Expression Status of Rap1 Pathway-Related Genes in Liver Metastases Compared with Corresponding Primary Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Clinical Samples
2.2. RNA Isolation from Fresh Specimens
2.3. RNA Isolation from FFPE Specimens
2.4. Quantitative Real-Time PCR
2.5. Western Blotting
2.6. Statistical Analysis
3. Results
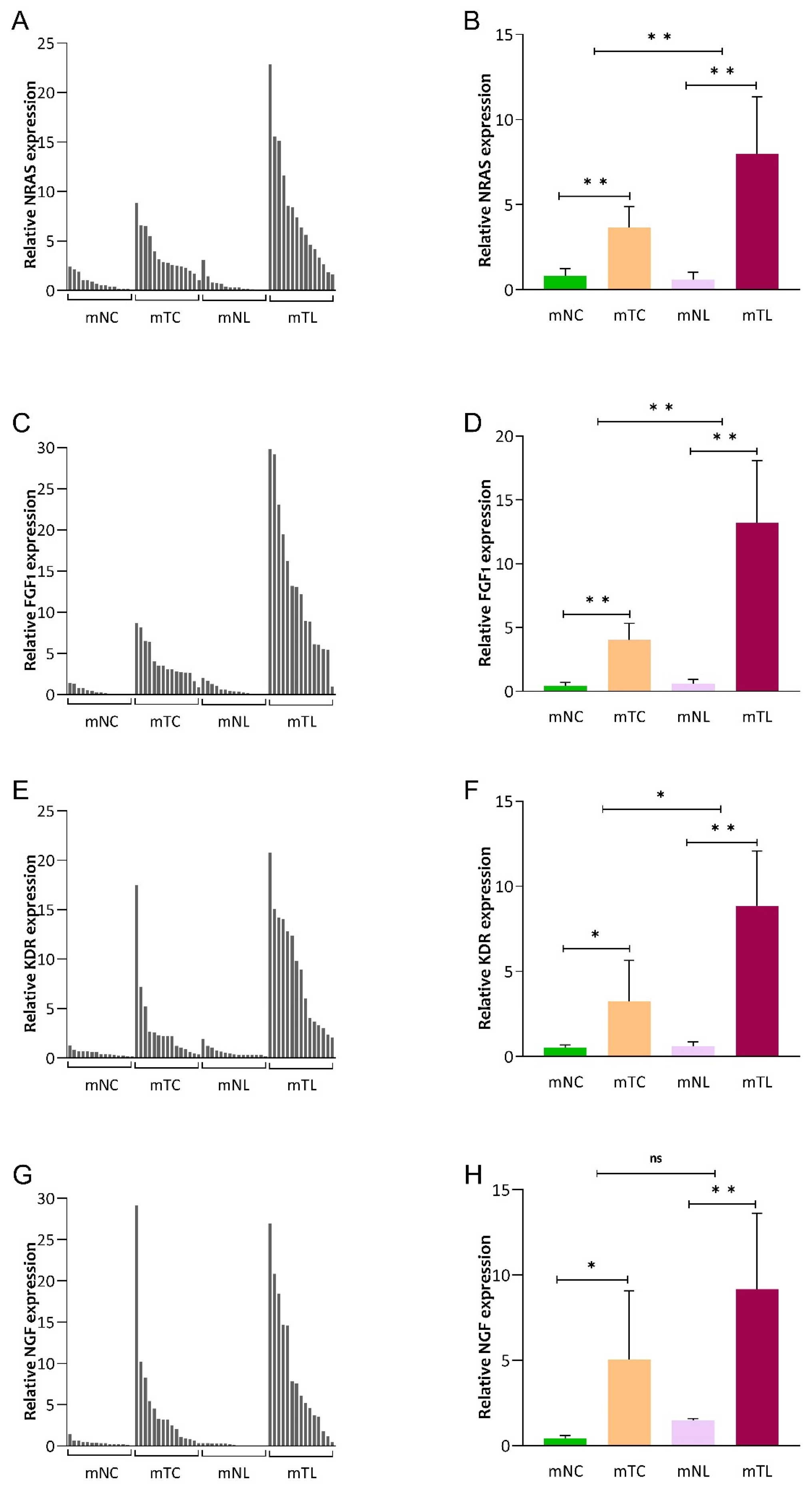
3.1. Expression of NRAS, FGF1, KDR, and NGF mRNA in CRC Tissues
3.2. Expression of NRAS, FGF1, KDR and NGF mRNA in CRLM Patients
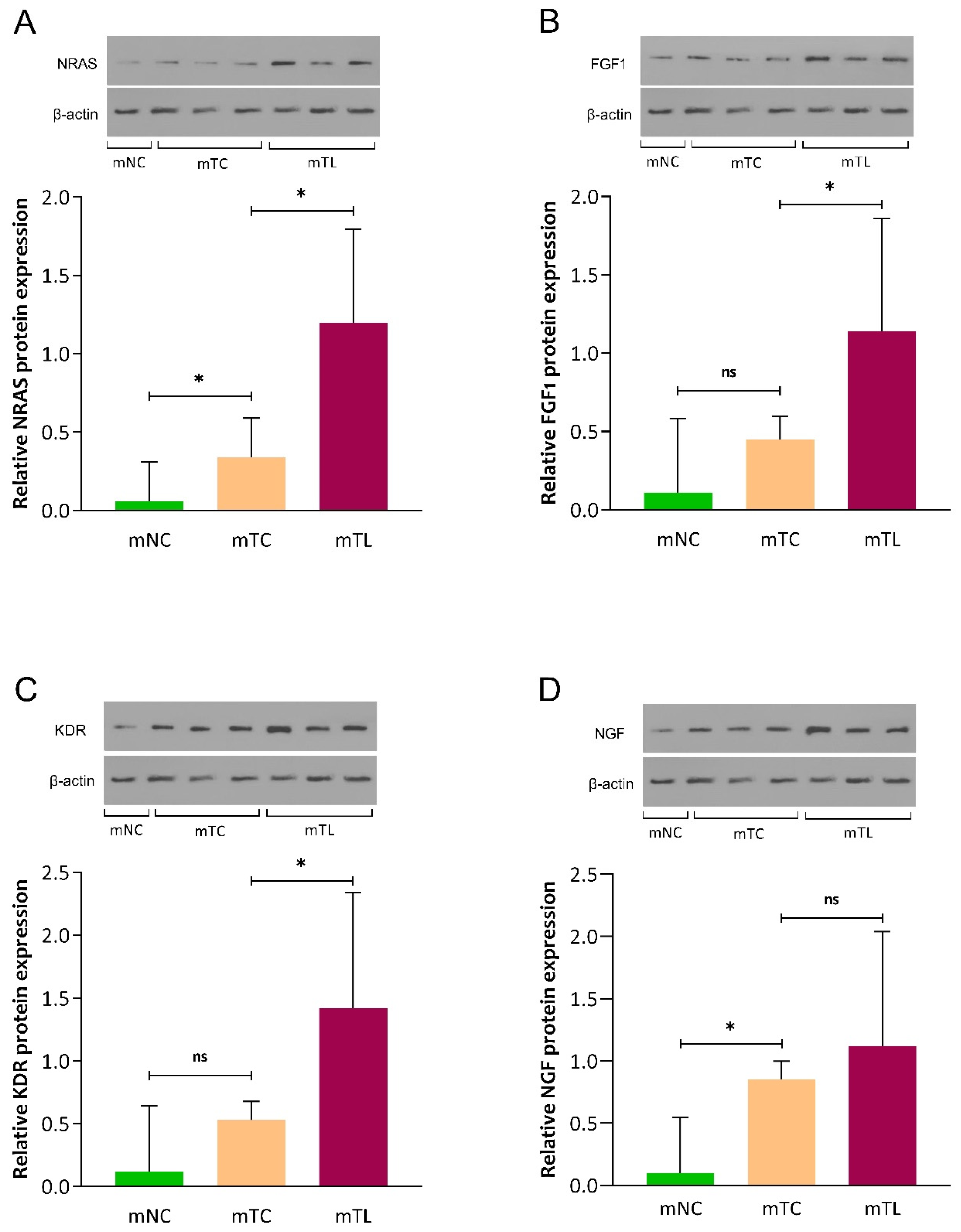
3.3. Expression of NRAS, FGF1, KDR, and NGF Protein in CRLM Patients
3.4. ROC Curve Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Engstrand, J.; Nilsson, H.; Strömberg, C.; Jonas, E.; Freedman, J. Colorectal cancer liver metastases—A population-based study on incidence, management and survival. BMC Cancer 2018, 18, 78. [Google Scholar] [CrossRef]
- Biller, L.H.; Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA 2021, 325, 669–685. [Google Scholar] [CrossRef]
- Stein, U.; Schlag, P.M. Clinical, biological, and molecular aspects of metastasis in colorectal cancer. In Targeted Therapies in Cancer; Springer: Berlin/Heidelberg, Germany, 2007; pp. 61–80. [Google Scholar]
- Ishaque, N.; Abba, M.L.; Hauser, C.; Patil, N.; Paramasivam, N.; Huebschmann, D.; Leupold, J.H.; Balasubramanian, G.P.; Kleinheinz, K.; Toprak, U.H. Whole genome sequencing puts forward hypotheses on metastasis evolution and therapy in colorectal cancer. Nat. Commun. 2018, 9, 4782. [Google Scholar] [CrossRef]
- Moisuc, D.C.; Marinca, M.V.; Matei, A.M.; Popovici, L.; Cianga, P. The Impact of Bevacizumab and Chemotherapy on Quality of Life in Metastatic Colorectal Cancer Patients. Healthcare 2023, 11, 591. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.-H.; Chen, Y.