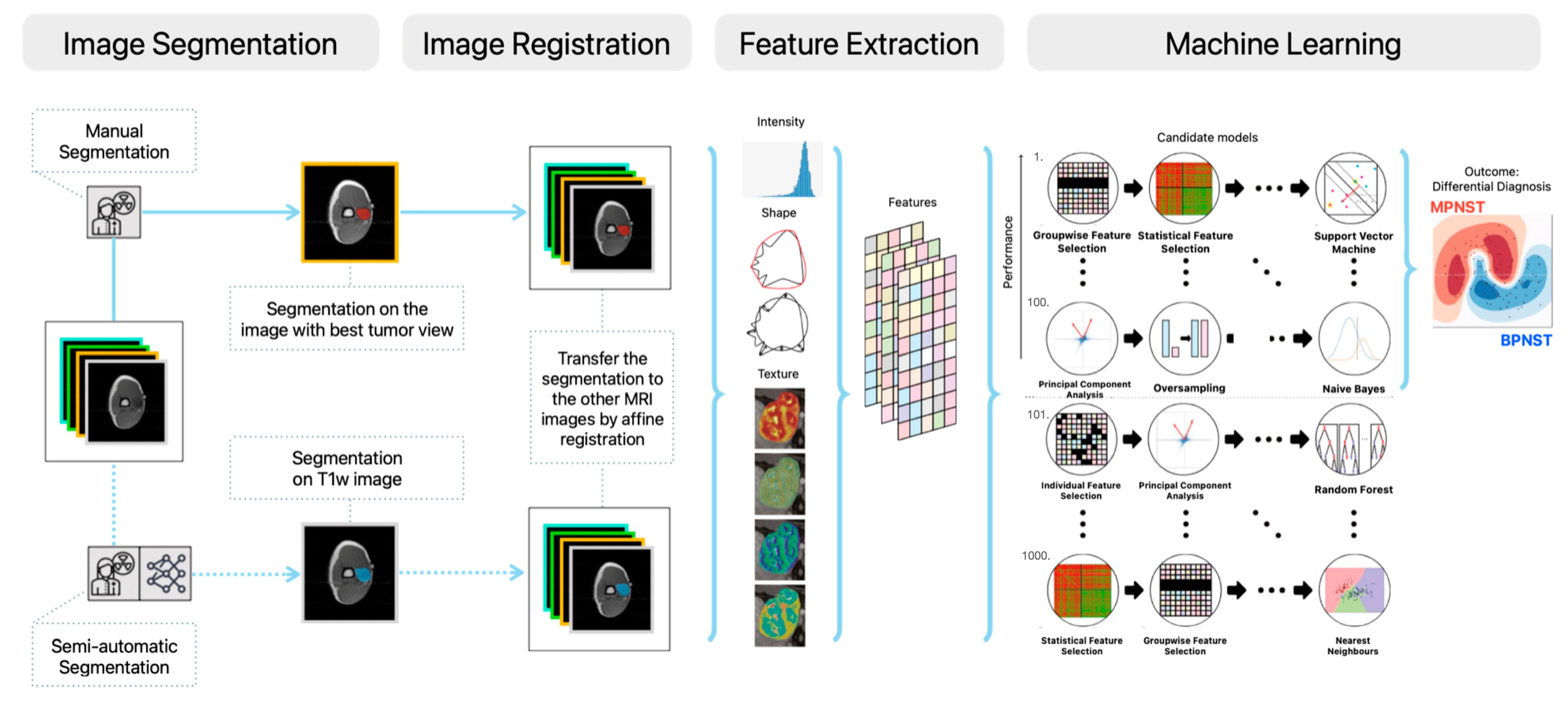
2. Materials and Methods
2.1. Study Population and Data Collection
2.2. Manual and Semi-Automatic Segmentation
2.3. Radiomics Feature Extraction
2.4. Decision Model Creation
2.5. Classification by Radiologists
2.6. Experimental Set-Up
2.7. Statistical Analysis and Evaluation
3. Results
3.1. Clinical Characteristics of Database
3.2. Evaluation of the Radiomics Models
3.3. Evaluation of the Radiologists
3.4. Evaluation of the Integrated Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levi, A.D.; Ross, A.L.; Cuartas, E.; Qadir, R.; Temple, H.T. The Surgical Management of Symptomatic Peripheral Nerve Sheath Tumors. Neurosurgery 2010, 66, 833–840. [Google Scholar] [CrossRef]
- Martin, E.; Geitenbeek, R.T.J.; Coert, J.H.; Hanff, D.F.; Graven, L.H.; Grünhagen, D.J.; Verhoef, C.; Taal, W. A Bayesian Approach for Diagnostic Accuracy of Malignant Peripheral Nerve Sheath Tumors: A Systematic Review and Meta-Analysis. Neuro Oncol. 2021, 23, 557. [Google Scholar] [CrossRef]
- Jack, A.S.; Huie, C.J.; Jacques, L.G. Epidemiology of Peripheral Nerve Sheath Tumors. In Diagnostic Assessment and Treatment of Peripheral Nerve Tumors; Springer: Cham, Switzerland, 2021; pp. 3–16. [Google Scholar]
- Ng, V.Y.; Scharschmidt, T.J.; Mayerson, J.L.; Fisher, J.L. Incidence and Survival in Sarcoma in the United States: A Focus on Musculoskeletal Lesions. Anticancer Res. 2013, 33, 2597–2604. [Google Scholar]
- Zou, C.; Smith, K.D.; Liu, J.; Lahat, G.; Myers, S.; Wang, W.-L.; Zhang, W.; McCutcheon, I.E.; Slopis, J.M.; Lazar, A.J.; et al. Clinical, Pathological, and Molecular Variables Predictive of Malignant Peripheral Nerve Sheath Tumor Outcome. Ann. Surg. 2009, 249, 1014–1022. [Google Scholar] [CrossRef]
- Stucky, C.-C.H.; Johnson, K.N.; Gray, R.J.; Pockaj, B.A.; Ocal, I.T.; Rose, P.S.; Wasif, N. Malignant Peripheral Nerve Sheath Tumors (MPNST): The Mayo Clinic Experience. Ann. Surg. Oncol. 2012, 19, 878–885. [Google Scholar] [CrossRef]
- Anghileri, M.; Miceli, R.; Fiore, M.; Mariani, L.; Ferrari, A.; Mussi, C.; Lozza, L.; Collini, P.; Olmi, P.; Casali, P.G. Malignant Peripheral Nerve Sheath Tumors: Prognostic Factors and Survival in a Series of Patients Treated at a Single Institution. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2006, 107, 1065–1074. [Google Scholar] [CrossRef]
- Miao, R.; Wang, H.; Jacobson, A.; Lietz, A.P.; Choy, E.; Raskin, K.A.; Schwab, J.H.; Deshpande, V.; Nielsen, G.P.; DeLaney, T.F. Radiation-Induced and Neurofibromatosis-Associated Malignant Peripheral Nerve Sheath Tumors (MPNST) Have Worse Outcomes than Sporadic MPNST. Radiother. Oncol. 2019, 137, 61–70. [Google Scholar] [CrossRef]
