Integrating Spatial and Morphological Characteristics into Melanoma Prognosis: A Computational Approach
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
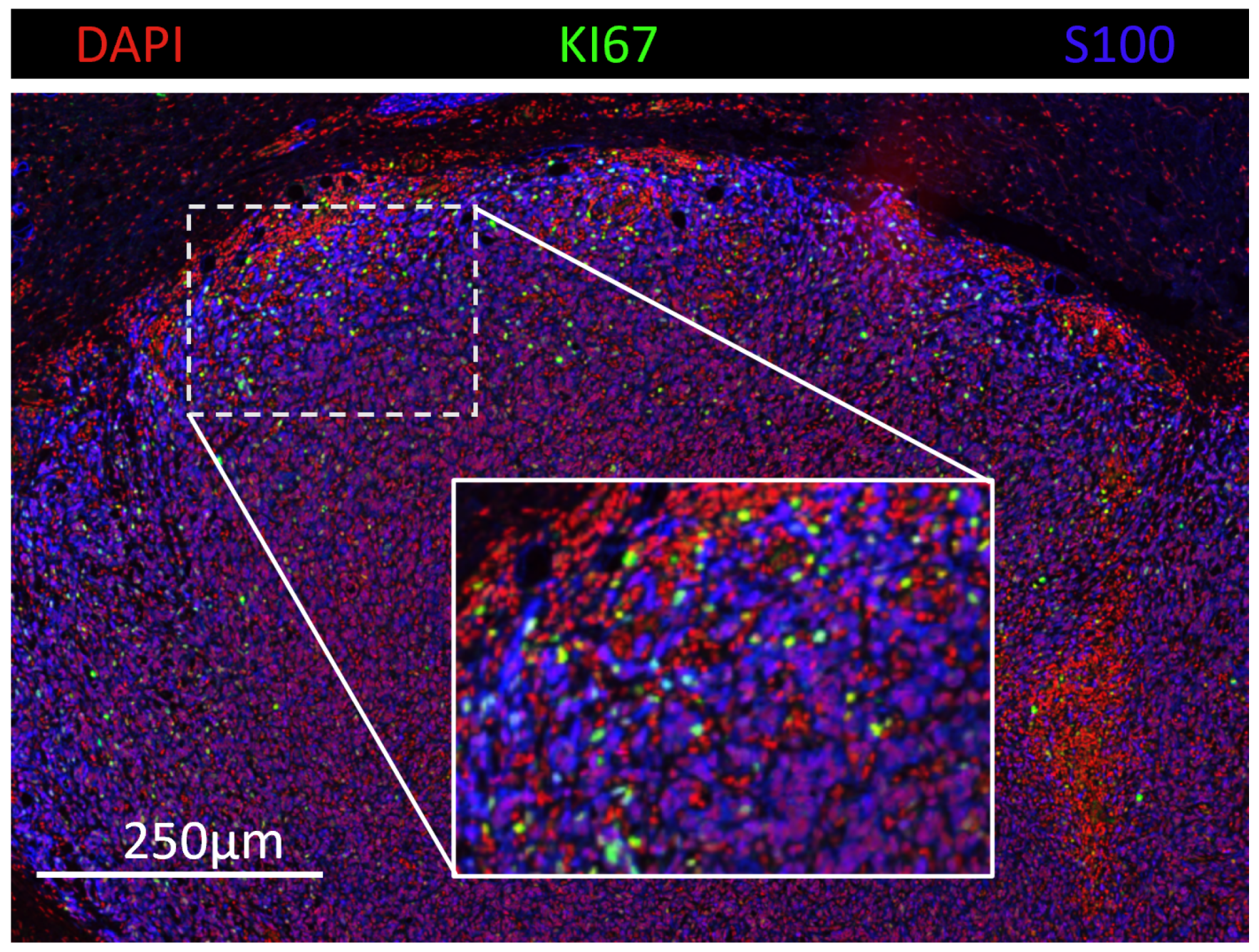
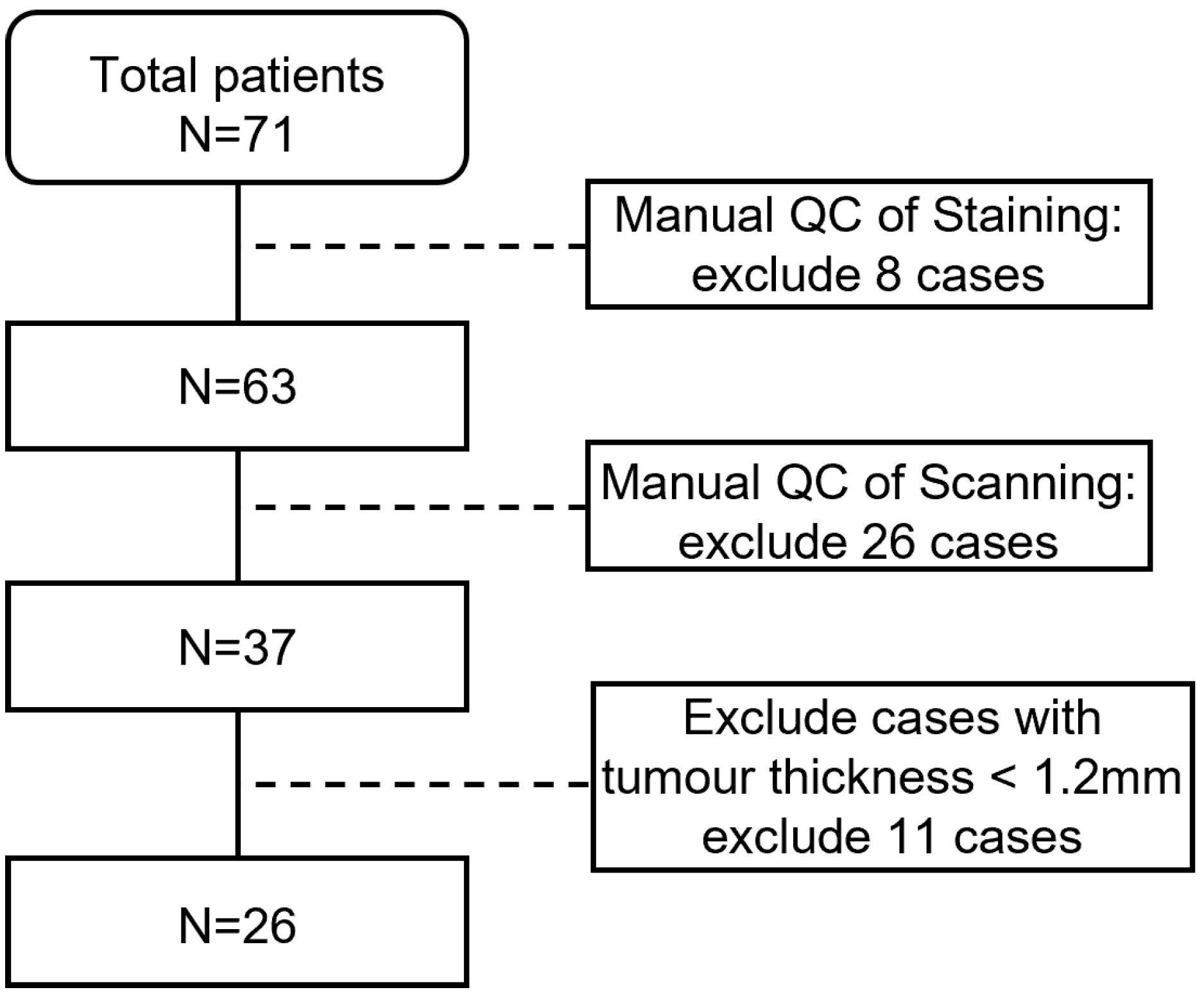
2.1. Dataset Collection
2.2. Cell Detection and Measuring
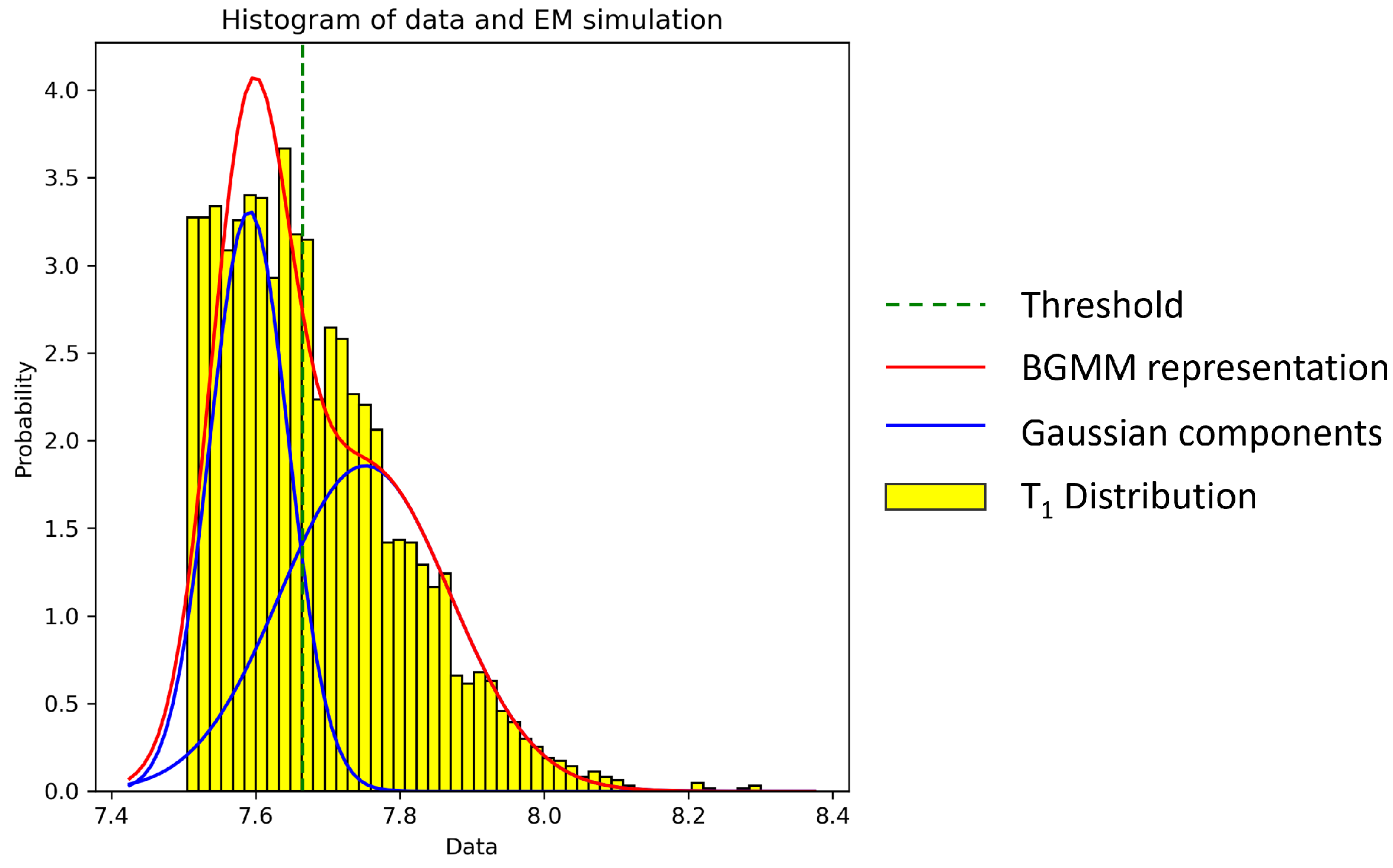
2.3. Cell Thresholding
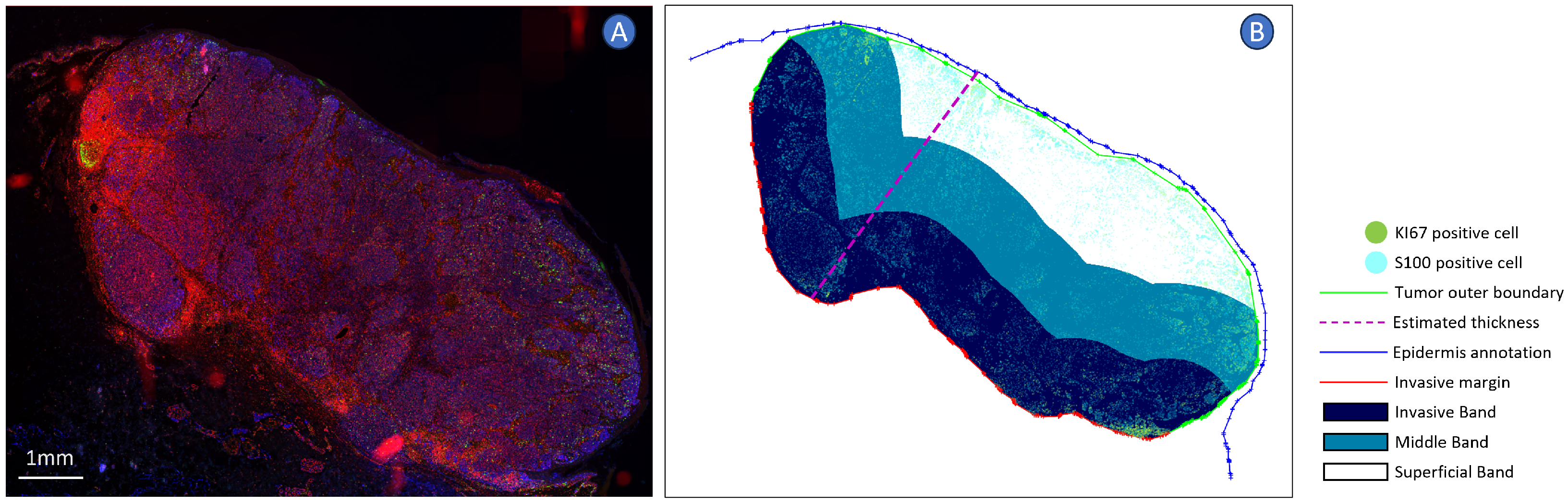
2.4. Spatial Analysis
2.5. Survival Analysis
3. Results
3.1. Clinical Characteristics
3.2. Risk Prediction
3.3. Factor Correlation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schadendorf, D.; van Akkooi, A.C.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Domingues, B.; Lopes, J.M.; Soares, P.; Pópulo, H. Melanoma treatment in review. Immunotargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Arrangoiz, R.; Dorantes, J.; Cordera, F.; Juarez, M.M.; Paquentin, E.M.; de León, E.L. Melanoma review: Epidemiology, risk factors, diagnosis and staging. J. Cancer Treat. Res. 2016, 4, 1005–1011. [Google Scholar] [CrossRef][Green Version]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. Ther. 2019, 20, 1366–1379. [Google Scholar] [CrossRef]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, P.; Curry, J.L.; Ning, J.; Piao, J.; Torres-Cabala, C.A.; Aung, P.P.; Ivan, D.; Ross, M.I.; Levenback, C.F.; Frumovitz, M.; et al. Tumor thickness and mitotic rate robustly predict melanoma-specific survival in patients with primary vulvar melanoma: A retrospective review of 100 cases. Clin. Cancer Res. 2017, 23, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Piñero-Madrona, A.; Ruiz-Merino, G.; Cerezuela Fuentes, P.; Martínez-Barba, E.; Rodríguez-López, J.; Cabezas-Herrera, J. Mitotic rate as an important prognostic factor in cutaneous malignant melanoma. Clin. Transl. Oncol. 2019, 21, 1348–1356. [Google Scholar] [CrossRef] [PubMed]