-X.; Fang, J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 2020, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Allgayer, H.; Leupold, J.H.; Patil, N. Defining the “Metastasome”: Perspectives from the genome and molecular landscape in colorectal cancer for metastasis evolution and clinical consequences. In Proceedings of the Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020; pp. 1–13. [Google Scholar]
- Nguyen, D.X.; Massagué, J. Genetic determinants of cancer metastasis. Nat. Rev. Genet. 2007, 8, 341–352. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef]
- Johnson, D.S.; Chen, Y.H. Ras family of small GTPases in immunity and inflammation. Curr. Opin. Pharmacol. 2012, 12, 458–463. [Google Scholar] [CrossRef]
- Pizon, V.; Baldacci, G. Rap1A protein interferes with various MAP kinase activating pathways in skeletal myogenic cells. Oncogene 2000, 19, 6074–6081. [Google Scholar] [CrossRef]
- Rezaei, F.M.; Hashemzadeh, S.; Gavgani, R.R.; Feizi, M.H.; Pouladi, N.; Kafil, H.S.; Rostamizadeh, L.; Oskooei, V.K.; Taheri, M.; Sakhinia, E. Dysregulated KDR and FLT1 gene expression in colorectal cancer patients. Rep. Biochem. Mol. Biol. 2019, 8, 244. [Google Scholar]
- Lei, Y.; He, X.; Huang, H.; He, Y.; Lan, J.; Yang, J.; Liu, W.; Zhang, T. Nerve growth factor orchestrates NGAL and matrix metalloproteinases activity to promote colorectal cancer metastasis. Clin. Transl. Oncol. 2022, 24, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Afrăsânie, V.-A.; Marinca, M.V.; Alexa-Stratulat, T.; Gafton, B.; Păduraru, M.; Adavidoaiei, A.M.; Miron, L.; Rusu, C. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer–practical implications for the clinician. Radiol. Oncol. 2019, 53, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Duan, T.; Zhou, D.; Yao, Y.; Shao, X. The Association of Aberrant Expression of FGF1 and mTOR-S6K1 in Colorectal Cancer. Front. Oncol. 2021, 11, 706838. [Google Scholar] [CrossRef] [PubMed]