- Evans, D.G.R. Malignant Peripheral Nerve Sheath Tumours in Neurofibromatosis 1. J. Med. Genet. 2002, 39, 311–314. [Google Scholar] [CrossRef]
- Ferner, R.E.; Huson, S.M.; Thomas, N.; Moss, C.; Willshaw, H.; Evans, D.G.; Upadhyaya, M.; Towers, R.; Gleeson, M.; Steiger, C. Guidelines for the Diagnosis and Management of Individuals with Neurofibromatosis 1. J. Med. Genet. 2007, 44, 81–88. [Google Scholar] [CrossRef]
- Wong, W.W.; Hirose, T.; Scheithauer, B.W.; Schild, S.E.; Gunderson, L.L. Malignant Peripheral Nerve Sheath Tumor: Analysis of Treatment Outcome. Int. J. Radiat. Oncol. Biol. Phys. 1998, 42, 351–360. [Google Scholar] [CrossRef]
- Zehou, O.; Fabre, E.; Zelek, L.; Sbidian, E.; Ortonne, N.; Banu, E.; Wolkenstein, P.; Valeyrie-Allanore, L. Chemotherapy for the Treatment of Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis 1: A 10-Year Institutional Review. Orphanet J. Rare Dis. 2013, 8, 127. [Google Scholar] [CrossRef]
- James, A.W.; Shurell, E.; Singh, A.; Dry, S.M.; Eilber, F.C. Malignant Peripheral Nerve Sheath Tumor. Surg. Oncol. Clin. 2016, 25, 789–802. [Google Scholar] [CrossRef]
- Hagi, T.; Nakamura, T.; Yokoji, A.; Matsumine, A.; Sudo, A. Medullary Metastasis of a Malignant Peripheral Nerve Sheath Tumor: A Case Report. Oncol. Lett. 2016, 12, 1906–1908. [Google Scholar] [CrossRef]
- Stark, A.M.; Buhl, R.; Hugo, H.H.; Mehdorn, H.M. Malignant Peripheral Nerve Sheath Tumours–Report of 8 Cases and Review of the Literature. Acta Neurochir. 2001, 143, 357–364. [Google Scholar] [CrossRef]
- Akshintala, S.; Mallory, N.C.; Lu, Y.; Ballman, K.V.; Schuetze, S.M.; Chugh, R.; Maki, R.G.; Reinke, D.K.; Widemann, B.C.; Kim, A. Outcome of Patients with Malignant Peripheral Nerve Sheath Tumors Enrolled on Sarcoma Alliance for Research Through Collaboration (SARC) Phase II Trials. Oncologist 2023, 28, 453–459. [Google Scholar] [CrossRef]
- Sobczuk, P.; Teterycz, P.; Czarnecka, A.M.; Świtaj, T.; Koseła-Paterczyk, H.; Kozak, K.; Falkowski, S.; Goryń, T.; Zdzienicki, M.; Morysiński, T. Malignant Peripheral Nerve Sheath Tumors–Outcomes and Prognostic Factors Based on the Reference Center Experience. Surg. Oncol. 2020, 35, 276–284. [Google Scholar] [CrossRef]
- Yamaguchi, U.; Hasegawa, T.; Hirose, T.; Chuman, H.; Kawai, A.; Ito, Y.; Beppu, Y. Low Grade Malignant Peripheral Nerve Sheath Tumour: Varied Cytological and Histological Patterns. J. Clin. Pathol. 2003, 56, 826–830. [Google Scholar] [CrossRef]
- Valentin, T.; Le Cesne, A.; Ray-Coquard, I.; Italiano, A.; Decanter, G.; Bompas, E.; Isambert, N.; Thariat, J.; Linassier, C.; Bertucci, F. Management and Prognosis of Malignant Peripheral Nerve Sheath Tumors: The Experience of the French Sarcoma Group (GSF-GETO). Eur. J. Cancer 2016, 56, 77–84. [Google Scholar] [CrossRef]