- Van Diest, P.J.; van der Wall, E.; Baak, J.P. Prognostic value of proliferation in invasive breast cancer: A review. J. Clin. Pathol. 2004, 57, 675. [Google Scholar] [CrossRef] [PubMed]
- Nagore, E.; Oliver, V.; Botella-Estrada, R.; Moreno-Picot, S.; Insa, A. Prognostic factors in localized invasive cutaneous melanoma: High value of mitotic rate, vascular invasion and microscopic satellitosis. Melanoma Res. 2005, 15, 169–177. [Google Scholar] [CrossRef]
- Nikolaou, V.; Stratigos, A. Emerging trends in the epidemiology of melanoma. Br. J. Dermatol. 2014, 170, 11–19. [Google Scholar] [CrossRef]
- Blum, A.; Brand, C.; Ellwanger, U.; Schlagenhauff, B.; Stroebel, W.; Rassner, G.; Garbe, C. Awareness and early detection of cutaneous melanoma: An analysis of factors related to delay in treatment. Br. J. Dermatol. 1999, 141, 783–787. [Google Scholar] [CrossRef]
- Blakely, A.M.; Cohen, J.T.; Comissiong, D.S.; Vezeridis, M.P.; Miner, T.J. Prognosis and management of thick and ultrathick melanoma. Am. J. Clin. Oncol. 2019, 42, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Marconi, A.; Quadri, M.; Farnetani, F.; Ciardo, S.; Palazzo, E.; Lotti, R.; Cesinaro, A.M.; Fabbiani, L.; Vaschieri, C.; Puviani, M.; et al. In vivo melanoma cell morphology reflects molecular signature and tumor aggressiveness. J. Investig. Dermatol. 2022, 142, 2205–2216. [Google Scholar] [CrossRef] [PubMed]
- Blessing, K.; Grant, J.; Sanders, D.; Kennedy, M.; Husain, A.; Coburn, P. Small cell malignant melanoma: A variant of naevoid melanoma. Clinicopathological features and histological differential diagnosis. J. Clin. Pathol. 2000, 53, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Wolberg, W.H.; Nick Street, W.; Mangasarian, O.L. Importance of nuclear morphology in breast cancer prognosis. Clin. Cancer Res. 1999, 5, 3542–3548. [Google Scholar] [PubMed]