- Schirripa, M.; Cremolini, C.; Loupakis, F.; Morvillo, M.; Bergamo, F.; Zoratto, F.; Salvatore, L.; Antoniotti, C.; Marmorino, F.; Sensi, E. Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int. J. Cancer 2015, 136, 83–90. [Google Scholar] [CrossRef]
- Keyes, J.; Ganesan, A.; Molinar-Inglis, O.; Hamidzadeh, A.; Zhang, J.; Ling, M.; Trejo, J.; Levchenko, A.; Zhang, J. Signaling diversity enabled by Rap1-regulated plasma membrane ERK with distinct temporal dynamics. Elife 2020, 9, e57410. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Zeng, Z.; Shen, N.; Wang, B.; Zhang, Y.; Shen, H.; Lu, W.; Wei, R.; Ma, W. Bioinformatic analysis identifying FGF1 gene as a new prognostic indicator in clear cell renal cell carcinoma. Cancer Cell Int. 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Nussinov, R.; Jang, H.; Zhang, M.; Tsai, C.-J.; Sablina, A.A. The mystery of Rap1 suppression of oncogenic Ras. Trends Cancer 2020, 6, 369–379. [Google Scholar] [CrossRef]
- Shimizu, A.; Zankov, D.P.; Kurokawa-Seo, M.; Ogita, H. Vascular endothelial growth factor-a exerts diverse cellular effects via small G proteins, Rho and Rap. Int. J. Mol. Sci. 2018, 19, 1203. [Google Scholar] [CrossRef]
- Dong, G.; Guo, X.; Fu, X.; Wan, S.; Zhou, F.; Myers, R.E.; Bao, G.; Burkart, A.; Yang, H.; Xing, J. Potentially functional genetic variants in KDR gene as prognostic markers in patients with resected colorectal cancer. Cancer Sci. 2012, 103, 561–568. [Google Scholar] [CrossRef]
- Shibuya, M.; Claesson-Welsh, L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp. Cell Res. 2006, 312, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhao, Y.; Song, X.; Song, X.; Niu, L.; Xie, L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J. Clin. Lab. Anal. 2019, 33, e23004. [Google Scholar] [CrossRef] [PubMed]