- Nelson, C.N.; Dombi, E.; Rosenblum, J.S.; Miettinen, M.M.; Lehky, T.J.; Whitcomb, P.O.; Hayes, C.; Scott, G.; Benzo, S.; Widemann, B.C.; et al. Safe Marginal Resection of Atypical Neurofibromas in Neurofibromatosis Type 1. J. Neurosurg. 2020, 133, 1516–1526. [Google Scholar] [CrossRef]
- Hajiabadi, M.M.; Campos, B.; Sedlaczek, O.; Khajeh, E.; Nikdad, M.; von Deimling, A.; Mehrabi, A.; Unterberg, A.; Ahmadi, R. Interdisciplinary Approach Allows Minimally Invasive, Nerve-Sparing Removal of Retroperitoneal Peripheral Nerve Sheath Tumors. Langenbecks Arch. Surg. 2020, 405, 199–205. [Google Scholar] [CrossRef]
- Dunn, G.P.; Spiliopoulos, K.; Plotkin, S.R.; Hornicek, F.J.; Harmon, D.C.; Delaney, T.F.; Williams, Z. Role of Resection of Malignant Peripheral Nerve Sheath Tumors in Patients with Neurofibromatosis Type 1: Clinical Article. J. Neurosurg. 2013, 118, 142–148. [Google Scholar] [CrossRef]
- Graham, D.S.; Russell, T.A.; Eckardt, M.A.; Motamedi, K.; Seeger, L.L.; Singh, A.S.; Bernthal, N.M.; Kalbasi, A.; Dry, S.M.; Nelson, S.D.; et al. Oncologic Accuracy of Image-Guided Percutaneous Core-Needle Biopsy of Peripheral Nerve Sheath Tumors at a High-Volume Sarcoma Center. Am. J. Clin. Oncol. 2019, 42, 739. [Google Scholar] [CrossRef]
- Vos, M.; Starmans, M.P.A.; Timbergen, M.J.M.; van der Voort, S.R.; Padmos, G.A.; Kessels, W.; Niessen, W.J.; van Leenders, G.J.L.H.; Grünhagen, D.J.; Sleijfer, S.; et al. Radiomics Approach to Distinguish between Well Differentiated Liposarcomas and Lipomas on MRI. Br. J. Surg. 2019, 106, 1800–1809. [Google Scholar] [CrossRef]
- Karsy, M.; Guan, J.; Ravindra, V.; Stilwill, S.; Mahan, M. Diagnostic Quality of Magnetic Resonance Imaging Interpretation for Peripheral Nerve Sheath Tumors: Can Malignancy Be Determined? J. Neurol. Surg. Part Cent. Eur. Neurosurg. 2016, 77, 495–504. [Google Scholar] [CrossRef]
- Wu, M.; Hu, Y.; Hang, J.; Peng, X.; Mao, C.; Ye, X.; Li, A. Qualitative and Quantitative Contrast-Enhanced Ultrasound Combined with Conventional Ultrasound for Predicting the Malignancy of Soft Tissue Tumors. Ultrasound Med. Biol. 2022, 48, 237–247. [Google Scholar] [CrossRef]
- Brinkman, M.; Jentjens, S.; Boone, K.; Anten, M.; Stumpel, C.T.R.M.; Nelemans, P.J.; van Kroonenburgh, M.J.P.G. Evaluation of the Most Commonly Used (Semi-)Quantitative Parameters of 18F-FDG PET/CT to Detect Malignant Transformation of Neurofibromas in Neurofibromatosis Type 1. Nucl. Med. Commun. 2018, 39, 961. [Google Scholar] [CrossRef]
- Derlin, T.; Tornquist, K.; Münster, S.; Apostolova, I.; Hagel, C.; Friedrich, R.E.; Wedegärtner, U.; Mautner, V.F. Comparative Effectiveness of 18F-FDG PET/CT Versus Whole-Body MRI for Detection of Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis Type 1. Clin. Nucl. Med. 2013, 38, e19. [Google Scholar] [CrossRef]