- Nakazato, Y.; Minami, Y.; Kobayashi, H.; Satomi, K.; Anami, Y.; Tsuta, K.; Tanaka, R.; Okada, M.; Goya, T.; Noguchi, M. Nuclear grading of primary pulmonary adenocarcinomas: Correlation between nuclear size and prognosis. Cancer 2010, 116, 2011–2019. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Kohashi, K.; Iwasaki, T.; Yamamoto, T.; Ishihara, S.; Toda, Y.; Yamamoto, H.; Nakashima, Y.; Oda, Y. Prognostic value of nuclear morphometry in myxoid liposarcoma. Cancer Sci. 2023, 114, 2178. [Google Scholar] [CrossRef] [PubMed]
- Karpińska-Kaczmarczyk, K.; Kram, A.; Kaczmarczyk, M.; Domagała, W. Prognostic significance of morphometric parameters of nucleoli and nuclei of invasive ductal breast carcinomas. Pol. J. Pathol. 2009, 60, 124–129. [Google Scholar] [PubMed]
- Fernandez-Lopez, F.; Paredes-Cotore, J.; Cadarso-Suárez, C.; Forteza-Vila, J.; Puente-Domínguez, J.; Potel-Lesquereux, J. Prognostic value of nuclear morphometry in colorectal cancer. Dis. Colon Rectum 1999, 42, 386–392. [Google Scholar] [CrossRef]
- Sørensen, F.B.; Gamel, J.W.; Jensen, O.A.; Ladekarl, M.; McCurdy, J. Prognostic value of nucleolar size and size pleomorphism in choroidal melanomas. APMIS 1993, 101, 358–368. [Google Scholar] [CrossRef]
- Tsakiroglou, A.M.; Fergie, M.; Oguejiofor, K.; Linton, K.; Thomson, D.; Stern, P.L.; Astley, S.; Byers, R.; West, C.M. Spatial proximity between T and PD-L1 expressing cells as a prognostic biomarker for oropharyngeal squamous cell carcinoma. Br. J. Cancer 2020, 122, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zang, X.; Yang, L.; Huang, J.; Liang, F.; Rodriguez-Canales, J.; Wistuba, I.I.; Gazdar, A.; Xie, Y.; Xiao, G. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J. Thorac. Oncol. 2017, 12, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Verghese, G.; Lennerz, J.K.; Ruta, D.; Ng, W.; Thavaraj, S.; Siziopikou, K.P.; Naidoo, T.; Rane, S.; Salgado, R.; Pinder, S.E.; et al. Computational pathology in cancer diagnosis, prognosis, and prediction–present day and prospects. J. Pathol. 