- Hanna, D.L.; Loupakis, F.; Yang, D.; Cremolini, C.; Schirripa, M.; Li, M.; Matsusaka, S.; Berger, M.D.; Miyamoto, Y.; Zhang, W. Prognostic value of ACVRL1 expression in metastatic colorectal cancer patients receiving first-line chemotherapy with bevacizumab: Results from the triplet plus bevacizumab (TRIBE) study. Clin. Color. Cancer 2018, 17, e471–e488. [Google Scholar] [CrossRef] [PubMed]
- Zarour, L.R.; Anand, S.; Billingsley, K.G.; Bisson, W.H.; Cercek, A.; Clarke, M.F.; Coussens, L.M.; Gast, C.E.; Geltzeiler, C.B.; Hansen, L. Colorectal cancer liver metastasis: Evolving paradigms and future directions. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 163–173. [Google Scholar] [CrossRef]
- Puccini, A.; Xiu, J.; Goldberg, R.M.; Grothey, A.; Shields, A.F.; Salem, M.E.; Seeber, A.; Battaglin, F.; Berger, M.D.; El-Deiry, W.S. Molecular differences between lymph nodes (LNs) and distant metastases (mets) in colorectal cancer (CRC). J. Clin. Oncol. 2019, 37, 3130. [Google Scholar] [CrossRef]
- Poturnajova, M.; Furielova, T.; Balintova, S.; Schmidtova, S.; Kucerova, L.; Matuskova, M. Molecular features and gene expression signature of metastatic colorectal cancer. Oncol. Rep. 2021, 45, 10. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, M.L.; Corchete, L.A.; Sarasquete, M.E.; del Mar Abad, M.; Bengoechea, O.; Fermiñán, E.; Anduaga, M.F.; Del Carmen, S.; Iglesias, M.; Esteban, C. Prognostic impact of a novel gene expression profile classifier for the discrimination between metastatic and non-metastatic primary colorectal cancer tumors. Oncotarget 2017, 8, 107685. [Google Scholar] [CrossRef][Green Version]
- Batelja-Vuletic, L.; Tomasovic-Loncaric, C.; Ceppi, M.; Bruzzone, M.; Fucic, A.; Krstanac, K.; Boras Vucicevic, V. Comparison of androgen receptor, VEGF, HIF-1, Ki67 and MMP9 expression between non-metastatic and metastatic stages in stromal and tumor cells of oral squamous cell carcinoma. Life 2021, 11, 336. [Google Scholar] [CrossRef]