- Reinert, C.P.; Schuhmann, M.U.; Bender, B.; Gugel, I.; la Fougère, C.; Schäfer, J.; Gatidis, S. Comprehensive Anatomical and Functional Imaging in Patients with Type I Neurofibromatosis Using Simultaneous FDG-PET/MRI. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 776–787. [Google Scholar] [CrossRef]
- Peeken, J.C.; Bernhofer, M.; Spraker, M.B.; Pfeiffer, D.; Devecka, M.; Thamer, A.; Shouman, M.A.; Ott, A.; Nüsslin, F.; Mayr, N.A.; et al. CT-Based Radiomic Features Predict Tumor Grading and Have Prognostic Value in Patients with Soft Tissue Sarcomas Treated with Neoadjuvant Radiation Therapy. Radiother. Oncol. 2019, 135, 187–196. [Google Scholar] [CrossRef]
- Peeken, J.C.; Spraker, M.B.; Knebel, C.; Dapper, H.; Pfeiffer, D.; Devecka, M.; Thamer, A.; Shouman, M.A.; Ott, A.; von Eisenhart-Rothe, R.; et al. Tumor Grading of Soft Tissue Sarcomas Using MRI-Based Radiomics. eBioMedicine 2019, 48, 332–340. [Google Scholar] [CrossRef]
- Wang, H.; Chen, H.; Duan, S.; Hao, D.; Liu, J. Radiomics and Machine Learning With Multiparametric Preoperative MRI May Accurately Predict the Histopathological Grades of Soft Tissue Sarcomas. J. Magn. Reson. Imaging 2020, 51, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Ly, I.; Liu, T.; Cai, W.; Michaels, O.; Viera, M.; Kwon, D.; Bredella, M.; Jordan, J.; Borcherding, D.; Boswell, D.; et al. Comparison of a STIR- and T1-Based Radiomics Model to Differentiate between Plexiform Neurofibromas and Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis Type 1 (NF1) (S17.010). Neurology 2023, 100, 4040. [Google Scholar] [CrossRef]
- Zhang, M.; Tong, E.; Hamrick, F.; Lee, E.H.; Tam, L.T.; Pendleton, C.; Smith, B.W.; Hug, N.F.; Biswal, S.; Seekins, J.; et al. Machine-Learning Approach to Differentiation of Benign and Malignant Peripheral Nerve Sheath Tumors: A Multicenter Study. Neurosurgery 2021, 89, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Ristow, I.; Madesta, F.; Well, L.; Shenas, F.; Wright, F.; Molwitz, I.; Farschtschi, S.; Bannas, P.; Adam, G.; Mautner, V.F.; et al. Evaluation of Magnetic Resonance Imaging-Based Radiomics Characteristics for Differentiation of Benign and Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis Type 1. Neuro Oncol. 2022, 24, 1790–1798. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Staring, M.; Murphy, K.; Viergever, M.A.; Pluim, J.P.W. Elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans. Med. Imaging 2010, 29, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Spaanderman, D.J.; Starmans, M.P.A.; van Erp, G.C.M.; Hanff, D.F.; Sluijter, J.H.; Schut, A.-R.W.; van Leenders, G.J.L.H.; Verhoef, C.; Grunhagen, D.J.; Niessen, W.J.; et al. Minimally Interactive Segmentation of Soft-Tissue Tumors on CT and MRI Using Deep Learning. arXiv 2024, arXiv:2402.07746. [Google Scholar]