2023, 260, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Bian, C.; Wang, Y.; Lu, Z.; An, Y.; Wang, H.; Kong, L.; Du, Y.; Tian, J. ImmunoAIzer: A deep learning-based computational framework to characterize cell distribution and gene mutation in tumor microenvironment. Cancers 2021, 13, 1659. [Google Scholar] [CrossRef]
- Song, A.H.; Jaume, G.; Williamson, D.F.; Lu, M.Y.; Vaidya, A.; Miller, T.R.; Mahmood, F. Artificial intelligence for digital and computational pathology. Nat. Rev. Bioeng. 2023, 1, 930–949. [Google Scholar] [CrossRef]
- Sauter, D.; Lodde, G.; Nensa, F.; Schadendorf, D.; Livingstone, E.; Kukuk, M. Deep learning in computational dermatopathology of melanoma: A technical systematic literature review. Comput. Biol. Med. 2023, 163, 107083. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, V.G.; Martinek, J.; Foroughi pour, A.; Zhou, J.; Palucka, K.; Chuang, J.H. Computational analysis of immune synapses in melanoma tumor microenvironment. Cancer Res. 2023, 83, 5883. [Google Scholar] [CrossRef]
- Lee, J.T.; Herlyn, M. Microenvironmental influences in melanoma progression. J. Cell. Biochem. 2007, 101, 862–872. [Google Scholar] [CrossRef]
- Yan, Y.; Leontovich, A.A.; Gerdes, M.J.; Desai, K.; Dong, J.; Sood, A.; Santamaria-Pang, A.; Mansfield, A.S.; Chadwick, C.; Zhang, R.; et al. Understanding heterogeneous tumor microenvironment in metastatic melanoma. PLoS ONE 2019, 14, e0216485. [Google Scholar] [CrossRef]
- Shi, J.Y.; Wang, X.; Ding, G.Y.; Dong, Z.; Han, J.; Guan, Z.; Ma, L.J.; Zheng, Y.; Zhang, L.; Yu, G.Z.; et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut 2020, 70, 951–961. [Google Scholar] [CrossRef]