- Golubnitschaja, O.; Sridhar, K.C. Liver metastatic disease: New concepts and biomarker panels to improve individual outcomes. Clin. Exp. Metastasis 2016, 33, 743–755. [Google Scholar] [CrossRef]
- Sayagués, J.M.; del Mar Abad, M.; Melchor, H.B.; Gutiérrez, M.L.; González-González, M.; Jensen, E.; Bengoechea, O.; Fonseca, E.; Orfao, A.; Muñoz-Bellvis, L. Intratumoural cytogenetic heterogeneity of sporadic colorectal carcinomas suggests several pathways to liver metastasis. J. Pathol. J. Pathol. Soc. Great Br. Irel. 2010, 221, 308–319. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, M.; Fontanillo, C.; Abad, M.; Gutierrez, M.; Mota, I.; Bengoechea, O.; Santos-Briz, A.; Blanco, O.; Fonseca ECiudad, J.; Fuentes, M.; et al. Identification of a characteristic copy number alteration profile by high-resolution single nucleotide polymorphism arrays associated with metastatic sporadic colorectal cancer. Cancer 2014, 120, 1948–1959. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, M.; Munoz-Bellvis, L.; Mackintosh, C.; Fontanillo, C.; Gutierrez, M.L.; Abad, M.M.; Bengoechea, O.; Teodosio, C.; Fonseca, E.; Fuentes, M. Prognostic Impact of del (17p) and del (22q) as assessed by interphase FISH in sporadic colorectal carcinomas. PLoS ONE 2012, 7, e42683. [Google Scholar] [CrossRef]
- Li, B.; Kang, H.; Xiao, Y.; Du, Y.; Xiao, Y.; Song, G.; Zhang, Y.; Guo, Y.; Yang, F.; He, F. LncRNA GAL promotes colorectal cancer liver metastasis through stabilizing GLUT1. Oncogene 2022, 41, 1882–1894. [Google Scholar] [CrossRef]
- Bröker, M.E.; Lalmahomed, Z.S.; Roest, H.P.; van Huizen, N.A.; Dekker, L.J.; Calame, W.; Verhoef, C.; IJzermans, J.N.; Luider, T.M. Collagen peptides in urine: A new promising biomarker for the detection of colorectal liver metastases. PLoS ONE 2013, 8, e70918. [Google Scholar] [CrossRef]