- Starmans, M.P.A.; van der Voort, S.R.; Phil, T.; Klein, S. Workflow for optimal radiomics classification (WORC). Zenodo. 2018. Available online: https://github.com/MStarmans91/WORC (accessed on 14 January 2024).
- van der Voort, S.R.; Starmans, M.P.A. Predict: A radiomics Extensive Digital Interchangable Classification Toolkit (PREDICT). Zenodo. 2018. Available online: https://github.com/Svdvoort/PREDICTFastr (accessed on 14 January 2024).
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Starmans, M.P.A.; van der Voort, S.R.; Phil, T.; Timbergen, M.J.M.; Vos, M.; Padmos, G.A.; Kessels, W.; Hanff, D.; Grunhagen, D.J.; Verhoef, C.; et al. Reproducible radiomics through automated machine learning validated on twelve clinical applications. arXiv 2021, arXiv:2108.08618. [Google Scholar]
- Nadeau, C.; Bengio, Y. Inference for the Generalization Error. In Advances in Neural Information Processing Systems; MIT Press; Cambridge, MA, USA, 1999; Volume 12. [Google Scholar]
- Efron, B.; Tibshirani, R. Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Rejoinder. Stat. Sci. 1986, 1, 77. [Google Scholar] [CrossRef]
- Demehri, S.; Belzberg, A.; Blakeley, J.; Fayad, L.M. Conventional and Functional MR Imaging of Peripheral Nerve Sheath Tumors: Initial Experience. Am. J. Neuroradiol. 2014, 35, 1615–1620. [Google Scholar] [CrossRef]
- Benz, M.R.; Czernin, J.; Dry, S.M.; Tap, W.D.; Allen-Auerbach, M.S.; Elashoff, D.; Phelps, M.E.; Weber, W.A.; Eilber, F.C. Quantitative F18-Fluorodeoxyglucose Positron Emission Tomography Accurately Characterizes Peripheral Nerve Sheath Tumors as Malignant or Benign. Cancer 2010, 116, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, M.; Spiridonov, S.; Yanik, E.L.; Jennings, J.W.; Hillen, T.; Ponisio, M.; McDonald, D.J.; Dehdashti, F.; Cipriano, C.A. How Effective Are Noninvasive Tests for Diagnosing Malignant Peripheral Nerve Sheath Tumors in Patients with Neurofibromatosis Type 1? Diagnosing MPNST in NF1 Patients. Sarcoma 2019, 2019, e4627521. [Google Scholar] [CrossRef] [PubMed]
- Salamon, J.; Derlin, T.; Bannas, P.; Busch, J.D.; Herrmann, J.; Bockhorn, M.; Hagel, C.; Friedrich, R.E.; Adam, G.; Mautner, V.F. Evaluation of Intratumoural Heterogeneity on 18F-FDG PET/CT for Characterization of Peripheral Nerve Sheath Tumours in Neurofibromatosis Type 1. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Tovmassian, D.; Abdul Razak, M.; London, K. The role of [18F]FDG-PET/CT in predicting malignant transformation of plexiform neurofbromas in neurofbromatosis-1. Int. J. Surg. Oncol. 2016, 2016, 6162182. [Google Scholar] [CrossRef]