- Clark, J.C.; Dass, C.R.; Choong, P.F. A review of clinical and molecular prognostic factors in osteosarcoma. J. Cancer Res. Clin. Oncol. 2008, 134, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Loughrey, M.B.; Bankhead, P.; Coleman, H.G.; Hagan, R.S.; Craig, S.; McCorry, A.M.; Gray, R.T.; McQuaid, S.; Dunne, P.D.; Hamilton, P.W.; et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology 2018, 73, 327–338. [Google Scholar] [CrossRef]
- Humphries, M.; Maxwell, P.; Salto-Tellez, M. QuPath: The global impact of an open source digital pathology system. Comput. Struct. Biotechnol. J. 2021, 19, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Usman, H.A.; Abidin, F.A.Z. Digital image analysis of immunohistochemistry KI-67 using QuPath software in breast cancer. J. Kedokt. Dan Kesehat. Indones. 2021, 12, 34–43. [Google Scholar] [CrossRef]
- Schmidt, U.; Weigert, M.; Broaddus, C.; Myers, G. Cell detection with star-convex polygons. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part II 11. Springer: Berlin/Heidelberg, Germany, 2018; pp. 265–273. [Google Scholar]
- Stevens, M.; Nanou, A.; Terstappen, L.W.; Driemel, C.; Stoecklein, N.H.; Coumans, F.A. StarDist image segmentation improves circulating tumor cell detection. Cancers 2022, 14, 2916. [Google Scholar] [CrossRef]
- Weigert, M.; Schmidt, U. Nuclei instance segmentation and classification in histopathology images with Stardist. In Proceedings of the 2022 IEEE International Symposium on Biomedical Imaging Challenges (ISBIC), Kolkata, India, 28–31 March 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–4. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Zhao, L.; Zheng, S.; Yang, W.; Wei, H.; Huang, X. An image thresholding approach based on Gaussian mixture model. Pattern Anal. Appl. 2019, 22, 75–88. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B (Methodol.) 1977, 39, 1–22. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Cox, D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B (Methodol.) 