- Vermaat, J.S.; Nijman, I.J.; Koudijs, M.J.; Gerritse, F.L.; Scherer, S.J.; Mokry, M.; Roessingh, W.M.; Lansu, N.; De Bruijn, E.; Van Hillegersberg, R. Primary colorectal cancers and their subsequent hepatic metastases are genetically different: Implications for selection of patients for targeted treatment. Clin. Cancer Res. 2012, 18, 688–699. [Google Scholar] [CrossRef]
- Kim, T.-M.; Jung, S.-H.; An, C.H.; Lee, S.H.; Baek, I.-P.; Kim, M.S.; Park, S.-W.; Rhee, J.-K.; Lee, S.-H.; Chung, Y.-J. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin. Cancer Res. 2015, 21, 4461–4472. [Google Scholar] [CrossRef]
- Pretzsch, E.; Bösch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M. Mechanisms of metastasis in colorectal cancer and metastatic organotropism: Hematogenous versus peritoneal spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef]
- Seedhouse, S.J.; Affronti, H.C.; Karasik, E.; Gillard, B.M.; Azabdaftari, G.; Smiraglia, D.J.; Foster, B.A. Metastatic phenotype in CWR22 prostate cancer xenograft following castration. Prostate 2016, 76, 359–368. [Google Scholar] [CrossRef]
- Lee, J.-W.; Ryu, Y.-K.; Ji, Y.-H.; Kang, J.H.; Moon, E.-Y. Hypoxia/reoxygenation-experienced cancer cell migration and metastasis are regulated by Rap1-and Rac1-GTPase activation via the expression of thymosin beta-4. Oncotarget 2015, 6, 9820. [Google Scholar] [CrossRef]
- Bahreini, F.; Saidijam, M.; Afshar, S.; Mousivand, Z.; Najafi, R. The Effect of miR-145-5p, DANCR and NRAS Expression Levels on the Survival Rate of Colorectal Cancer Patients. Asian Pac. J. Cancer Prev. APJCP 2021, 22, 4043. [Google Scholar] [CrossRef]
- Mao, Y.; Chen, B.; Wang, H.; Zhang, Y.; Yi, X.; Liao, W.; Zhao, L. Diagnostic performance of magnetic resonance imaging for colorectal liver metastasis: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 1969. [Google Scholar] [CrossRef]
- Mainenti, P.P.; Romano, F.; Pizzuti, L.; Segreto, S.; Storto, G.; Mannelli, L.; Imbriaco, M.; Camera, L.; Maurea, S. Non-invasive diagnostic imaging of colorectal liver metastases. World J. Radiol. 2015, 7, 157. [Google Scholar] [CrossRef]