- Ahlawat, S.; Blakeley, J.O.; Rodriguez, F.J.; Fayad, L.M. Imaging Biomarkers for Malignant Peripheral Nerve Sheath Tumors in Neurofibromatosis Type 1. Neurology 2019, 93, e1076–e1084. [Google Scholar] [CrossRef]
- Mazal, A.T.; Ashikyan, O.; Cheng, J.; Le, L.Q.; Chhabra, A. Diffusion-Weighted Imaging and Diffusion Tensor Imaging as Adjuncts to Conventional MRI for the Diagnosis and Management of Peripheral Nerve Sheath Tumors: Current Perspectives and Future Directions. Eur. Radiol. 2019, 29, 4123–4132. [Google Scholar] [CrossRef]

| Variable | Benign (N = 74) | Malignant (N = 35) | p-Value |
|---|---|---|---|
| Mean Age (SD) | 44 yr (19 yr) | 43 yr (25 yr) | 0.78 |
| Gender Male Female | 22 (30%) 52 (70%) | 15 (43%) 20 (57%) | 0.26 |
| Neurogenetic diagnosis None NF1 Schwannomatosis | 35 (47%) 38 (52%) 1 (1%) | 16 (47%) 18 (53%) 0 (0%) | 0.79 |
| Spontaneous pain No Yes Unknown | 28 (41%) 40 (59%) 6 | 10 (30%) 23 (70%) 2 | 0.40 |
| Pre-operative motor deficits No Yes Unknown | 60 (88%) 8 (12%) 6 | 22 (67%) 11 (33%) 2 | 0.02 |
| Mean Volume (SD) | 54 cm3 (141 cm3) | 208 cm3 (324 cm3) | 0.01 |
| Models | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Models with manual segmentations | ||||
| T1w (n = 90) | 0.71 [0.56, 0.86] | 0.75 [0.67, 0.84] | 0.30 [0.10, 0.50] | 0.93 [0.84, 1.00] |
| T2w (n = 87) | 0.70 [0.56, 0.84] | 0.65 [0.56, 0.75] | 0.33 [0.14, 0.52] | 0.83 [0.71, 0.95] |
| T1w, T2w (n = 97) | 0.68 [0.54, 0.83] | 0.72 [0.64, 0.79] | 0.29 [0.11, 0.46] | 0.91 [0.82, 0.99] |
| T1w, T2w, T1w-FS-GD OR T1w-SPIR-GD (n = 99) | 0.70 [0.57, 0.83] | 0.72 [0.66, 0.79] | 0.26 [0.09, 0.42] | 0.92 [0.85, 1.00] |
| T1w, T2w, T2w-FS OR T2w-STIR (n = 103) | 0.66 [0.55, 0.77] | 0.67 [0.60, 0.74] | 0.25 [0.08, 0.42] | 0.88 [0.78, 0.98] |
| T1w, T1w-FS-GD OR T1w-SPIR-GD (n = 93) | 0.70 [0.56, 0.83] | 0.73 [0.66, 0.80] | 0.26 [0.11, 0.41] | 0.95 [0.87, 1.00] |
| T2w, T2w-FS OR T2w-STIR (n = 94) | 0.66 [0.56, 0.77] | 0.68 [0.60, 0.75] | 0.26 [0.10, 0.42] | 0.87 [0.76, 0.97] |
| Models with semi-automatic segmentations using InteractiveNet | ||||
| T1w_Interactive model (n = 87) | 0.68 [0.56, 0.79] | 0.70 [0.63, 0.77] | 0.23 [0.07, 0.38] | 0.94 [0.85, 1.00] |
| T1w_Interactive_Sufficient model (n = 66) | 0.64 [0.48, 0.79] | 0.69 [0.59, 0.79] | 0.20 [0.01, 0.38] | 0.89 [0.76, 1.00] |
| Models | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Imaging + Clinical features | ||||
| T1w + clinical model (n = 90) | 0.74 [0.60, 0.88] | 0.75 [0.69, 0.82] | 0.31 [0.12, 0.50] | 0.92 [0.84, 1.00] |