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Van Herck, Y.; Antoranz, A.; Andhari, M.D.; Milli, G.; Bechter, O.; De Smet, F.; Bosisio, F.M. Multiplexed immunohistochemistry and digital pathology as the foundation for next-generation pathology in melanoma: Methodological comparison and future clinical applications. Front. Oncol. 2021, 11, 636681. [Google Scholar] [CrossRef] [PubMed]
- Onega, T.; Barnhill, R.L.; Piepkorn, M.W.; Longton, G.M.; Elder, D.E.; Weinstock, M.A.; Knezevich, S.R.; Reisch, L.M.; Carney, P.A.; Nelson, H.D.; et al. Accuracy of digital pathologic analysis vs traditional microscopy in the interpretation of melanocytic lesions. JAMA Dermatol. 2018, 154, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Caie, P.D.; Dimitriou, N.; Arandjelović, O. Precision medicine in digital pathology via image analysis and machine learning. In Artificial Intelligence and Deep Learning in Pathology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 149–173. [Google Scholar]


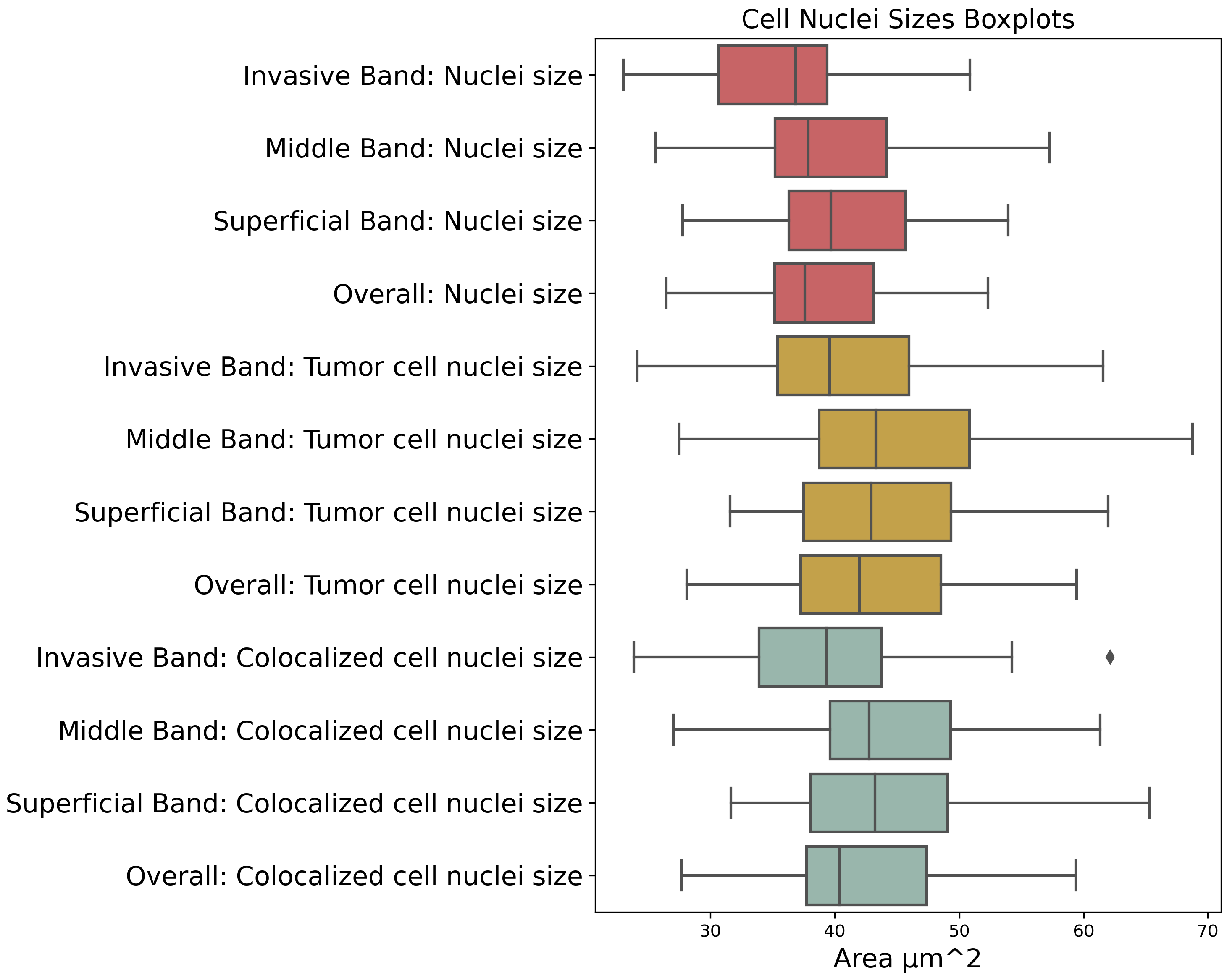
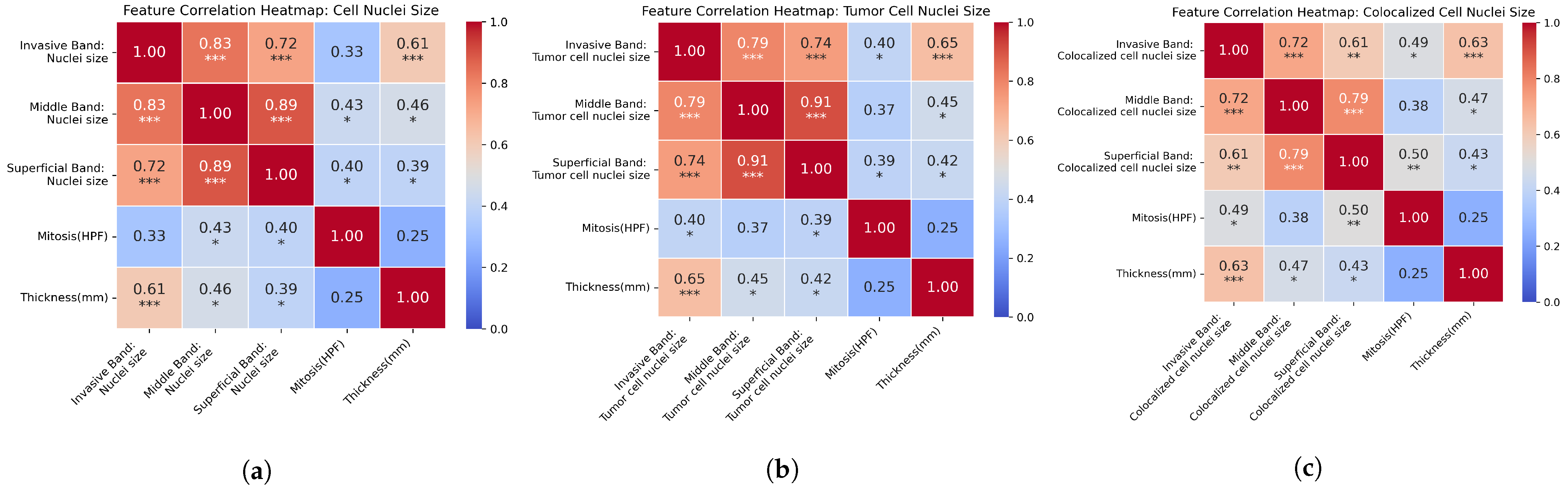
| Prognostic Factor | Band |
|---|---|
| Mitosis (HPF) | N/A |
| Thickness (mm) | N/A |
| Estimated Thickness (mm) | N/A |
| Age | N/A |
| Colocalized-to-Total Cell Ratio | Invasive, Middle, Superficial, Overall |
| Colocalized-to-Tumor Cell Ratio | Invasive, Middle, Superficial, Overall |
| Nuclei Size | Invasive, Middle, Superficial, Overall |
| Tumor Cell Nuclei Size | Invasive, Middle, Superficial, Overall |
| Colocalized Cell Nuclei Size | Invasive, Middle, Superficial, Overall |
| Characteristic | Value | No. | % |
|---|---|---|---|
| Median Age, years | 62.5 | ||
| Age Range, years | 24–78 | ||
| Gender | Male | 20 | 76.9 |
| Female | 6 | 23.1 | |
| OS: event | True | 16 | 61.5 |
| False | 10 | 38.5 | |
| Median OS: months | 35.5 | ||
| PFS: event | True | 18 | 69.2 |
| False | 8 | 30.8 | |
| Median PFS months | 27.5 |
| Variables | HR | 95% CI | p Value | |