- Renzulli, M.; Clemente, A.; Ierardi, A.M.; Pettinari, I.; Tovoli, F.; Brocchi, S.; Peta, G.; Cappabianca, S.; Carrafiello, G.; Golfieri, R. Imaging of colorectal liver metastases: New developments and pending issues. Cancers 2020, 12, 151. [Google Scholar] [CrossRef]
- Zheng, W.; Wu, F.; Fu, K.; Sun, G.; Sun, G.; Li, X.; Jiang, W.; Cao, H.; Wang, H.; Tang, W. Emerging mechanisms and treatment progress on liver metastasis of colorectal cancer. OncoTargets Ther. 2021, 14, 3013–3036. [Google Scholar] [CrossRef]
- Nassar, F.J.; Msheik, Z.S.; Itani, M.M.; Helou, R.E.; Hadla, R.; Kreidieh, F.; Bejjany, R.; Mukherji, D.; Shamseddine, A.; Nasr, R.R. Circulating miRNA as biomarkers for colorectal cancer diagnosis and liver metastasis. Diagnostics 2021, 11, 341. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.; Leupold, J.; Colburn, N.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef]
- Mudduluru, G.; Abba, M.; Batliner, J.; Patil, N.; Scharp, M.; Lunavat, T.R.; Leupold, J.H.; Oleksiuk, O.; Juraeva, D.; Thiele, W.; et al. A Systematic Approach to Defining the microRNA Landscape in Metastasis. Cancer Res 2015, 75, 3010–3019. [Google Scholar] [CrossRef]
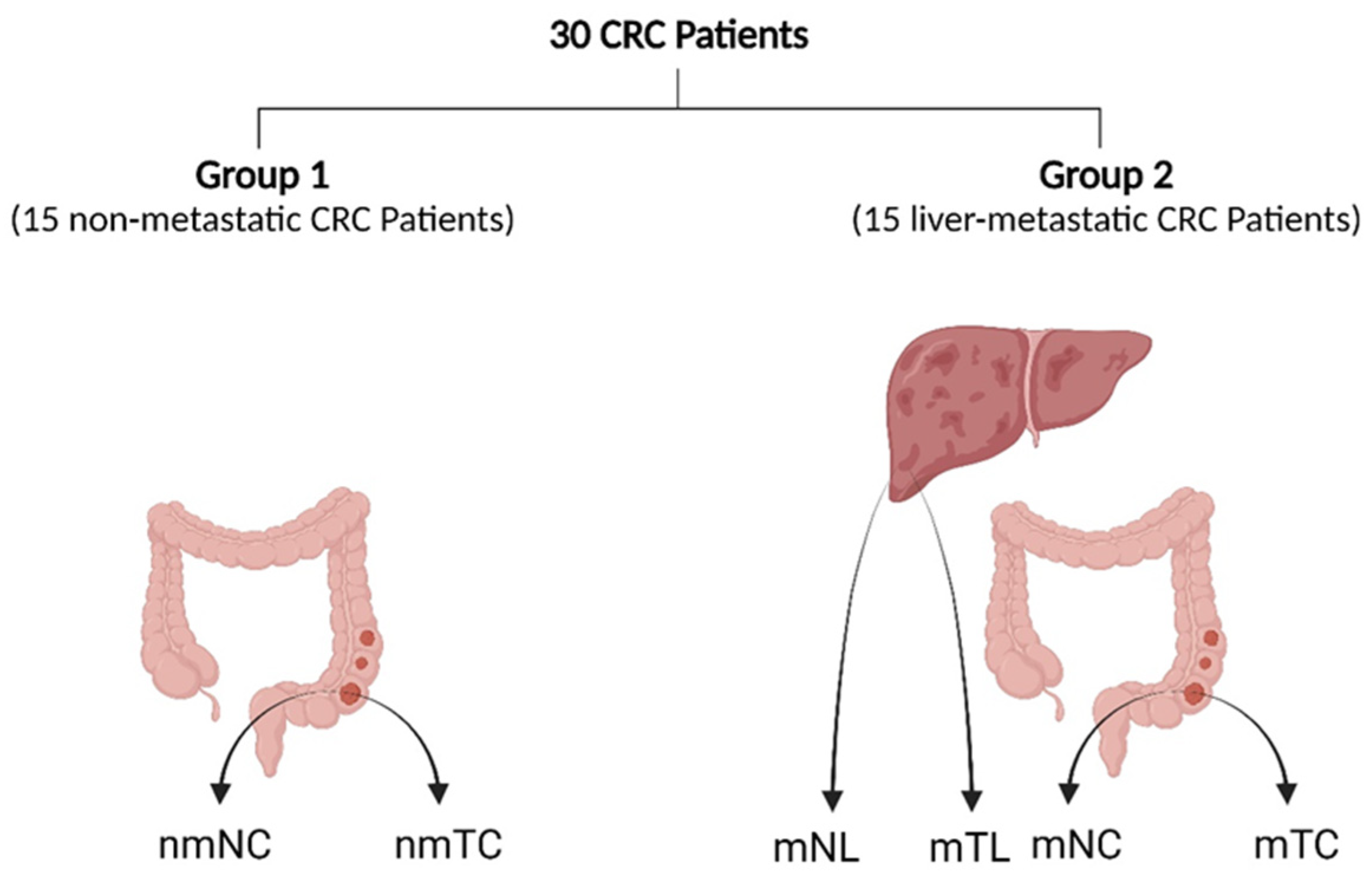



| Clinical Characteristics | Group 1 (n = 15) | Group 2 (n = 15) | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age at Surgery (year) | ≤60 | 5 | 33.4 | 10 | 66.6 |
| >60 | 10 | 66.6 | 5 | 33.4 | |
| Gender | male | 9 | 60 | 11 | 73.4 |
| female | 6 | 40 | 4 | 26.7 | |
| Serum Albumin (g/dL) | normal: 3.5–5.4 | 13 | 86.7 | 12 | 80 |
| low: <3.5 | 2 | 13.3 | 3 | 20 | |
| Total Bilirubin (mg/dL) | normal: <2 | 10 | 66.6 | 11 | 73.3 |
| high: >2 | 5 | 33.4 | 4 | 26.7 | |
| Tumor Size | ≤5 cm | 11 | 73.3 | 4 | 26.7 |
| >5 cm | 4 | 26.7 | 11 | 73.4 | |