| Radiologist 1 with clinical features (n = 108) | 0.75 [0.65, 0.85] | 0.72 [0.64, 0.80] | 0.71 [0.56, 0.86] | 0.73 [0.63, 0.83] |
| Radiologist 2 with clinical features (n = 108) | 0.66 [0.55, 0.77] | 0.62 [0.53, 0.71] | 0.60 [0.43, 0.77] | 0.63 [0.52, 0.74] |
| Imaging only | ||||
| T1w model (n = 90) | 0.71 [0.56, 0.86] | 0.75 [0.67, 0.84] | 0.30 [0.10, 0.50] | 0.93 [0.84, 1.00] |
| Radiologist 1 (n = 108) | 0.78 [0.68, 0.88] | 0.79 [0.71, 0.87] | 0.71 [0.56, 0.86] | 0.82 [0.73, 0.91] |
| Radiologist 2 (n = 108) | 0.68 [0.58, 0.78] | 0.64 [0.55, 0.73] | 0.60 [0.42, 0.78] | 0.66 [0.55, 0.77] |
| Clinical features only | ||||
| Clinical model (n = 90) | 0.60 [0.46, 0.73] | 0.71 [0.64, 0.78] | 0.26 [0.07, 0.44] | 0.88 [0.80, 0.96] |
| Volume only | ||||
| Volume model (n = 90) | 0.64 [0.50, 0.78] | 0.71 [0.63, 0.80] | 0.28 [0.07, 0.49] | 0.88 [0.75, 1.00] |
| Metrics | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| OR operation | ||||
| T1w + clinical model OR Radiologists 1 (n = 90) | N/A | 0.68 [0.65, 0.70] | 0.74 [0.70, 0.78] | 0.65 [0.63, 0.68] |
| T1w + clinical model OR Radiologists 2 (n = 90) | N/A | 0.63 [0.61, 0.65] | 0.71 [0.67, 0.75] | 0.61 [0.58, 0.63] |
| AND operation | ||||
| T1w + clinical model AND Radiologist 1 (n = 90) | N/A | 0.78 [0.76, 0.79] | 0.21 [0.18, 0.25] | 0.96 [0.96, 0.97] |
| T1w + clinical model AND Radiologist 2 (n = 90) | N/A | 0.75 [0.74, 0.76] | 0.20 [0.17, 0.24] | 0.93 [0.91, 0.94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jansma, C.Y.M.N.; Wan, X.; Acem, I.; Spaanderman, D.J.; Visser, J.J.; Hanff, D.; Taal, W.; Verhoef, C.; Klein, S.; Martin, E.; et al. Preoperative Classification of Peripheral Nerve Sheath Tumors on MRI Using Radiomics. Cancers 2024, 16, 2039. https://doi.org/10.3390/cancers16112039
Jansma CYMN, Wan X, Acem I, Spaanderman DJ, Visser JJ, Hanff D, Taal W, Verhoef C, Klein S, Martin E, et al. Preoperative Classification of Peripheral Nerve Sheath Tumors on MRI Using Radiomics. Cancers. 2024; 16(11):2039. https://doi.org/10.3390/cancers16112039
Chicago/Turabian StyleJansma, Christianne Y. M. N., Xinyi Wan, Ibtissam Acem, Douwe J. Spaanderman, Jacob J. Visser, David Hanff, Walter Taal, Cornelis Verhoef, Stefan Klein, Enrico Martin, and et al. 2024. "Preoperative Classification of Peripheral Nerve Sheath Tumors on MRI Using Radiomics" Cancers 16, no. 11: 2039. https://doi.org/10.3390/cancers16112039
APA StyleJansma, C. Y. M. N., Wan, X., Acem, I., Spaanderman, D. J., Visser, J. J., Hanff, D., Taal, W., Verhoef, C., Klein, S., Martin, E., & Starmans, M. P. A. (2024). Preoperative Classification of Peripheral Nerve Sheath Tumors on MRI Using Radiomics. Cancers, 16(11), 2039. https://doi.org/10.3390/cancers16112039