|---|---|---|---|---|
| Lower | Upper | |||
| Mitosis (HPF) | 1.15 | 1.06 | 1.24 | <0.005 |
| Thickness (mm) | 1.70 | 1.23 | 2.35 | <0.005 |
| Estimated Thickness (mm) | 6.01 | 1.59 | 22.67 | 0.01 |
| Age | 1.01 | 0.98 | 1.05 | 0.41 |
| Invasive Band: Colocalized Cell Density | 0.90 | 0.45 | 1.82 | 0.77 |
| Middle Band: Colocalized Cell Density | 0.88 | 0.51 | 1.52 | 0.64 |
| Superficial Band: Colocalized Cell Density | 0.84 | 0.53 | 1.32 | 0.44 |
| Overall: Colocalized Cell Density | 0.71 | 0.03 | 16.63 | 0.83 |
| Invasive Band: Cell Density | 0.82 | 0.47 | 1.42 | 0.48 |
| Middle Band: Cell Density | 0.85 | 0.48 | 1.51 | 0.59 |
| Superficial Band: Cell Density | 0.99 | 0.66 | 1.49 | 0.98 |
| Overall: Cell Density | 0.78 | 0.40 | 1.52 | 0.46 |
| Invasive Band: nuclei size | 1.10 | 1.03 | 1.18 | <0.005 |
| Middle Band: nuclei size | 1.08 | 1.01 | 1.15 | 0.02 |
| Superficial Band: nuclei size | 1.10 | 1.02 | 1.19 | 0.01 |
| Overall: nuclei size | 1.11 | 1.03 | 1.20 | 0.01 |
| Invasive Band: tumor cell nuclei size | 1.07 | 1.02 | 1.13 | <0.005 |
| Middle Band: tumor cell nuclei size | 1.05 | 1.00 | 1.10 | 0.04 |
| Superficial Band: tumor cell nuclei size | 1.07 | 1.01 | 1.13 | 0.01 |
| Overall: tumor cell nuclei size | 1.08 | 1.02 | 1.14 | 0.01 |
| Invasive Band: colocalized cell nuclei size | 1.09 | 1.04 | 1.16 | <0.005 |
| Middle Band: colocalized cell nuclei size | 1.08 | 1.01 | 1.14 | 0.01 |
| Superficial Band: colocalized cell nuclei size | 1.07 | 1.02 | 1.12 | 0.01 |
| Overall: colocalized cell nuclei size | 1.11 | 1.04 | 1.18 | <0.005 |
| Variables | HR | 95% CI | p Value | C-Index | LOOCV C-Index | |
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Base model | ||||||
| Mitosis (HPF) | 1.18 | 1.07 | 1.29 | <0.005 | ||
| Thickness (mm) | 2.01 | 1.31 | 3.07 | <0.005 | 0.77 (0.61, 0.91) | 0.78 (0.61, 0.93) |
| Nuclei Size | ||||||