| Stage | I–III | 15 | 100 | 0 | 0 |
| IV | 0 | 0 | 15 | 100 | |
| pT | T 1–2 | 9 | 60 | 0 | 0 |
| T 3–4 | 6 | 40 | 15 | 100 | |
| pN | N0 | 14 | 93.4 | 1 | 6.6 |
| N1 | 1 | 6.6 | 14 | 93.4 | |
| M | M0 | 15 | 100 | 0 | 0 |
| M1 | 0 | 0 | 15 | 100 | |
| Genes | NRAS | FGF1 | KDR | NGF |
|---|---|---|---|---|
| Cut-off point | 1.91 | 2.43 | 2 | 1.89 |
| AUC | 0.81 | 0.83 | 0.71 | 0.71 |
| p-value | 0.003 | 0.001 | 0.051 | 0.044 |
| 95% CI | 0.65–0.94 | 0.65–0.94 | 0.51–0.85 | 0.51–0.85 |
| Sensitivity(%) | 86.67 | 86.67 | 60 | 66.67 |
| Specificity(%) | 80 | 80 | 80 | 66.67 |
| LR+ | 4.33 | 4.33 | 3 | 2 |
| LR− | 0.16 | 0.16 | 0.50 | 0.49 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbastabar, M.; Allgayer, H.; Sepidarkish, M.; Sadeghi, F.; Ghasemi, M.; Pour-bagher, R.; Parsian, H. Expression Status of Rap1 Pathway-Related Genes in Liver Metastases Compared with Corresponding Primary Colorectal Cancer. Cancers 2024, 16, 171. https://doi.org/10.3390/cancers16010171
Abbastabar M, Allgayer H, Sepidarkish M, Sadeghi F, Ghasemi M, Pour-bagher R, Parsian H. Expression Status of Rap1 Pathway-Related Genes in Liver Metastases Compared with Corresponding Primary Colorectal Cancer. Cancers. 2024; 16(1):171. https://doi.org/10.3390/cancers16010171
Chicago/Turabian StyleAbbastabar, Maryam, Heike Allgayer, Mahdi Sepidarkish, Farzin Sadeghi, Maryam Ghasemi, Roghayeh Pour-bagher, and Hadi Parsian. 2024. "Expression Status of Rap1 Pathway-Related Genes in Liver Metastases Compared with Corresponding Primary Colorectal Cancer" Cancers 16, no. 1: 171. https://doi.org/10.3390/cancers16010171
APA StyleAbbastabar, M., Allgayer, H., Sepidarkish, M., Sadeghi, F., Ghasemi, M., Pour-bagher, R., & Parsian, H. (2024). Expression Status of Rap1 Pathway-Related Genes in Liver Metastases Compared with Corresponding Primary Colorectal Cancer. Cancers, 16(1), 171. https://doi.org/10.3390/cancers16010171