| Invasive Band: Nuclei size | 1.02 | 0.92 | 1.12 | 0.75 | ||
| Mitosis (HPF) | 1.17 | 1.06 | 1.29 | <0.005 | 0.78 (0.62, 0.91) | 0.78 (0.62, 0.91) |
| Thickness (mm) | 1.90 | 1.11 | 3.25 | 0.02 | ||
| Tumor Cell Nuclei Size | ||||||
| Invasive Band: Tumor Cell Nuclei size | 1.00 | 0.93 | 1.08 | 0.92 | ||
| Mitosis (HPF) | 1.18 | 1.06 | 1.30 | <0.005 | 0.79 (0.65, 0.92) | 0.78 (0.61, 0.92) |
| Thickness (mm) | 1.97 | 1.16 | 3.37 | 0.01 | ||
| Colocalized Cell Nuclei Size | ||||||
| Invasive Band: Colocalized Cell Nuclei size | 1.03 | 0.95 | 1.11 | 0.54 | ||
| Mitosis (HPF) | 1.17 | 1.05 | 1.29 | <0.005 | 0.79 (0.64, 0.93) | 0.78 (0.61, 0.93) |
| Thickness (mm) | 1.86 | 1.13 | 3.05 | 0.01 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, C.; Ashton, G.; Grant, M.; Rodriguez, V.P.; Martin, I.P.; Tsakiroglou, A.M.; Cook, M.; Fergie, M. Integrating Spatial and Morphological Characteristics into Melanoma Prognosis: A Computational Approach. Cancers 2024, 16, 2026. https://doi.org/10.3390/cancers16112026
Bian C, Ashton G, Grant M, Rodriguez VP, Martin IP, Tsakiroglou AM, Cook M, Fergie M. Integrating Spatial and Morphological Characteristics into Melanoma Prognosis: A Computational Approach. Cancers. 2024; 16(11):2026. https://doi.org/10.3390/cancers16112026
Chicago/Turabian StyleBian, Chang, Garry Ashton, Megan Grant, Valeria Pavet Rodriguez, Isabel Peset Martin, Anna Maria Tsakiroglou, Martin Cook, and Martin Fergie. 2024. "Integrating Spatial and Morphological Characteristics into Melanoma Prognosis: A Computational Approach" Cancers 16, no. 11: 2026. https://doi.org/10.3390/cancers16112026
APA StyleBian, C., Ashton, G., Grant, M., Rodriguez, V. P., Martin, I. P., Tsakiroglou, A. M., Cook, M., & Fergie, M. (2024). Integrating Spatial and Morphological Characteristics into Melanoma Prognosis: A Computational Approach. Cancers, 16(11), 2026. https://doi.org/10.3390/cancers16112026







